 Open Access Article
Open Access ArticleProtein-free ribosomal RNA folds to a near-native state in the presence of Mg2+†
Timothy K. Lenz,
Ashlyn M. Norris,
Nicholas V. Hud and
Loren Dean Williams *
*
Department of Chemistry & Biochemistry, Georgia Institute of Technology, Atlanta, GA 30332, USA. E-mail: loren.williams@chemistry.gatech.edu
First published on 30th November 2017
Abstract
The assembled bacterial ribosome contains around 50 proteins and many counterions. Here, focusing on rRNA from the large ribosomal subunit, we demonstrate that Mg2+ causes structural collapse in the absence of ribosomal proteins. The collapsed rRNA forms many native-like RNA–RNA interactions, similar to those observed in the assembled ribosome. We assayed rRNA structure by chemical footprinting in the presence and absence of Mg2+. Our results indicate that Mg2+-dependent conformational change is focused in non-helical regions, consistent with tertiary interactions. In the presence of Mg2+, the large subunit rRNA adopts a state that includes the core inter-domain architecture of the assembled ribosome. We infer that the rRNA–Mg2+ state represents the core architecture of the LSU which, while not catalytically active, positions the residues of the LSU rRNA in such a way as to promote native interactions with rProteins to ultimately form a functional LSU.
Introduction
Cations accumulate around RNAs and facilitate folding.1,2 Monovalent cations promote formation of RNA secondary structure (2° structure). Mg2+ is often necessary for adoption of native states, characterized by long-range tertiary interactions. Mg2+ can increase local RNA density and rigidity by packing RNA functional groups in its first coordination shell.Ribosomal structure is maintained by complex networks of rRNA–rRNA, rRNA–protein and rRNA–cation interactions.3,4 Mg2+ is required for peptidyl transferase activity of the ribosome.5 Over a hundred Mg2+ ions are observed in the X-ray structure of the large ribosomal subunit (LSU) of H. marismortui.3,6 The LSU contains four dinuclear Mg2+ complexes7 and over 20 bidentate Mg2+ clamps.8 Nucleotides that directly contact Mg2+ are dispersed throughout the 2° structure, in all seven domains of the LSU rRNA (Fig. S3†).
Focusing on the LSU, we infer roles of Mg2+ in the formation of native rRNA structure, and determine the degree to which rRNA sequence governs folding in the absence of ribosomal proteins (rProteins). We investigate collapse of rRNA from a 2° structural state in the presence of Na+ alone into a compacted state upon addition of Mg2+. We characterize rRNA folding at nucleotide resolution in the absence of rProteins, 5S rRNA, or base modifications.
Results and discussion
Chemical footprinting
We assayed rRNA folding by comparing footprinting data obtained in the presence and absence of Mg2+. SHAPE (Selective 2′-Hydroxyl Acylation analyzed by Primer Extension) provides quantitative information about nucleotide flexibility and base pairing.9 SHAPE was used previously to characterize 2° and tertiary interactions in crystalline P4–P6 domain of the tetrahymena ribozyme.10 SHAPE has been performed previously on intact yeast ribosomes11 and deproteinized E. coli, C. difficile, and H. volcanii rRNAs.12Comparison of SHAPE data collected under various conditions can be used to characterize structural transitions. SHAPE has been used previously to monitor RNA structural transitions induced by cations, small molecules, or proteins.9,13 We have previously utilized this technique to monitor Mg2+-induced transitions of domains and subdomains of LSU rRNA.14–16
Here SHAPE was performed, in the absence and presence of Mg2+, on the intact ∼2900 nt T. thermophilus 23S rRNA (LSU rRNA). Experiments were performed in the presence of 250 mM Na+ (abbreviated Na+ conditions), which favors formation of 2° structure,1,2 and in 250 mM Na+ plus 10 mM Mg2+ (Na+/Mg2+ conditions), which favors formation of tertiary interactions. SHAPE data were collected for 2890 of the 2911 LSU rRNA nucleotides (>99%). Na+ conditions yielded low SHAPE reactivities for helical nucleotides (Fig. S2, ESI†). Reactive sites are observed at non-helical regions of the canonical LSU rRNA 2° structure.17,18
SHAPE data under different conditions were quantitatively compared showing broadly distributed changes in reactivity upon addition of Mg2+ to the LSU rRNA (Fig. 1). For this work we have collected, quantitatively analyzed and visualized an extensive amount of data. Na+ and Na+/Mg2+ data were normalized, and Na+ SHAPE reactivities were subtracted from Na+/Mg2+ reactivities at each nucleotide position to obtain ΔMg2+ data (Fig. 1 and ESI† Dataset 1). In analysis of Mg2+-dependent SHAPE changes, values >0.3 SHAPE units (positive or negative) are termed ‘ΔMg2+ sites’, i.e., sites which experience a significant Mg2+-dependent alteration in SHAPE reactivity; ∼7.5% of LSU rRNA nucleotides exceed this threshold. Overall, the locations of ΔMg2+ sites are consistent with Mg2+-induced formation of tertiary interactions observed in the three-dimensional structure of the ribosome. The extent and distribution of ΔMg2+ sites suggest a global transition from a state comprising 2° structure with limited tertiary interactions (the rRNA–Na+ state) to a state containing a significant extent of tertiary interactions, plus additional local interactions (the rRNA–Mg2+ state).
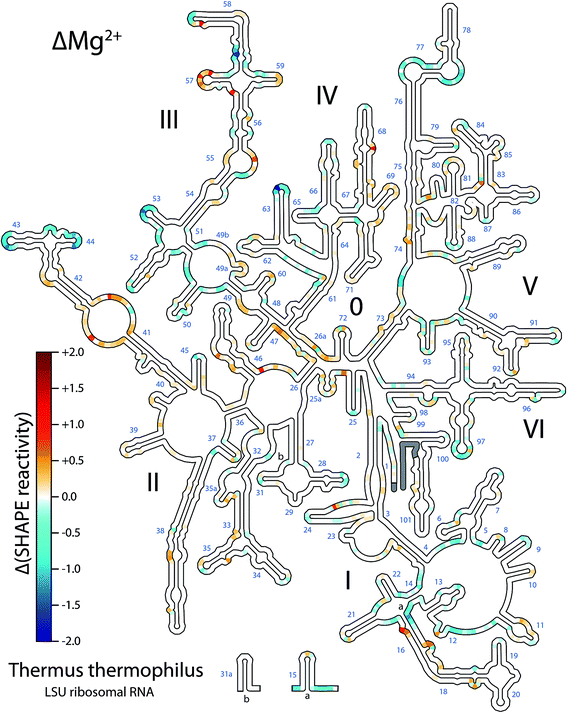 | ||
| Fig. 1 T. thermophilus LSU rRNA exhibits Mg2+-dependent changes in structure. Changes in SHAPE reactivity caused by the addition of Mg2+ (ΔMg2+ reactivity) are mapped onto LSU rRNA 2° structure. Positive values (red) indicate increased SHAPE reactivity upon addition of 10 mM Mg2+. Negative values (blue) indicate decreased reactivity. Grey; data not available for those nucleotides. Domain numbers and helix numbers are indicated. Methods are described in the ESI† and full numeric SHAPE data are available in ESI† Dataset 1. This figure was generated using RiboVision; a full-page version with greater detail is provided in the ESI (Fig. S4†). | ||
Addition of Mg2+ causes changes in SHAPE reactivity that are dispersed throughout the LSU rRNA. However, nucleotides within helical regions in the 2° structure19 are excluded from these changes (Fig. 1). The overall absence of ΔMg2+ sites from helices confirms that addition of Mg2+ does not change RNA 2° structure of the LSU rRNA.
By contrast, nucleotides found in loops, bulges and otherwise unpaired regions in the 2° structure are responsive to Mg2+ (Fig. 1 and S2†). The observed changes in SHAPE reactivity are consistent with a model in which Mg2+ induces native-like tertiary interactions. Six lines of evidence support this hypothesis: (i) ΔMg2+ sites are highly focused in rRNA loops, bulges, and other non-helical regions, which in general are expected to form tertiary interactions, (ii) ΔMg2+ sites cluster at or near nucleotides involved in tertiary interactions in the assembled LSU, (iii) ΔMg2+ sites cluster at or near rRNA regions involved in inter-domain interactions, (iv) ΔMg2+ sites are absent from regions that do not form tertiary interactions, (v) ΔMg2+ sites do not cluster exclusively at or near sites of first-shell RNA–Mg2+ interactions (ESI†), obviating the possibility that the signal arises predominantly from direct Mg2+ interactions, and (vi) it has been demonstrated previously that Mg2+ effects on SHAPE reactivity are related to formation of tertiary interactions.9,20
Structural analysis
Using the RiboVision visualization suite,21 we compared RNA tertiary interactions22 in the assembled T. thermophilus ribosome4 to ΔMg2+ sites observed here by SHAPE. Regions with the greatest density of ΔMg2+ sites were manually inspected in both two and three dimensions. The ΔMg2+ sites correlate with locations of tertiary interactions in the assembled LSU (Fig. 2 and 3); nearly all ΔMg2+ sites occur at or directly adjacent to one or more nucleotides that participate in tertiary interactions in the LSU. Conversely, loops that do not participate in RNA–RNA tertiary interactions in the assembled LSU are not responsive to Mg2+ (ESI†).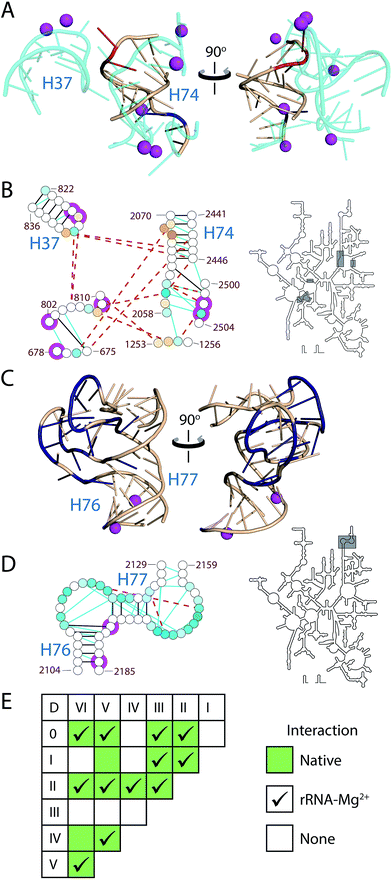 | ||
| Fig. 2 Mg2+-induced changes in SHAPE reactivity in LSU rRNA are consistent with formation of tertiary interactions. (A) The 3D structure of H74 and the surrounding rRNA (cyan). H74 nucleotides are colored by ΔMg2+ values (red, increased reactivity; dark blue, decreased reactivity; beige, little/no change in reactivity). (B) 2° Structure, tertiary interactions and Mg2+ interactions of H74 in the assembled ribosome. (C) 3D structure of the uL1 protuberance. (D) 2° Structure, tertiary interactions and Mg2+ interactions of the uL1 protuberance. Nucleotides of the uL1 protuberance are colored by ΔMg2+ values as in Fig. 1. (E) Inter-domain interactions within the LSU rRNA. Green boxes indicate domain pairs that interact through long-range RNA–RNA contacts in the assembled LSU. Checkmarks indicate where data supports presence of inter-domain interactions in the rRNA–Mg2+ state. In (B and D), lines indicate RNA–RNA interactions in the 3D structure. Solid black lines are 2° hydrogen bonding interactions. Dashed brown lines are tertiary hydrogen bonding interactions. Cyan lines are stacking interactions. Magenta circles are nucleotides in first-shell interactions with Mg2+ ions. Gray boxes on intact 2° structure (right) highlight the region displayed on the left. Three dimensional figures were generated with PyMol using the T. thermophilus ribosome (PDB entries 2J00 and 2J01). | ||
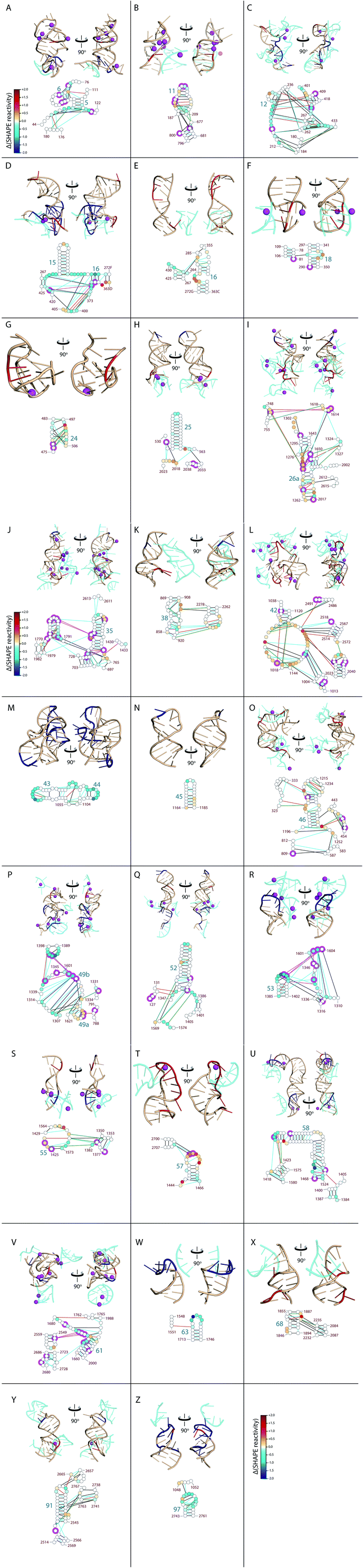 | ||
| Fig. 3 ΔMg2+ regions of the LSU rRNA. LSU rRNA with high density of ΔMg2+ sites is shown with regions of rRNA that interact at or near the ΔMg2+ sites. A–Z Top panels: 3D structures of selected regions of LSU rRNA (adjacent ΔMg2+ sites) from the T. thermophilus ribosome crystal structure (PDB IDs: 2J00 and 2J01). Nucleotides are colored by ΔMg2+ values (red, increased reactivity; dark blue, decreased reactivity; wheat, little to no change in reactivity). LSU rRNA segments that form tertiary interactions at or near these ΔMg2+ sites are colored cyan. Mg2+ cations that form first shell interactions with RNA (2.4 Å cut-off) are represented by magenta spheres. RNA is in cartoon representation. A–Z Bottom panels: Mg2+-induced changes in SHAPE reactivity mapped onto LSU rRNA 2° structure. Nucleotides are represented as circles, colored as in Fig. 1. Lines represent RNA–RNA interactions observed in the T. thermophilus ribosome structure (determined by FR3D; black, base–base; orange, base–phosphate; blue, base–stacking; and green, base–sugar). Magenta halos indicate nucleotides that interact directly with Mg2+ ions. The same nucleotides are displayed in corresponding 3D and 2D representations. Helix and nucleotide numbers are indicated. | ||
Relationships between ΔMg2+ sites and local and tertiary interactions are illustrated in Fig. 2 and 3. Helix 74 (H74), which lies at the functional and structural core of the LSU, contains ΔMg2+ sites consistent with formation of native tertiary interactions (Fig. 2A and B). In the assembled LSU, H74 interacts with RNA from Domain 0 (D0), DII and DV, in addition to Mg2+ ions to form a portion of the LSU functional core, the peptidyl transferase center. ΔMg2+ sites in H74 correlate with nucleotides involved in tertiary interactions that connect DV to distant nucleotides of D0 and DII (ESI†) that are remote from H74 in the primary and 2° structure. Formation of these tertiary interactions would necessarily cause collapse of the global RNA structure, decreasing the radius of gyration, as previously observed upon addition of Mg2+ to the LSU rRNA.23
ΔMg2+ sites in the uL1 protuberance are consistent with Mg2+-induced formation of loop–loop interactions between nucleotides 2109–2119 and 2161–2173 (Fig. 2C and D). The uL1 protuberance extends from the exterior of the LSU and interacts with rProtein uL1.24 The loop regions of the uL1 protuberance become unreactive to SHAPE upon introduction of Mg2+ (Fig. S2†). In the same way, virtually all ΔMg2+ sites in the LSU rRNA occur at or adjacent to nucleotides involved in tertiary interactions in the assembled LSU (Fig. 3). The locations of ΔMg2+ sites suggest formation of an intricate tertiary interaction network, corresponding with global collapse of the LSU rRNA into a native-like rRNA–Mg2+ state. Further analysis of ΔMg2+ sites is provided in the ESI.†
In sum, the ΔMg2+ data support formation of native-like inter-domain architecture of protein-free LSU rRNA in the presence of Mg2+. Most domain pairs observed in the assembled LSU are inferred to interact in the rRNA–Mg2+ state (Fig. 2E). In the assembled ribosome, long-range inter-domain tertiary interactions connect disparate 2° regions of LSU rRNA.4 Mg2+ has been implicated previously in facilitating these inter-domain contacts,6 whereas moderate monovalent cation concentrations induce 2° structure only.25 SHAPE results here support formation of 12 out of 14 native domain pair interactions (Fig. 2E and Table S5, ESI†). The network of inter-domain interactions in the rRNA–Mg2+ state supports formation of the core domain architecture of the LSU, in the absence of other interaction partners.
In the rRNA–Mg2+ state, tertiary interactions not supported by correlation of ΔMg2+ sites to assembled LSU-interacting regions must fall into one of three classes; (i) already present in Na+ alone, (ii) present only in Na+/Mg2+, but undetectable by comparative SHAPE, or (iii) absent or unstable in Na+/Mg2+, in which case their formation is dependent on agents other than or in addition to Mg2+. Inter-domain interactions that are not indicated by ΔMg2+ sites may be pre-organized for protein-mediated association. The extent of supported Mg2+-induced tertiary interactions (Fig. 2 and 3) suggests that most nucleotides involved in the absent interactions are pre-organized for association, contingent upon addition of a mediating rProtein (see ESI, Table S6†). In this model, formation of certain native tertiary interactions is more favorable in the rRNA–Mg2+ state than in presence of Na+ alone.
A role for rProteins is suggested by RNA–RNA interactions that appear to be absent from the rRNA–Mg2+ state. These absent interactions occur at or adjacent to regions of native rRNA interaction with rProteins uL2, uL3, uL4, uL15, uL16 and bL20 (Table S6†). Based on this, we suggest that these rProteins foster formation of certain inter-domain RNA–RNA interactions in the assembled ribosome. Five of the six genes coding for these rProteins (uL2, uL3, uL4, uL16, and bL20) are essential for survival,26 and all six rProteins exhibit strong interconnectivity in LSU assembly maps. The correlation between functional and structural importance of this set of rProteins implies that the interactions they mediate foster formation of the active, native LSU. It appears that the rRNA–Mg2+ state represents the core architecture of the LSU which, while not catalytically active,27 positions the residues of the LSU rRNA in such a way as to promote native interactions with rProteins to ultimately form a functional LSU.
Conclusions
The LSU rRNA interacts in vivo with the 5S rRNA and over 30 LSU proteins.4 Our experiments involve in vitro-transcribed LSU rRNA in solutions containing only buffer and ions, within reasonable physiological ranges, with no additional components. We have used comparative SHAPE to provide single-nucleotide information regarding Mg2+-induced structural changes. We propose a model of the near-native LSU state in the presence of Mg2+ based on correlations between our data and three-dimensional ribosomal structures. Our model, termed the rRNA–Mg2+ state, is collapsed with a considerably reduced radius, and includes core inter-domain architecture similar to that in the native LSU, but is not expected to be catalytically competent. Protein-free rRNA is not capable of peptidyl transferase, even in presence of Mg2+.27 Mg2+ interactions may be cooperative with rProtein interactions in formation of the native LSU. Certain rProtein–rRNA interactions are cation-mediated,6 underscoring the interdependence of rProteins and cations in the structure of the assembled LSU.The results presented here are consistent with previous results, in which removal of Mg2+ results in global unfolding of the LSU rRNA, as detected by several biophysical techniques.23 Virtually all aspects of ribosome structure and function involve Mg2+ to some degree.28 Ribosome function can survive significant deproteinization, but is completely abolished upon exposure to divalent cation-chelating compounds.5 We suggest that Mg2+ is crucial to the formation of local interactions, inter-domain contacts and core LSU architecture. When the assembled LSU is depleted of Mg2+, this core architecture is lost, resulting in dissociation of domains and their unfolding, and loss of ribosomal function. Our results support a model in which the LSU rRNA requires only Mg2+ to fold to a well-defined, collapsed near-native state, and elements such as rProteins induce local structural changes that foster formation of the remaining interactions to form a fully catalytic LSU. This model is consistent with the findings of Woodson and coworkers, who used chemical footprinting and molecular dynamics simulations of the small ribosomal subunit to show that ribosomal proteins dampen rRNA fluctuations at subdomain interfaces and cause switching of conformation of select regions of the rRNA.29
Conflicts of interest
The authors of this paper declare that they have no conflict of interest.Acknowledgements
We thank J. C. Bowman and Drs S. C. Harvey, C. D. Okafor, and R. M. Wartell for helpful discussions, and Dr M. A. D. Goodisman for use of the CE instrument. This work was supported by the NASA Astrobiology Institute (NNX16AJ28G).References
- P. Brion and E. Westhof, Annu. Rev. Biophys. Biomol. Struct., 1997, 26, 113–137 CrossRef CAS PubMed.
- J. C. Bowman, T. K. Lenz, N. V. Hud and L. D. Williams, Curr. Opin. Struct. Biol., 2012, 22, 262–272 CrossRef CAS PubMed.
- N. Ban, P. Nissen, J. Hansen, P. B. Moore and T. A. Steitz, Science, 2000, 289, 905–920 CrossRef CAS PubMed.
- M. Selmer, C. M. Dunham, F. V. Murphy, A. Weixlbaumer, S. Petry, A. C. Kelley, J. R. Weir and V. Ramakrishnan, Science, 2006, 313, 1935–1942 CrossRef CAS PubMed.
- H. F. Noller, V. Hoffarth and L. Zimniak, Science, 1992, 256, 1416–1419 CAS.
- D. J. Klein, P. B. Moore and T. A. Steitz, RNA, 2004, 10, 1366–1379 CrossRef CAS PubMed.
- C. Hsiao and L. D. Williams, Nucleic Acids Res., 2009, 37, 3134–3142 CrossRef CAS PubMed.
- A. S. Petrov, J. C. Bowman, S. C. Harvey and L. D. Williams, RNA, 2011, 17, 291–297 CrossRef CAS PubMed.
- S. A. Mortimer and K. M. Weeks, J. Am. Chem. Soc., 2007, 129, 4144–4145 CrossRef CAS PubMed.
- Q. Vicens, A. R. Gooding, A. Laederach and T. R. Cech, RNA, 2007, 13, 536–548 CrossRef CAS PubMed.
- J. A. Leshin, R. Heselpoth, A. T. Belew and J. D. Dinman, RNA Biol., 2011, 8, 478–487 CrossRef CAS PubMed.
- C. A. Lavender, R. Lorenz, G. Zhang, R. Tamayo, I. L. Hofacker and K. M. Weeks, PLoS Comput. Biol., 2015, 11, e1004126 Search PubMed.
- C. E. Dann III, C. A. Wakeman, C. L. Sieling, S. C. Baker, I. Irnov and W. C. Winkler, Cell, 2007, 130, 878–892 CrossRef PubMed.
- S. S. Athavale, J. J. Gossett, C. Hsiao, J. C. Bowman, E. O'Neill, E. Hershkovitz, T. Preeprem, N. V. Hud, R. M. Wartell, S. C. Harvey and L. D. Williams, RNA, 2012, 18, 752–758 CrossRef CAS PubMed.
- C. Hsiao, T. K. Lenz, J. K. Peters, P. Y. Fang, D. M. Schneider, E. J. Anderson, T. Preeprem, J. C. Bowman, E. B. O'Neill, L. Lie, S. S. Athavale, J. J. Gossett, C. Trippe, J. Murray, A. S. Petrov, R. M. Wartell, S. C. Harvey, N. V. Hud and L. D. Williams, Nucleic Acids Res., 2013, 41, 3373–3385 CrossRef CAS PubMed.
- K. A. Lanier, S. S. Athavale, A. S. Petrov, R. M. Wartell and L. D. Williams, Biochemistry, 2016, 55, 4603–4613 CrossRef CAS PubMed.
- A. S. Petrov, C. R. Bernier, B. Gulen, C. C. Waterbury, E. Hershkovitz, C. Hsiao, S. C. Harvey, N. V. Hud, G. E. Fox, R. M. Wartell and L. D. Williams, PLoS One, 2014, 9, e88222 Search PubMed.
- H. F. Noller, J. Kop, V. Wheaton, J. Brosius, R. R. Gutell, A. M. Kopylov, F. Dohme, W. Herr, D. A. Stahl, R. Gupta and C. R. Woese, Nucleic Acids Res., 1981, 9, 6167–6189 CrossRef CAS PubMed.
- A. S. Petrov, C. R. Bernier, E. Hershkovitz, Y. Xue, C. C. Waterbury, M. A. Grover, S. C. Harvey, N. V. Hud, R. M. Wartell and L. D. Williams, Nucleic Acids Res., 2013, 41, 7522–7535 CrossRef CAS PubMed.
- S. Blouin, R. Chinnappan and D. A. Lafontaine, Nucleic Acids Res., 2011, 39, 3373–3387 CrossRef CAS PubMed.
- C. Bernier, A. S. Petrov, C. Waterbury, J. Jett, F. Li, L. E. Freil, B. Xiong, L. Wang, A. Le, B. L. Milhouse, E. Hershkovitz, M. Grover, Y. Xue, C. Hsiao, J. C. Bowman, S. C. Harvey, J. Z. Wartel and L. D. Williams, Faraday Discuss., 2014, 169, 195–207 RSC.
- M. Sarver, C. L. Zirbel, J. Stombaugh, A. Mokdad and N. B. Leontis, J. Math. Biol., 2008, 56, 215–252 CrossRef PubMed.
- Q. M. Yi and K. P. Wong, Biochem. Biophys. Res. Commun., 1982, 104, 733–739 CrossRef CAS PubMed.
- A. Nikulin, I. Eliseikina, S. Tishchenko, N. Nevskaya, N. Davydova, O. Platonova, W. Piendl, M. Selmer, A. Liljas, D. Drygin, R. Zimmermann, M. Garber and S. Nikonov, Nat. Struct. Mol. Biol., 2003, 10, 104–108 CAS.
- J. K. Frederiksen, N. S. Li, R. Das, D. Herschlag and J. A. Piccirilli, RNA, 2012, 18, 1123–1141 CrossRef CAS PubMed.
- S. Shoji, C. M. Dambacher, Z. Shajani, J. R. Williamson and P. G. Schultz, J. Mol. Biol., 2011, 413, 751–761 CrossRef CAS PubMed.
- P. Khaitovich, T. Tenson, A. S. Mankin and R. Green, RNA, 1999, 5, 605–608 CrossRef CAS PubMed.
- K. Trappl and N. Polacek, Met. Ions Life Sci., 2011, 9, 253–275 CAS.
- S. C. Abeysirigunawardena, H. Kim, J. Lai, K. Ragunathan, M. C. Rappé, Z. Luthey-Schulten, T. Ha and S. A. Woodson, Nat. Commun., 2017, 8, 492 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Describing experimental methods and structural analysis. See DOI: 10.1039/c7ra08696b |
| This journal is © The Royal Society of Chemistry 2017 |
