Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Hydrophilic cell-derived extracellular matrix as a niche to promote adhesion and differentiation of neural progenitor cells†
Lingyan Yangab,
Ziyun Jianga,
Linhong Zhouc,
Keli Zhaoa,
Xun Maa and
Guosheng Cheng *ab
*ab
aCAS Key Laboratory of Nano-Bio Interface, Suzhou Institute of Nano-Tech and Nano-Bionics, Chinese Academy of Sciences, Jiangsu 215123, China. E-mail: gscheng2006@sinano.ac.cn
bSchool of Nano Technology and Nano Bionics, University of Science and Technology of China, Anhui 230026, China
cDepartment of Pharmacy, School of Medicine, Xi'an Jiaotong University, Shaanxi 710061, China
First published on 27th September 2017
Abstract
The natural extracellular matrix (ECM) offers a dynamic and intricate microenvironment, which serves as a structural support and regulates cell phenotype and its function. Recently, ECM has been revealed as a favorable and biocompatible architecture for stem cell adhesion and growth in cellular therapy to treat various diseases. However, cell-derived ECM is rarely used as a culture substrate for anchorage-dependent cells, such as neural progenitor cells (NPCs), which have the potential to differentiate into basal forebrain cholinergic neurons (BFCNs) for Alzheimer's disease. Here, we report mouse embryonic fibroblast (MEF)-derived ECM, with an appropriately hydrophilic property (water contact angle of 66.8°) to mimic the neural niche for NPCs. In addition, MEF-derived ECM possesses a nanotopological surface, plentiful kinds of components and exhibits excellent adhesion properties for anchoring NPCs. Compared with a laminin-coated plate, MEF-derived ECM promotes NPC proliferation and differentiation into BFCNs by ∼1.6 fold and ∼3.1 fold, respectively, consequently enhancing the production of acetylcholine by ∼2.0 fold. This MEF-derived ECM could be a favorable cell culture carrier for NPC attachment, with great potential for applications in stem cell therapy for Alzheimer's disease.
Introduction
Alzheimer's disease is a very serious neurodegenerative disorder and is primarily characterized by progressive deficits in spatial learning and memory, which are related to the early substantial loss of basal forebrain cholinergic neurons (BFCNs) and the absence of cortical cholinergic input.1 The developed primate and rodent models have demonstrated the roles of BFCNs in memory function, hippocampal neurogenesis, and functional plasticity of the cortex.2 The transplant of exogenous BFCNs to ameliorate memory deficits through stable engraftment of cells in the adult cortex has become a potential therapeutic strategy.3 However, due to the limitations in acquiring sufficient BFCNs from precursor cells, the ability to promote the differentiation of stem cells into BFCNs would be a significant step toward a cell replacement therapy.4 Recently, neural progenitor cells (NPCs)-based therapy has emerged as a promising approach to restore the original BFCN function in the cortical and hippocampal regions for treatment of Alzheimer's disease.5 NPCs can proliferate and differentiate into BFCNs when they attach onto a matrix under the appropriate conditions, and thus, they can be considered as anchorage-dependent cells.6 Anchorage-dependent cells require a good cell carrier to spread, reside, proliferate, differentiate and maintain other cellular functions. Especially, stem cell functions require activation of intrinsic transcription and interaction with a specific extracellular microenvironment niche.The extracellular matrix (ECM) is a natural cell-growth microenvironment, containing a variety of biological components secreted by the resident cells in tissues and organs.7 It has been proved that ECM provides a mechanical support and signaling cues to regulate cell survival, proliferation, differentiation and metastasis via the interaction of cells and ECM.8–11 Given the advantageous features, native tissue-derived ECM, mainly derived from allogeneic and xenogeneic tissues treated with chemical or thermal decellularization, has been utilized extensively toward promoting tissue engineering and regenerative medicine.12 However, tissue-derived ECM has several limitations, such as limited donors, inflammatory reactions, pathogens transfer disease and uncontrollable degradation.13 Recently, electrostatic spinning, hydrogels and three-dimensional bio-printing technologies have been used to fabricate scaffolds with synthetic material or a limited set of natural polymer blends, but these fail to adequately mimic the complex morphology and composition of natural ECM.14 In addition, purified ECM components, including fibrin, laminin, fibronectin, collagen and hyaluronan etc., serve important roles to mimic the native microenvironment for the engraftment of anchorage-dependent cells and have been employed in various types of models in vitro and in vivo.6 Moreover, natural ECM can also promote tissue-specific stem cell differentiation, whereas individual components, such as laminin and collagen, have no significant effects on cell proliferation and differentiation.15 Therefore, individual ECM components are not enough to build an ideal niche because different components act for different functions.16
To more fully replicate the biological molecules and surface characteristics of ECM in natural tissues, a great deal of research has attempted to develop an approach that involves the fabrication of cell-derived ECM. Similar to tissue-derived ECM, cell-derived ECM also has highly advantageous biophysical and biochemical properties. More importantly, cell-derived ECM can eliminate the possibility of inflammatory reactions and pathogen transfer.17 Therefore, cell-derived ECM has been extensively applied to tissue engineering and regenerative medicine, instead of the tissue-derived ECM.13,18–20 Meanwhile, cell-derived ECM as cell attachment carrier has gained increasing interests in vitro applications. In previous studies, cell-derived ECM was mainly contributed to culture dorsal root ganglion neurons,20 mesenchymal stem cells21 and Schwann cells,16 which are anchorage-independent cells, with less strict matrix requirements since those cells can adhere and grow on commercialized tissue culture polystyrenes without any modifications. To date, there are scarcely reports regarding the use of cell-derived ECM as a niche for anchorage-dependent cells, such as neural stem/progenitor cells.
Here, MEF (mouse embryonic fibroblast) was selected to fabricate a cell-derived ECM model. Which has been evaluated as a neural niche for cultured NPC. Importantly, this MEF-derived ECM exhibits excellent adhesion performance for NPC anchoring and residency, while promote NPC proliferation and generation into BFCN. Our results reveal that MEF-derived ECM could be a promising candidate for neural niche, providing a novel technique for developing cell-derived ECM for NPC application into clinical therapy of Alzheimer's diseases.
Experimental
Materials
Gelatin, L-ascorbic acid, sodium ascorbate, deoxycholate, DNase, Triton X-100 and DAPI were provided from Sigma-Aldrich, USA. Glutaraldehyde and tertiary butanol were ordered from Alfa Aesar, USA. DMEM, FBS, penicillin/streptomycin, PBS, TRIzol reagent, AmplexRed Acetylcholine/Acetylcholinesterase Detection Kit, EdU Labeling/Detection Kit and ECL Western blot Substrate Kit were offered from Thermo Fisher, USA. Mouse anti-fibronectin antibody, rabbit anti-laminin antibody, mouse anti-nestin antibody, rabbit anti-vimentin antibody, mouse anti-Map-2 antibody, goat anti-ChAT antibody, rabbit anti-p75 antibody, rabbit anti-VAChT antibody and secondary antibodies were supplied from Abcam, USA. First Strand cDNA Synthesis Kit and SYBR Green Real-time PCR Master Mix were purchased from Applied Biosystems, USA.Formation of MEF-derived ECM
Tissue culture plates were pre-coated with 0.5% gelatin overnight at 37 °C and exposed to ultraviolet rays for 2 h to enhance crosslinking. MEFs were first cultured in DMEM with 10% FBS for 3–5 d until 100% confluence. The cells were continually cultured in DMEM with 20% FBS and 50 μg mL−1 L-ascorbic acids and 100 μg mL−1 sodium ascorbate for 14 d to stimulate ECM formation, and the medium was routinely changed every 48 h.22 For decellularization, cells were washed with deionized water at 37 °C for 20 min and then dried, followed by treatments with 0.5% Triton X-100 plus 1% deoxycholate for 10 min and with 100 U mL−1 DNase for 30 min at 37 °C. After washing for three times with PBS, the MEF-derived ECM was stored in PBS containing 100 U mL−1 penicillin/streptomycin at 4 °C.Scanning electron microscopy imaging
The samples were washed three times with PBS, and fixed with 2.5% glutaraldehyde for 30 min at 4 °C, followed by post-fixation in 1% osmium tetroxide. The samples were then dehydrated in gradient concentrations of ethanol (50% to 100%) for 10 min, followed by replacement with 100% tertiary butanol, and finally, lyophilized using a vacuum drier. After the samples were coated with gold, they were observed using a scanning electron microscope (Quanta 400 FEG, FEI, USA).Immunofluorescence staining
The samples were fixed with 4% paraformaldehyde for 20 min at room temperature, washed with PBS, and then blocked with 5% normal goat serum for 1 h. The samples were incubated with the relevant primary antibodies overnight at 4 °C. After washing with PBS, samples were incubated with the appropriate fluorescently labelled secondary antibodies for 2 h in the dark at room temperature, followed by nuclear staining with 5 μg mL−1 DAPI. Fluorescence microscopy images were collected using fluorescence microscopy (Nikon, Japan).Wettability assessment
The interaction force between a water droplet and the MEF-derived ECM interface was assessed by a high-sensitivity microelectromechanical balance system (Dataphysics DCAT11, Germany), and the droplet contact angle was measured using the captive droplet method (Dataphysics OCA20).23 The volume of the droplet was approximately 3 μL for each test. Glass without any modifications and a laminin coated surface were used as controls. All the experiments were repeated more than five times.EdU/Hoechst 33342 double staining
Based on the instructions from the EdU Labelling/Detection Kit, NPCs were plated into a 24-well plate with containing MEF-derived ECM or laminin coated and cultured for 7 d. Then the cells incubated in medium containing 10 μM EdU for an additional 24 h at 37 °C in 5% CO2. Subsequently, the NPCs were fixed with 4% paraformaldehyde for 20 min. After rinsing with PBS, cells were incubated with 1× Apollo reaction buffer for 30 min in the dark, permeated with 0.5% Triton X-100 in PBS, and stained with 5 μg mL−1 Hoechst 33342 dye for 30 min. Images were obtained with a fluorescence microscope (Nikon, Japan). The percentage of EdU-positive cells was calculated from five random fields in each well (five wells per group).Real-time polymerase chain reaction (RT-PCR)
After the NPCs were cultured on MEF-derived ECM or laminin pre-coated plate for 24 h (for adhesion), 7 d (for proliferation) or 21 d (for differentiation), total RNA was extracted using TRIzol reagent. cDNA was synthesized from total RNA using a First Strand cDNA Synthesis Kit according to the manufacturer's instructions. The primer sequences for genes are shown in Table S1.† SYBR Green Real-time PCR Master mix was used to quantify mRNA expression according to the manufacturer's instructions. Reaction mixtures were incubated at 95 °C for 15 sec and 60 °C for 45 sec for 40 cycles. The 2−ΔΔCt method was used to analyze the relative mRNA expression. β-Actin was used as an endogenous control.Western blot assay
After NPCs were cultured on MEF-derived ECM or laminin for 24 h (for adhesion), 7 d (for proliferation) or 21 d (for differentiation), cells were harvested with 0.25% trypsin in 0.03% EDTA and lysed in RIPA buffer. The collected proteins were loaded onto a 10% SDS-PAGE gel, separated by gel electrophoresis, and transferred onto a PVDF membrane. The membrane were blocked for 1 h and incubated with primary antibodies overnight. Subsequently, the membranes were incubated with HRP-conjugated secondary antibodies for 2 h. After washing with TBS-T, the membranes were reacted with the ECL western blot substrate before exposure. β-Actin was used as an endogenous control.Residency and motion assay
The residency and motion of NPCs cultured on ECM or laminin pre-coated plates were assessed by monitoring cells for 24 h and taking pictures every 30 min using a live cell imaging system (Cytation 3, BioTek, USA). For the cell tracking analysis, each individual cell position (x, y) was tracked, and the cell tracking plots, accumulated distance, Euclidean distance and velocity were generated using Chemotaxis Tool freeware (ibidi, Germany).24Quantification of acetylcholine
After the NPCs were cultured on MEF-derived ECM or laminin with differentiation medium for 21 d, quantification of the ACh levels in the cultured neurons was performed with an AmplexRed Acetylcholine/Acetylcholinesterase Detection Kit and correlated with protein levels determined using the Protein Quantification Kit according to the manufacturer's manual.Statistical analysis
All quantitative data are expressed as the mean ± SD. Statistical analysis was performed using a two-sample t-test with Origin Pro 8 software. *p < 0.05 was considered statistically significant; **p < 0.01 was considered highly significant. All experimental data shown represent experiments performed in triplicate.Results and discussion
Morphology and composition of MEF-derived ECM
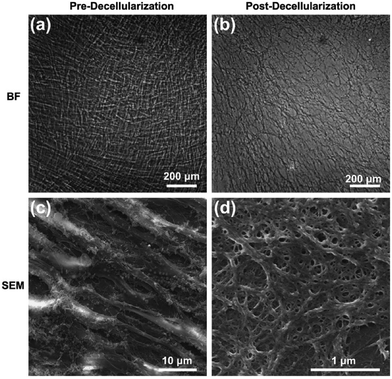
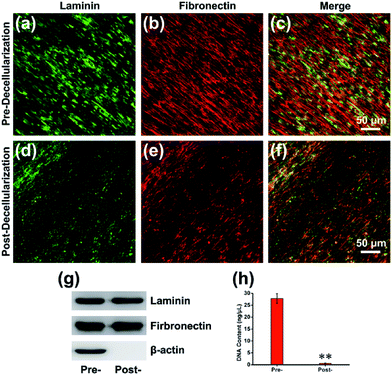
The detailed process of our experiment is presented in Scheme 1. Fibroblasts are able to synthesize larger quantities of ECM biomolecules, including collagenous matrix and elastin networks.25 MEFs were applied to fabricate the cell-derived ECM in this study (ESI, Fig. S1†), which can be easily isolated and cultured up passage 14 without a decrease in collagen synthesis or a reduction in the rate of growth. Stimulated with L-ascorbic acid and sodium ascorbate for 14 d, MEFs were embedded in secreted ECM deposition and interweaved together (Fig. 1a and c). After decellularization, the cells were lysed, and most of the adhesive molecules and proteins were removed, thus clearly exposing the protein fibers. The remaining ECM consisted of a large number of interconnected filaments distributed into a network (Fig. 1b), and high resolution observation showed that post-decellularized ECM exhibited a filamentous, nanoscale (∼80 nm in diameter), porous network with a fabric-style ultrastructural appearance (Fig. 1d).The composition of the MEF-derived ECM was first examined by immunofluorescence staining, which presented that the ECM was abundant with structural proteins such as laminin (Fig. 2a and d) and fibronectin (Fig. 2b and e) before or after decellularization. Western blot analysis with β-actin as an internal control revealed that the amounts of laminin and fibronectin were maintained. However, β-actin, as a cytoskeletal protein, was absent after decellularization (Fig. 2g). To examine the presence of any nuclear contamination following decellularization, DNA content was quantified with an assay kit as described previously.26 The results of the DNA quantification indicated that almost of cellular DNA (>98%) was removed after decellularization (Fig. 2h). To further determine the composition of the ECM, the proteins were analyzed using nano-liquid chromatography tandem mass spectrometry (nLC-MS/MS) analysis after the efficient decellularization.10 The obtained results indicated the reliable detection and relative quantification of 809 different proteins, and the 10 most abundant proteins are listed according to counts of peptides (ESI, Table S2†). Within those 809 kinds of components, many of the proteins constituting the matrix may play key roles in maintaining the superior biocompatibility. For instance, collagen is the major structural protein in the ECM and imparts mechanical properties to natural tissues.27 Meanwhile, collagen is a critical component for cell anchorage in the NPC niche, and the interaction of cells with collagen influences adhesion, residency, cell–cell communication, and cell survival. Another important component, Tenascin C (TNC), is an extracellular matrix glycoprotein that is highly expressed by NPCs located in the brain and spinal cord during development and in the adults. Consistent with the dynamic interplay of factors within the NPC niche, the TNC-mediated alterations in growth factor responsiveness may be due, in part, to secondary alteration heparan sulfate proteoglycan alterations.28
Wettability of MEF-derived ECM
Previous investigations have shown that the material with the best hydrophilicity was favorable for cell adhesion.29 However, recent research has proposed that the adhesion activity is related to the degree of hydrophilicity, and the primary reason for this is that different surface hydrophilicities can absorb a variety of types of proteins and molecules. It is generally accepted that hydrophobic surfaces adsorb more protein, whereas hydrophilic surfaces do not facilitate protein adsorption. The adsorption of proteins to scaffold surface has been reported as highest one with water contact angle of 60–80°.30 In this work, the hydrophilicity of MEF-derived ECM was determined by the water contact angle measured using the captive droplet method. The contact angle of the droplet with ECM, laminin and glass were approximately 66.8° ± 4.55°, 15.4° ± 3.85° and 120.4° ± 4.16°, respectively (Fig. 3). The appropriate hydrophilicity is determined by the components, nanotopology and porous surface. More importantly, the different hydrophilicities lead to different adhesive properties. The adhered proteins can transmit signals to the cells through cell adhesion receptors and thereby affect cell survival, growth and differentiation.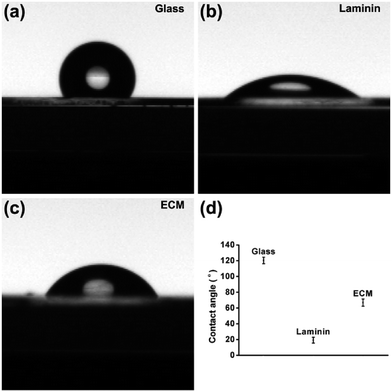 | ||
| Fig. 3 Digital images showing water droplet wettability on (a) glass without any modifications, (b) laminin coated plate and (c) MEF-derived ECM. (d) The contact angle statistics. | ||
Adhesion and residency performance of MEF-derived ECM
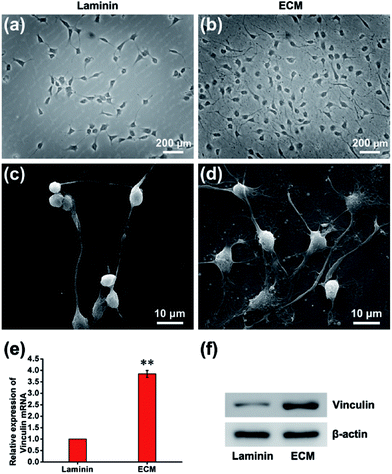
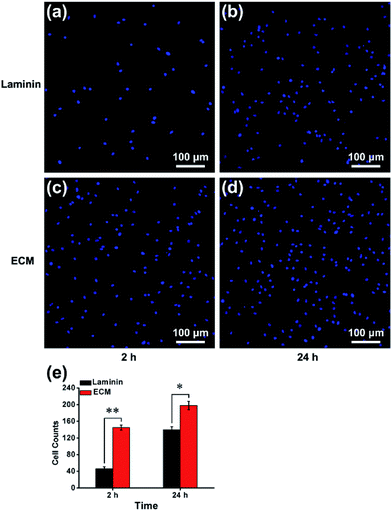
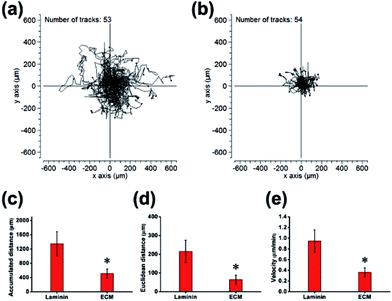
In many previous studies, to facilitate anchorage-dependent cell adhesion, individual ECM components, including laminin, fibronectin, collagen and other bioactive molecules, have been used to modify plates or glass surfaces.6 In this work, laminin pre-coated was adopted as the control group, and no obvious difference in micromorphology was observed between them. Pre-coated with laminin is a standard protocol for surface treatment, and it is a widely used biomolecule for culturing neural stem cells. As investigated using live/dead assay kit and immunofluorescence staining with anti-nestin antibody (ESI, Fig. S3 and S4†), MEF-derived ECM had excellent biocompatibility and maintained the stemness properties. NPCs were then cultured for 24 h, and the cell adhesion performance on ECM was observed. SEM images revealed that NPCs anchored on the surface of ECM had an extensive spreading morphology and formed a strong interaction with the ECM, exhibiting excellent cell adhesion performance (Fig. 4a–d). Additionally, vinculin is a key component of cellular adhesion plaques and adherents junctions. Our real-time PCR and western blot results indicated significantly increased mRNA and protein expression levels of vinculin in NPCs cultured on the ECM (Fig. 4e and f). Further experiments were performed to re-validate cell anchorage efficiency on ECM. Cells were fixed after culturing for 2 or 24 h and then stained with DAPI for fluorescence microscopy images. In Fig. 5, it can be observed that after 2 h of culture, most of the NPCs were adhered on the ECM, and only a few cells were adhered on the laminin. After 24 h of culture, the number of cells adhered to the ECM was greater than that of the laminin group. This indicated that rapid, efficient anchorage was more favorable on ECM than on laminin.Cell residency and motion are a complex process involving cell adhesion, polarization and forward movement.31 As to the residency of NPCs on the neural niche, we observed the cell adhesion and motion continuously for 24 h using a live cell imaging system (ESI, Movie S1 and S2†). The individual cell tracks were plotted with cell initial positions (point 0, 0), and the results showed that cells cultured on laminin (Fig. 6a) moved faster than that those cultured on MEF-derived ECM (Fig. 6b). The statistics histogram of the accumulated distance, Euclidean distance, and velocity calculated from individual cell tracks (Fig. 6c–e) confirmed that cell anchorage on ECM had a low movement ratio. Except for diverse composition, the appropriately hydrophilic surface of ECM is believed to adsorb sufficient nutrition to meet the metabolic demands of NPCs, which can facilitate cell adhesion and residency. More important, NPCs moved absolutely randomly without any preferential direction on the ECM and laminin (Fig. 6a and b and Movies in ESI†), and from this, it can be inferred that the ECM was a relatively homogeneous material.
Cell proliferation performance of MEF-derived ECM
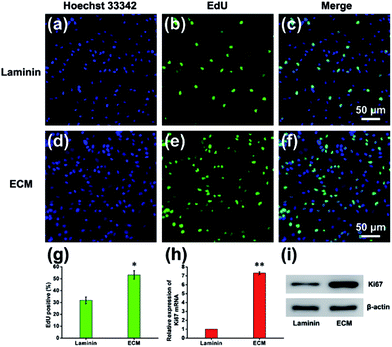
Cell proliferation is a process that results in an increase in the number of cells and defined by the balance between cell division and cell loss through cell death or differentiation. The proliferation of NPCs on MEF-derived ECM was examined by measuring the ratio of EdU-positive cells after NPCs were maintained in proliferation medium for 7 d. The EdU/Hoechst 33342 immunofluorescence results showed that ECM promoted NPCs proliferation (Fig. 7a–f). The percentage of EdU-positive cells in the ECM group was significantly higher than that of the laminin group (53.25% ± 4.54% vs. 32.83% ± 2.86%, Fig. 7g). NPC proliferation was also verified by measuring the expression of Ki67 mRNA and protein. As expected, the mRNA and protein expression levels of Ki67 were significantly higher in NPCs cultured on ECM than in those cultured on laminin pre-coated plates (Fig. 7h and i). According to the movies using live cell imaging system, cell division was observed within 24 h after NPC anchorage on the ECM, whereas rarely observed in the laminin group. This indicated that cells cultured on the ECM entered into proliferation state as soon as they adhered. How does the ECM facilitate this process? First, the appropriate hydrophilicity and nanotopgraphy of the ECM surface can lead to the adsorption of more protein and other growth cytokines from the proliferation medium and nearby cells, which provides more nutrients for cell growth and division. In addition, our MEF-derived ECM contains many glycan-binding proteins and basal lamina, for instance, filamin and a galectin, which can promote the proliferation of NPCs in vitro and in vivo.32Cell differentiation performance of MEF-derived ECM
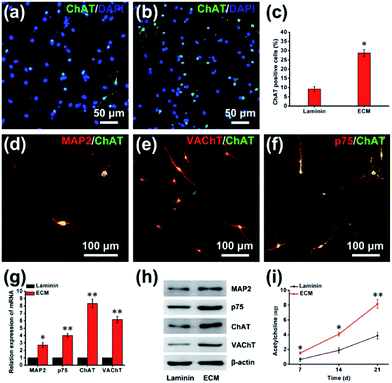
The successful differentiation of NPCs is a significant step for effective stem cell-mediated treatment of Alzheimer's disease. After 21 d of differentiation, immunofluorescence staining showed a vast increase in the number of cells positive for choline acetyltransferase (ChAT) from 9.23% ± 1.25% on the laminin to 28.75% ± 1.82% on the ECM (Fig. 8a–c). ChAT catalyzes the formation of ACh and is expressed by cholinergic neurons of both the basal forebrain and the motor system. In previous studies, ChAT positive neurons differentiation from human pluripotent stem cell on laminin coated was 15% at 32 d.2 In addition, immunofluorescence staining with double labeling confirmed that ChAT-immunopositive cells were almost all microtubule-associated protein 2 (MAP2)-, vesicular acetylcholine transport (VAChT)- and neurotrophin receptor (p75NTR, p75)-positive (Fig. 8d–f). For further quantitative analysis, cells were harvested and subjected to RT-PCR and western blot assay. As shown in Fig. 8g and h, compared with the cells on laminin pre-coated plates, a large and significant increase in the expression of markers for the BFCN lineage, including ChAT, MAP2, VAChT and p75. The functionality of the BFCN was confirmed through direct detection of ACh, which plays a significant role in synaptic transmission, mediating fast excitatory neurotransmission by binding to ACh receptors. As a neural niche, MEF-derived ECM for cholinergic neurons markedly increases ACh levels: 8.15 ± 0.57 ng ACh per microgram of protein, and 3.89 ± 0.45 ng ACh per microgram of protein for the laminin group at day 21, comparatively (Fig. 8i). This differentiation effect was almost certainly due to a complex interplay between the chemical and physical properties of the ECM and the cells. The ECM was composed of rich variety of protein, such as proteoglycans and glycoproteins. Heparan sulfate proteoglycans (HSPGs) are one of the main components of MEF-derived ECM according to our proteomic analysis (ESI, Table S2†), it's been noted that HSPGs could help drive the differentiation of neural cells by promoting FGF and BMP signaling.33 BMP-9 is a critical exogenous cytokines for the differentiation of NPCs into BFCNs in our method. Perlecan (HSPG2) is an HSPG, highly expressed in the basal lamina of the developing neuroepithelium. The growth cytokines binding ability of HSPG2, including its binding to FGF8 and SHH, is considered to be the primary mechanism by which HSPG2 regulates NPC proliferation and differentiation.34In summary, although the microenvironment that determines the fate of stem cells has been extensively reported, these were scarcely studies used cell-derived ECM as a neural niche for neural stem/progenitor cells, as anchorage-dependent cells, and there was also not any data about cell-derived ECM affecting the differentiation of neural stem/progenitor cells, especially. Therefore, we utilized MEF-derived ECM as a cell culture carrier for NPC in vitro. It was found that ECM not only maintains the viability and stemness of NPCs, but also exhibits an excellent adhesion performance. Meanwhile, the ECM promoted efficient proliferation and differentiation of cells into BFCNs. There might be three aspects for interpretation. (i) Nanotopographical surface: biomaterials with nanoscale topography effectively mimic surface characteristics of natural tissues and may significantly affect protein adsorption, cell adhesion. (ii) Diverse composition: MEF-derived ECM has a rich variety of components, including proteoglycans, glycoproteins and HSPGs. (iii) Appropriate hydrophilicity: appropriate hydrophilicity of MEF-derive ECM ensure the enhancement of anchorage of NPCs, and promotion of efficient protein adsorption of nutrition to meet the metabolic demands of NPCs, facilitating cell adhesion, residence and proliferation within the ECM.
Conclusions
Our findings revealed the MEF-derived ECM, as a neural niche for NPCs with excellent biocompatible properties, it possesses the abilities to maintain stemness, adhesion, residence, proliferation and differentiation. Nanotopographical structure, the variety of components, and appropriate hydrophilicity of MEF-derived ECM were proposed. It's important that MEF-derived ECM has been confirmed as a neural niche with great potential applications in neural stem cell therapy for Alzheimer's disease.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Basic Research Program of China (973 Program grant no. 2014CB965003).Notes and references
- C. J. Bissonnette, L. Lyass, B. J. Bhattacharyya, A. Belmadani, R. J. Miller and J. A. Kessler, Stem Cells, 2011, 29, 802–811 CrossRef PubMed.
- Y. Hu, Z. Y. Qu, S. Y. Cao, Q. Li, L. Ma, R. Krencik, M. Xu and Y. Liu, J. Neurosci. Methods, 2016, 266, 42–49 CrossRef CAS PubMed.
- W. Yue, Y. Li, T. Zhang, M. Jiang, Y. Qian, M. Zhang, N. Sheng, S. Feng, K. Tang, X. Yu, Y. Shu, C. Yue and N. Jing, Stem Cell Rep., 2015, 5, 776–790 CrossRef CAS PubMed.
- L. Duan, B. J. Bhattacharyya, A. Belmadani, L. Pan, R. J. Miller and J. A. Kessler, Mol. Neurodegener., 2014, 9, 3 CrossRef PubMed.
- G. Tincer, V. Mashkaryan, P. Bhattarai and C. Kizil, Yale J. Biol. Med., 2016, 89, 23–35 Search PubMed.
- N. Li, Q. Zhang, S. Gao, Q. Song, R. Huang, L. Wang, L. W. Liu, J. W. Dai, M. L. Tang and G. S. Cheng, Sci. Rep., 2013, 3, 1604 CrossRef PubMed.
- S. Yi, F. Ding, L. Gong and X. Gu, Curr. Stem Cell Res. Ther., 2017, 12, 233–246 CrossRef CAS PubMed.
- P. Y. Hwang, J. Chen, L. Jing, B. D. Hoffman and L. A. Setton, J. Biomech. Eng., 2014, 136, 021010 CrossRef PubMed.
- S. V. Glavey, A. Naba, S. Manier, K. Clauser, S. Tahri, J. Park, M. R. Reagan, M. Moschetta, Y. Mishima, M. Gambella, A. Rocci, A. Sacco, M. E. O'Dwyer, J. M. Asara, A. Palumbo, A. M. Roccaro, R. O. Hynes and I. M. Ghobrial, Leukemia, 2017 DOI:10.1038/leu.2017.102.
- R. Santhakumar, P. Vidyasekar and R. S. Verma, PLoS One, 2014, 9, e114697 Search PubMed.
- C. C. Cheng, D. J. Lee and J. K. Chen, Acta Biomater., 2017, 50, 476–483 CrossRef CAS PubMed.
- H. Lu, T. Hoshiba, N. Kawazoe and G. Chen, Biomaterials, 2011, 32, 2489–2499 CrossRef CAS PubMed.
- J. Liao, X. Guo, K. J. Grande-Allen, F. K. Kasper and A. G. Mikos, Biomaterials, 2010, 31, 8911–8920 CrossRef CAS PubMed.
- S. F. Badylak, D. O. Freytes and T. W. Gilbert, Acta Biomater., 2009, 5, 1–13 CrossRef CAS PubMed.
- D. Philip, S. S. Chen, W. Fitzgerald, J. Orenstein, L. Margolis and H. K. Kleinman, Stem Cells, 2005, 23, 288–296 CrossRef PubMed.
- B. Xiao, F. Rao, Z. Y. Guo, X. Sun, Y. G. Wang, S. Y. Liu, A. Y. Wang, Q. Y. Guo, H. Y. Meng, Q. Zhao, J. Peng, Y. Wang and S. B. Lu, Neural Regener. Res., 2016, 11, 1172–1179 CrossRef PubMed.
- S. M. Ruff, S. Keller, D. E. Wieland, V. Wittmann, G. E. Tovar, M. Bach and P. J. Kluger, Acta Biomater., 2017, 52, 159–170 CrossRef CAS PubMed.
- H. Lu, T. Hoshiba, N. Kawazoe, I. Koda, M. Song and G. Chen, Biomaterials, 2011, 32, 9658–9666 CrossRef CAS PubMed.
- Y. Gu, Z. Li, J. Huang, H. Wang, X. Gu and J. Gu, J. Tissue Eng. Regener. Med., 2017, 11, 2250–2260 CrossRef CAS PubMed.
- Y. Gu, J. Zhu, C. Xue, Z. Li, F. Ding, Y. Yang and X. Gu, Biomaterials, 2014, 35, 2253–2263 CrossRef CAS PubMed.
- M. Pei, F. He and V. L. Kish, Tissue Eng., Part A, 2011, 17, 3067–3076 CrossRef CAS PubMed.
- J. M. Bourget, R. Gauvin, D. Larouche, A. Lavoie, R. Labbe, F. A. Auger and L. Germain, Biomaterials, 2012, 33, 9205–9213 CrossRef CAS PubMed.
- Z. Y. Lu, W. Zhu, X. Y. Yu, H. C. Zhang, Y. J. Li, X. M. Sun, X. W. Wang, H. Wang, J. M. Wang, J. Luo, X. D. Lei and L. Jiang, Adv. Mater., 2014, 26, 2683–2687 CrossRef CAS PubMed.
- X. R. Li, H. Liang, J. Sun, Y. Zhuang, B. Xu and J. W. Dai, Adv. Healthcare Mater., 2015, 4, 1869–1876 CrossRef CAS PubMed.
- Q. Xing, C. Vogt, K. W. Leong and F. Zhao, Adv. Funct. Mater., 2014, 24, 3027–3035 CrossRef CAS PubMed.
- L. Wang, J. A. Johnson, D. W. Chang and Q. Zhang, Biomaterials, 2013, 34, 2641–2654 CrossRef CAS PubMed.
- Q. J. Lu, K. Ganesan, D. T. Simionescu and N. R. Vyavahare, Biomaterials, 2004, 25, 5227–5237 CrossRef CAS PubMed.
- M. Karus, B. Denecke, C. Ffrench-Constant, S. Wiese and A. Faissner, Development, 2011, 138, 5321–5331 CrossRef CAS PubMed.
- J. M. Goddard and J. H. Hotchkiss, Prog. Polym. Sci., 2007, 32, 698–725 CrossRef CAS.
- Y. Tamada and Y. Ikada, J. Colloid Interface Sci., 1993, 155, 334–339 CrossRef CAS.
- D. Guarnieri, A. De Capua, M. Ventre, A. Borzacchiello, C. Pedone, D. Marasco, M. Ruvo and P. A. Netti, Acta Biomater., 2010, 6, 2532–2539 CrossRef CAS PubMed.
- G. W. Lian, J. Lu, J. J. Hu, J. P. Zhang, S. H. Cross, R. J. Ferland and V. L. Sheen, J. Neurosci., 2012, 32, 7672–7684 CrossRef CAS PubMed.
- D. C. Kraushaar, S. Rai, E. Condac, A. Nairn, S. Y. Zhang, Y. Yamaguchi, K. Moremen, S. Dalton and L. C. Wang, J. Biol. Chem., 2012, 287, 22691–22700 CrossRef CAS PubMed.
- A. Wade, A. McKinney and J. J. Phillips, Biochim. Biophys. Acta, 2014, 1840, 2520–2525 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: MEFs culture; NPCs culture and differentiation; primers used for real-time PCR analysis; list of the 10 most abundant proteins according to the nLC-MS/MS analysis; cell viability of NPCs c by a live/dead assay; anti-nestin immunofluorescence images of NPCs. Movies of NPC behavior after seeding 24 h. See DOI: 10.1039/c7ra08273h |
| This journal is © The Royal Society of Chemistry 2017 |