Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A polysaccharide derived from Lentinus edodes impairs the immunosuppressive function of myeloid-derived suppressor cells via the p38 pathways†
Jun Duab,
Ruijie Wanga,
Wensheng Zhangb,
Chao Zhangb,
Xiao Lib,
Xiaodong Shib,
Minghua Huc,
Fangli Mac,
ChungWah Mac,
Xiaohong Wangd,
Ning Tao *b and
Zhihai Qin*b
*b and
Zhihai Qin*b
aInstitute of Biotechnology, The Key Laboratory of Chemical Biology and Molecular Engineering of Ministry of Education, Shanxi University, Taiyuan 030006, China
bProtein and Peptide Pharmaceutical Laboratory, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China. E-mail: zhihai@ibp.ac.cn; tao@ibp.ac.cn
cInfinitus Chinese Herbal Immunity Research Centre, Guangzhou, China
dZhejiang Province Cancer Hospital, China
First published on 21st July 2017
Abstract
We have previously reported that a novel polysaccharide, MPSSS, from Lentinus edodes can reverse the function of myeloid immune suppressor cell-mediated T cell inhibition and improve the efficacy of cancer therapy, but the mechanisms remained unknown. Here, an immortalized myeloid immune suppressor cell line (MSC2) was used to determine the molecular mechanisms of MPSSS-treatment. The results showed that MPSSS eliminated the immortalized myeloid suppressor cell line (MSC2)-mediated T cell inhibition through downregulating the activation of arginase and decreasing the cell-membrane receptor TNFR2. A p38 inhibitor almost completely prevented the MPSSS-impaired MSC2-mediated T cell inhibition by increasing arginase activity and TNFR2. MPSSS-treated MSC2 cells exhibited increased mRNA levels of TNFα and NOS2 and decreased mRNA levels of TGFβ1, c/EBPβ and HO1. The levels of TNFα were significantly increased after MPSSS stimulation of the MSC2 cells compared with the p38 inhibitor pretreatment group or the control group by ELISA analysis, while TGFβ1 induced the opposite effect under the same treatment condition. Together, these results suggest that MPSSS may reverse the function of the MSC2 cells through p38 activation and ERK suppression and provide a novel anti-cancer strategy by targeting myeloid immune suppressor cells.
1. Introduction
Polysaccharides isolated from Lentinus edodes have been widely studied and show potential anti-tumor activity.1 A β-1,3-glucan extracted from L. edodes has been used as an adjuvant drug for immune-modulators in the treatment of cancer.2,3 Our previous study found that a novel polysaccharide, MPSSS, from L. edodes can reduce the immunological suppression of myeloid-derived suppressor cells (MDSCs) in tumors, and its polymer fraction mainly consists of a β-1,6-linked glucan branched at C-4 with side chains, which are also β-1,6-linked glucans.4In tumor-bearing mice, MDSCs are generated in the bone marrow and migrate to the peripheral lymphoid organs and tumor tissues.5 The major function of the MDSCs during tumor progression is to inhibit T cell activity and promote tumor growth.6–9 To further investigate the nature and properties of these cells, myeloid suppressor cells were immortalized using a retrovirus encoding the v-myc and v-raf oncogenes. The MSC2 immortalized cells expressed monocyte/macrophage markers and still had the capacity to inhibit T lymphocyte activation.10
In this study, MSC2 cells were used to determine the underlying mechanism of the reduction of MDSC-mediated T cell inhibition by MPSSS. Here, we found that MPSSS significantly suppressed the arginase activity and TNFR2 of the MSC2 cells, which were necessary for MDSC survival and inhibited T cell proliferation. Moreover, MPSSS impaired the immunosuppressive function of the MSC2 cells through p38 activation. These results showed that MPSSS treatment caused the MSC2 cells to differentiate into M1-type macrophages, which are regarded as tumor-inhibiting.11 Thus, our work demonstrates that MPSSS regulates the p38 activation of MSC2 cells and promotes these cells to differentiate into M1-type macrophages and then reduces the immune suppression of the MSC2 cells. These results indicate that MPSSS is a potential candidate for an anti-tumor treatment that targets MDSCs.
2. Materials and methods
2.1. Chemicals and reagents
Carboxyfluoresceinsuccinimidyl ester (CFSE), L-arginine and α-isonitrosopropiophenone were purchased from Sigma-Aldrich (Darmstadt, Germany). The p38 inhibitor SB203580 and RIPA lysis buffer were purchased from Beyotime Biotechnology (Suzhou, China). DMEM and RPMI-1640 medium were purchased from HyClone (Logan, Utah, USA). Fetal bovine serum (FBS) was purchased from PAN (Aidenbach, Germany). Penicillin/streptomycin and TRIzol reagent were purchased from Invitrogen (Carlsbad, CA, USA). PrimeScript RT Master Mix and SYBR Green II Mix were purchased from Takara (Kusatsu, Shiga, Japan). TNFα and TGF-β1 ELISA kits were purchased from eBioscience (San Diego, CA, USA). PVDF membranes were purchased from Millipore (Darmstadt, Germany). The primary antibodies Phospho(P)-p38, p38, p-ERK, ERK, p-STAT1 and STAT1 were purchased from Cell Signaling Technology (Massachusetts, USA). GAPDH was purchased from Sanjian (Tianjin, China). Secondary anti-mouse and anti-rabbit antibodies were obtained from ZSGB-bio (Beijing, China). Antibodies including APC-anti-CD4, PE-anti-CD8α, PE-anti-TNFR1 and PE-anti-TNFR2 were purchased from Biolegend (Cambridge, UK). All other chemicals and reagents used in this study were of analytical grade.2.2. Cell culture
MSC2 cells were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in an incubator with a humidified atmosphere of 5% CO2.2.3. T cell proliferation assay
T cell proliferation was measured using the intracellular dye carboxyfluoresceinsuccinimidyl ester (CFSE). Splenocytes were isolated from Balb/c mice and incubated in RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in an incubator with a humidified atmosphere of 5% CO2 for 4 hours. Suspension cells were then collected and labelled with 1 μM CFSE at 25 °C for 2 minutes, followed by quenching in 10% FBS-RPMI 1640 medium. CFSE-labeled splenocytes were co-cultured at a 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio with MSC2 cells, stimulated with 1 μg ml−1 ConA, and sorted by the FACS Aria III (BD Biosciences San Diego, CA, USA). After 3 or 5 days, the cells were stained with APC-anti-CD4 and PE-anti-CD8α, and the CFSE signal of the gated lymphocytes was analyzed. Animal experiments were carried out in accordance with the Guidelines for the Care and Use of Laboratory Animals of the National Institute of Health, and were approved by the Biological Research Ethics Committee, Institute of Biophysics, Chinese Academy of Sciences.
3 ratio with MSC2 cells, stimulated with 1 μg ml−1 ConA, and sorted by the FACS Aria III (BD Biosciences San Diego, CA, USA). After 3 or 5 days, the cells were stained with APC-anti-CD4 and PE-anti-CD8α, and the CFSE signal of the gated lymphocytes was analyzed. Animal experiments were carried out in accordance with the Guidelines for the Care and Use of Laboratory Animals of the National Institute of Health, and were approved by the Biological Research Ethics Committee, Institute of Biophysics, Chinese Academy of Sciences.
2.4. Arginase activity
Arginase activity was determined by measuring the amount of urea generated from the hydrolysis of L-arginine, as previously described.12 Briefly, 106 MSC2 cells were solubilized with 100 μl lysis solution (0.1% Triton X-100, 10 mM MnCl2, and 25 mM Tris–HCl). The lysate was activated at 56 °C for 10 min, supplemented with 200 μl 500 mM L-arginine (pH 9.7) and then incubated at 37 °C for 60 min. The reaction was stopped by 800 μl H2SO4/H3PO4/H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 v/v/v). Subsequently, 40 μl α-isonitrosopropiophenone (dissolved in ethanol) was added, followed by another heating at 100 °C for 30 min. The absorbance of the concentration of urea was measured at 540 nm. One unit of enzyme activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol of urea per minute.
7 v/v/v). Subsequently, 40 μl α-isonitrosopropiophenone (dissolved in ethanol) was added, followed by another heating at 100 °C for 30 min. The absorbance of the concentration of urea was measured at 540 nm. One unit of enzyme activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol of urea per minute.
2.5. Flow cytometry
Cells were labelled for immunofluorescence and analyzed by flow cytometry for cell surface molecules. Antibodies including APC-anti-CD4, PE-anti-CD8α, PE-anti-TNFR1 and PE-anti-TNFR2 were diluted in PBS with 2% FBS.2.6. Protein extraction and immunoblotting assay
MPSSS-treated MSC2 cells were collected and lysed by a RIPA solution supplemented with 100 μM phenylmethanesulfonyl fluoride, 25 μg ml−1 aprotinin, 1 mM sodium orthovanadate and 50 mM NaF. The total protein concentrations were determined by the BCA protein assay. An equal amount of proteins was resolved on a 10% SDS-PAGE gel and then transferred to PVDF membranes. The primary antibodies used were: Phospho(P)-p38, p38, p-ERK, ERK, p-STAT1 and STAT1, which were diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, and GAPDH was diluted at 1
1000, and GAPDH was diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8000. HRP-conjugated goat anti-mouse or goat anti-rabbit IgG were used as secondary antibodies. ImageJ software was used to quantify the band density.
8000. HRP-conjugated goat anti-mouse or goat anti-rabbit IgG were used as secondary antibodies. ImageJ software was used to quantify the band density.
2.7. RNA extraction and real-time PCR
Total RNA was extracted from the MSC2 cells using TRIzol reagent and quantified on an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Total RNA was reverse transcribed with the PrimeScript RT Master Mix. The amounts of TNFα, TGF-β1, NOS2, HO1 and c/EBPβ mRNA were determined using the SYBR Green II Mix on an Applied Biosystems 7500. The internal control used was 18S rRNA. The primers sequences were listed in ESI Table S1.†2.8. Cytokine detection
TNFα and TGF-β1 in the culture supernatants were detected with ELISA kits.2.9. Statistical analysis
Quantitative data are presented as the mean ± standard deviation (SD) from three or more independent experiments. Student's t test was used to assess the differences between two groups, and P < 0.05 was considered statistically significant.3. Results
3.1. MPSSS can impair the immunosuppressive function of MSC2 cells
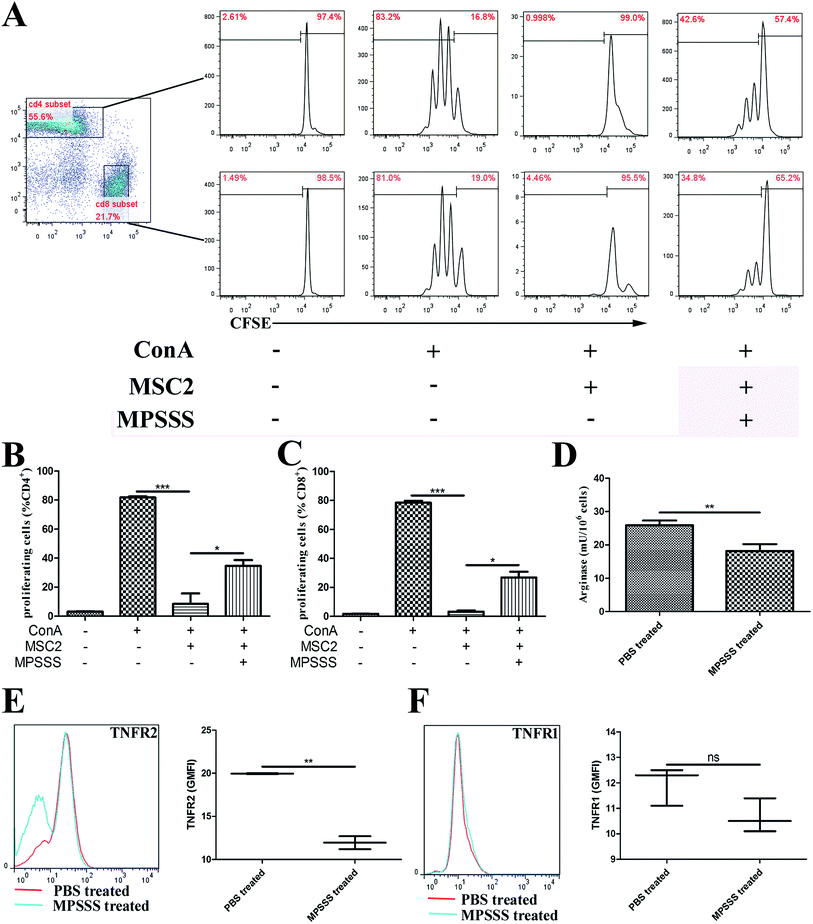
MSC2 cells were treated with or without MPSSS or lentinan. After 24 hours, the pretreated MSC2 cells were co-cultured with a suspension of splenocytes from Balb/c mice, labelled with CFSE, and stimulated with 1 μg ml−1 ConA. As shown in Fig. 1A, the CD4+ (Fig. 1B) or CD8+ (Fig. 1C) T cell proliferation upon ConA stimulation was drastically inhibited by the MSC2 cells. However, when the MSC2 cells were treated with MPSSS, the immunosuppressive activity of the MSC2 cells was impaired. Lentinan had no similar effect on MSC2 cells (ESI Fig. S1†). MPSSS had no effect on T cells and T cells proliferation induced by ConA (ESI Fig. S2†). Compared with the absence of MPSSS treatment, the activities of arginase in the MSC2 cells treated with MPSSS were downregulated (Fig. 1D). In addition, the cell-membrane receptor TNFR2 was decreased after MPSSS treatment (Fig. 1E), but TNFR1 was unaffected by the MPSSS treatment (Fig. 1F).3.2. MPSSS promoted MSC2 cell differentiation into M1-type cells
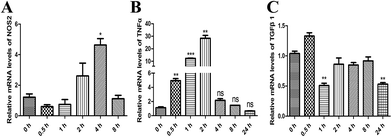
To investigate the effect of MPSSS on the differentiation of the MSC2 cells, the mRNA levels of NOS2, TNFα and TGF-β1 were analyzed by real-time PCR. After MPSSS treatment of the MSC2 cells over different time periods, the mRNA levels of NOS2 (Fig. 2A) and TNFα (Fig. 2B) were increased, and the maximum mRNA level of TNFα was 30 fold higher than that of untreated cells (Fig. 2B). In contrast to TNFα, the mRNA level of TGF-β1 was down regulated in the presence of MPSSS treatment compared to that in the absence of MPSSS treatment (Fig. 2C). Together, the results showing the downregulation of arginase and upregulation of NOS2 (Fig. 2A) indicated that MPSSS promoted the differentiation of the MSC2 cells into M1-type cells.3.3. MPSSS activated the p38 MAPK pathway
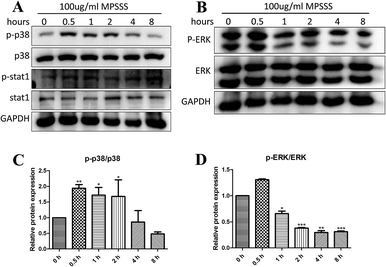
To investigate the effects of MPSSS on the activation of MAPKs, the MSC2 cells were treated with 100 μg ml−1 MPSSS for different time periods up to 8 hours. Western blot results showed that p38 was activated (phosphorylated) at maximum levels 0.5–2 h after MPSSS treatment (Fig. 3A and C). In addition, the levels of phosphorylated ERK were decreased after MPSSS treatment (Fig. 3B and D). As shown in Fig. 3A, the transcription factor STAT-1 was activated, which is an essential step for NOS2 induction in most cells.3.4. MPSSS impairs the inhibitory function of MSC2 cells though the p38 pathway
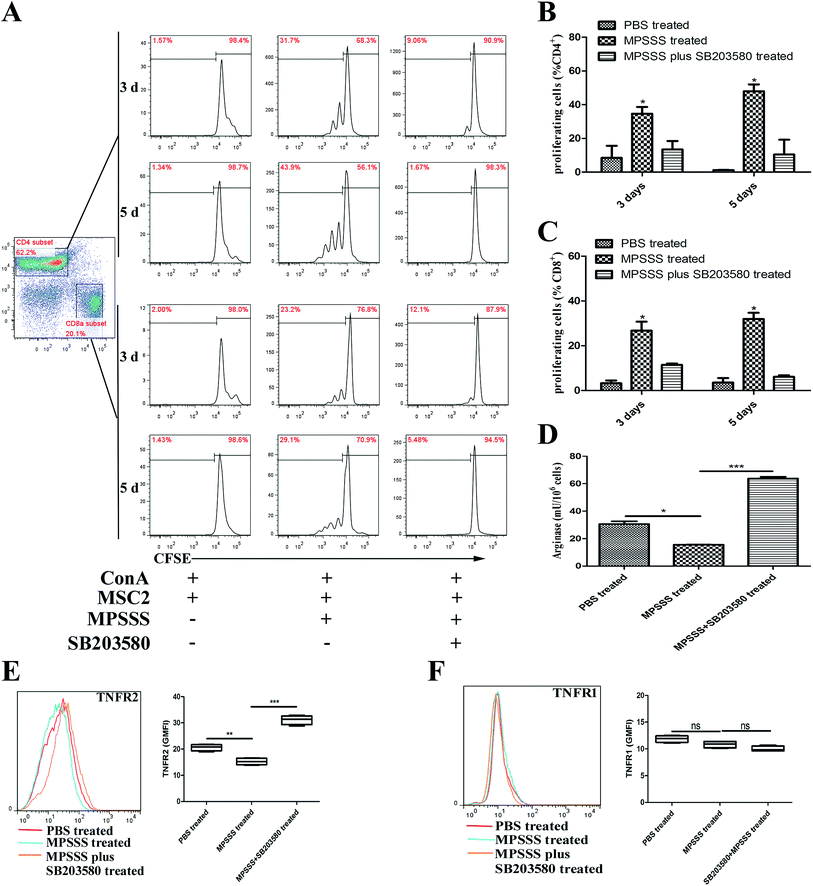
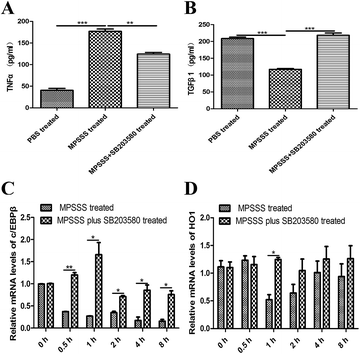
To investigate the role of p38 in the T cell proliferation and induction of arginase following MPSSS treatment, the MSC2 cells were pretreated with a p38 inhibitor SB203580 at 1 hour before MPSSS treatment. The results showed that MPSSS impaired the immunosuppressive function of the MSC2 cells, which was eliminated after the SB203580 pretreatment (Fig. 4A–C). In addition, when the p38 pathways were blocked, the activities of arginase in the MSC2 cells treated with MPSSS rapidly increased (Fig. 4D). In the presence of the SB203580 treatment, TNFR2 in the MPSSS-treated MSC2 cells was upregulated compared to that in the absence of the SB203580 treatment (Fig. 4E). With the same treatment, TNFR1 in the MSC2 cells was unaffected (Fig. 4F).3.5. Inhibition of p38 blocked the differentiation of the MSC2 cells and recovered the expression of c/EBPβ and HO1 induced by MPSSS
To investigate the effect of p38 on the differentiation of MSC2, we evaluated the effect of MPSSS on the MSC2 cells, which were pretreated with or without SB203580. The pretreated MSC2 cells were stimulated with MPSSS 24 hours. Then, the supernatant was used to detect TNFα and TGF-β1 by ELISA. Interestingly, the expression of TNFα was increased significantly after MPSSS treatment compared to that of the untreated cells or the SB203580 pretreatment cells (Fig. 5A). Meanwhile, the expression of TGF-β1 was reduced after MPSSS stimulation (Fig. 5B).The transcriptional factor c/EBPβ can activate the arginase promoter. We therefore assayed the mRNA level of c/EBPβ. The expression levels of c/EBPβ was decreased under simulation with MPSSS compared with those of the untreated MSC2 cells or the MPSSS plus SB203580-treated MSC2 cells (Fig. 5C). Depletion of arginine by arginase plays a major role in the MDSC-mediated T cell inhibition. In addition, the MPSSS-treated MSC2 cells can downregulate the expression of HO-1, and the SB203580 pretreatment of the MSC2 cells reversed this phenomenon (Fig. 5D).
4. Discussion
Our previous study isolated a polysaccharide, MPSSS, with a molecular weight of 5.772 × 105 daltons (Da) from L. edodes. Analysis of the monosaccharide composition of MPSSS showed that this polysaccharide fraction was mainly composed of glucose (75.0%), galactose (11.7%) and mannose (7.8%). The anti-cancer effect of MPSSS has been demonstrated; MPSSS reduced MDSC numbers and eliminated the MDSC-mediated T cell inhibition.4MDSCs aid tumor development by providing molecules and factors essential for angiogenesis and tumor metastasis and exert a profound inhibitory effect on T lymphocytes.13 Depletion of arginine by arginase plays a major role in the MDSC-mediated T cell inhibition.14 Furthermore, we have previously demonstrated that TNFR2 signaling promotes MDSC survival and accumulation and helps tumor cells evade the immune system.15,16 It has been reported that transmembrane TNFα but not secretory TNFα can activate MDSCs with enhanced suppressive activities via TNFR2 and promote tumor progression.16 In the present study, the results showed that MPSSS inhibited the suppressive function of the MSC2 cells, with significantly downregulated arginase activity (Fig. 1D) and decreased TNFR2 (Fig. 1E) but did not affect TNFR1 (Fig. 1F) and impaired the inhibition of T cell proliferation (Fig. 1A–C).
Induction of arginase and NOS2 is critical for macrophage polarization and function.17 Classical activation of macrophages by IFN-γ induces M1 polarization and results in the upregulation of TNF-α and induction of NOS2.18 On the other hand, the alternative activation of macrophages by Th2 cytokines induces M2 polarization and results in the upregulation of TGFβ and arginase.11,19,20 In all cases, the M1 phenotype cells are regarded as tumor-inhibiting, and the M2 phenotype cells are considered tumor-promoting.11 Our study demonstrated that MPSSS promoted MSC2 differentiation into M1-type cells, including elevated mRNA levels of NOS2 (Fig. 2A) and TNFα (Fig. 2B), with significantly decreased TGFβ1 mRNA (Fig. 2C). ERK activation is needed for arginase induction in activated macrophages and the M2 phenotype, while NOS2 induction depends on p38 activation.17 Mitogen and stress activated kinases 1 and 2 are activated by the p38 pathway and activate the transcription factor STAT1.21,22 Activation of the transcription factor STAT1 and the activation of the NOS2 promoter seem to be an essential step for NOS2 induction in most cells.23 Here, we found that p38 and STAT-1 were activated after MPSSS treatment in the MSC2 cells (Fig. 3A). Additionally, activation of ERK was clearly downregulated (Fig. 3B).
Hence, we wondered whether p38 activation plays a crucial role in the MPSSS-eliminated MSC2-mediated T cell inhibition. SB203580 is a p38 inhibitor that was identified to bind competitively at the ATP-binding pocket and has been widely used to study p38 functions.24–27 In this study, we found that MPSSS induced an ∼4-fold increase in the secretion of TNFα, which was blocked by SB203580 (Fig. 5A). As expected, inhibition of p38 completely reversed the effects of the MPSSS treatment and effectively decreased TGFβ1 secretion (Fig. 5B), arginase activity (Fig. 4D) and the expression of TNFR2 (Fig. 4E). Furthermore, MPSSS-stimulated MSC2 cells, which were pretreated with SB203580, could not prevent the inhibition of T cell proliferation (Fig. 4A–C).
It has been reported that c/EBPβ appears to play a major role in the induction of arginase expression,28 which acts downstream of ERK.29 Here, we found that MPSSS inhibited the expression of c/EBPβ (Fig. 5C) and reduced p-ERK (Fig. 3B). Interestingly, blocking the p-38 pathway showed that the expression of c/EBPβ and activation of arginase (Fig. 4D) were unaffected after MPSSS treatment. Some studies have shown that p38 MAPK signaling causes rapid inactivation of the ERK pathway mediated by PP2A.21,30 In addition, direct interaction between ERK and p38 MAPKs might also inhibit ERK phosphorylation.30 A previous study revealed that heme oxygenase-1 (HO-1) is one of the key mediators of the MDSC-associated suppression mechanism in transplantation,31 which leads to inhibition of T-cell proliferation through carbon monoxide (CO) production.32 In this study, we found that MPSSS-treated MSC2 cells could downregulate the expression of HO-1, and SB203580 pretreatment of the MSC2 cells reversed the phenomenon (Fig. 5D).
5. Conclusions
In conclusion, we found that a novel polysaccharide, MPSSS, obtained from L. edodes induced the differentiation of MSC2 cells and impaired their immunosuppressive function by suppressing arginase activity and the expression of TNFR2 via activation of p38 and dephosphorylation of ERK. The possible mechanisms involved in this process are illustrated in Fig. 6. Altogether, our results revealed that MPSSS may serve as a potential anti-cancer agent though reversing T cell inhibition by MDSCs (Fig. 6).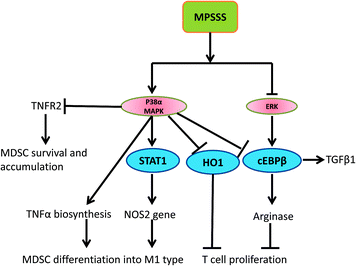 | ||
| Fig. 6 Schematic representation of the potential mechanisms involved in the MPSSS-induced inhibitory immunosuppressive function of MDSCs. | ||
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31400765, 31370910 and 31100646), Zhejiang Medical and Health Research Program (No. 2013KYA023), Shanxi Province Science Foundation for Youths (201601D202064), and Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (2015117).References
- X. Xu, H. Yan, J. Tang, J. Chen and X. Zhang, Polysaccharides in Lentinus edodes: isolation, structure, immunomodulating activity and future prospective, Crit. Rev. Food Sci. Nutr., 2014, 54, 474–487 CrossRef CAS PubMed.
- K. Oba, M. Kobayashi, T. Matsui, Y. Kodera and J. Sakamoto, Individual patient based meta-analysis of lentinan for unresectable/recurrent gastric cancer, Anticancer Res., 2009, 29, 2739–2745 Search PubMed.
- Y. Zhang, Q. Li, J. Wang, F. Cheng, X. Huang, Y. Cheng and K. Wang, Polysaccharide from Lentinus edodes combined with oxaliplatin possesses the synergy and attenuation effect in hepatocellular carcinoma, Cancer Lett., 2016, 377, 117–125 CrossRef CAS PubMed.
- H. Wu, N. Tao, X. Liu, X. Li, J. Tang, C. Ma, X. Xu, H. Shao, B. Hou, H. Wang and Z. Qin, Polysaccharide from Lentinus edodes inhibits the immunosuppressive function of myeloid-derived suppressor cells, PLoS One, 2012, 7, e51751 CAS.
- V. Kumar, S. Patel, E. Tcyganov and D. I. Gabrilovich, The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment, Trends Immunol., 2016, 37, 208–220 CrossRef CAS PubMed.
- B. Almand, J. I. Clark, E. Nikitina, J. van Beynen, N. R. English, S. C. Knight, D. P. Carbone and D. I. Gabrilovich, Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer, J. Immunol., 2001, 166, 678–689 CrossRef CAS.
- D. I. Gabrilovich, V. Bronte, S. H. Chen, M. P. Colombo, A. Ochoa, S. Ostrand-Rosenberg and H. Schreiber, The terminology issue for myeloid-derived suppressor cells, Cancer Res., 2007, 67, 425–426 CrossRef CAS PubMed , author reply.
- G. Gallina, L. Dolcetti, P. Serafini, C. De Santo, I. Marigo, M. P. Colombo, G. Basso, F. Brombacher, I. Borrello, P. Zanovello, S. Bicciato and V. Bronte, Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells, J. Clin. Invest., 2006, 116, 2777–2790 CAS.
- L. Yang, L. M. DeBusk, K. Fukuda, B. Fingleton, B. Green-Jarvis, Y. Shyr, L. M. Matrisian, D. P. Carbone and P. C. Lin, Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis, Cancer Cell, 2004, 6, 409–421 CrossRef CAS PubMed.
- E. Apolloni, V. Bronte, A. Mazzoni, P. Serafini, A. Cabrelle, D. M. Segal, H. A. Young and P. Zanovello, Immortalized Myeloid Suppressor Cells Trigger Apoptosis in Antigen-Activated T Lymphocytes, J. Immunol., 2000, 165, 6723–6730 CrossRef CAS.
- J. C. Becker, M. H. Andersen, D. Schrama and P. thor Straten, Immune-suppressive properties of the tumor microenvironment, Cancer Immunol. Immunother., 2013, 62, 1137–1148 CrossRef CAS PubMed.
- J. Jiang, Z. Wang, Z. Li, J. Zhang, C. Wang, X. Xu and Z. Qin, Early exposure of high-dose interleukin-4 to tumor stroma reverses myeloid cell-mediated T-cell suppression, Gene Ther., 2010, 17, 991–999 CrossRef CAS PubMed.
- P. Serafini, I. Borrello and V. Bronte, Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression, Semin. Cancer Biol., 2006, 16, 53–65 CrossRef CAS PubMed.
- P. C. Rodriguez and A. C. Ochoa, Arginine regulation by myeloid derived suppressor cells and tolerance in cancer: mechanisms and therapeutic perspectives, Immunol. Rev., 2008, 222, 180–191 CrossRef CAS PubMed.
- X. Zhao, L. Rong, X. Li, X. Liu, J. Deng, H. Wu, X. Xu, U. Erben, P. Wu, U. Syrbe, J. Sieper and Z. Qin, TNF signaling drives myeloid-derived suppressor cell accumulation, J. Clin. Invest., 2012, 122, 4094–4104 CAS.
- X. Hu, B. Li, X. Li, X. Zhao, L. Wan, G. Lin, M. Yu, J. Wang, X. Jiang, W. Feng, Z. Qin, B. Yin and Z. Li, Transmembrane TNF-alpha promotes suppressive activities of myeloid-derived suppressor cells via TNFR2, J. Immunol., 2014, 192, 1320–1331 CrossRef CAS PubMed.
- Y. Jin, Y. Liu and L. D. Nelin, Extracellular signal-regulated kinase mediates expression of arginase II but not inducible nitric-oxide synthase in lipopolysaccharide-stimulated macrophages, J. Biol. Chem., 2015, 290, 2099–2111 CrossRef CAS PubMed.
- S. Gordon, Alternative activation of macrophages, Nat. Rev. Immunol., 2003, 3, 23–35 CrossRef CAS PubMed.
- A. Mantovani, S. Sozzani, M. Locati, P. Allavena and A. Sica, Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes, Trends Immunol., 2002, 23, 549–555 CrossRef CAS PubMed.
- K. Ckless, A. Lampert, J. Reiss, D. Kasahara, M. E. Poynter, C. G. Irvin, L. K. Lundblad, R. Norton, A. van der Vliet and Y. M. Janssen-Heininger, Inhibition of arginase activity enhances inflammation in mice with allergic airway disease, in association with increases in protein S-nitrosylation and tyrosine nitration, J. Immunol., 2008, 181, 4255–4264 CrossRef CAS.
- A. Cuadrado and A. R. Nebreda, Mechanisms and functions of p38 MAPK signalling, Biochem. J., 2010, 429, 403–417 CrossRef CAS PubMed.
- J. S. Arthur, MSK activation and physiological roles, Front. Biosci., Landmark Ed., 2008, 13, 5866–5879 CrossRef CAS.
- H. Kleinert, P. M. Schwarz and U. Forstermann, Regulation of the expression of inducible nitric oxide synthase, Biol. Chem., 2003, 384, 1343–1364 CrossRef CAS PubMed.
- S. T. Wrobleski and A. M. Doweyko, Structural comparison of p38 inhibitor-protein complexes: a review of recent p38 inhibitors having unique binding interactions, Curr. Top. Med. Chem., 2005, 5, 1005–1016 CrossRef CAS PubMed.
- J. Zhang, B. Shen and A. Lin, Novel strategies for inhibition of the p38 MAPK pathway, Trends Pharmacol. Sci., 2007, 28, 286–295 CrossRef CAS PubMed.
- S. Kumar, J. Boehm and J. C. Lee, p38 MAP kinases: key signalling molecules as therapeutic targets for inflammatory diseases, Nat. Rev. Drug Discovery, 2003, 2, 717–726 CrossRef CAS PubMed.
- L. R. Coulthard, D. E. White, D. L. Jones, M. F. McDermott and S. A. Burchill, p38(MAPK): stress responses from molecular mechanisms to therapeutics, Trends Mol. Med., 2009, 15, 369–379 CrossRef CAS PubMed.
- M. J. Gray, M. Poljakovic, D. Kepka-Lenhart and S. M. Morris Jr, Induction of arginase I transcription by IL-4 requires a composite DNA response element for STAT6 and C/EBPbeta, Gene, 2005, 353, 98–106 CrossRef CAS PubMed.
- F. A. Simard, A. Cloutier, T. Ear, H. Vardhan and P. P. McDonald, MEK-independent ERK activation in human neutrophils and its impact on functional responses, J. Leukocyte Biol., 2015, 98, 565–573 CrossRef CAS PubMed.
- M. R. Junttila, S. P. Li and J. Westermarck, Phosphatase-mediated crosstalk between MAPK signaling pathways in the regulation of cell survival, FASEB J., 2008, 22, 954–965 CrossRef CAS PubMed.
- Y. Zhao, T. Wu, S. Shao and B. Shi, Phenotype, development, and biological function of myeloid-derived suppressor cells, OncoImmunology, 2016, 5, e1004983 CrossRef PubMed.
- V. De Wilde, N. Van Rompaey, M. Hill, J. F. Lebrun, P. Lemaitre, F. Lhomme, C. Kubjak, B. Vokaer, G. Oldenhove, L. M. Charbonnier, M. C. Cuturi, M. Goldman and A. Le Moine, Endotoxin-induced myeloid-derived suppressor cells inhibit alloimmune responses via heme oxygenase-1, Am. J. Transplant., 2009, 9, 2034–2047 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra06789e |
| This journal is © The Royal Society of Chemistry 2017 |