Open Access Article
Open Access ArticlePolarity-extended quantitative analysis of bear bile and its analogues using serially coupled reversed phase-hydrophilic interaction liquid chromatography-tailored multiple reaction monitoring†
Qingqing Songa,
Wenjing Liua,
Yu Yanab,
Peng Lic,
Jun Lia,
Pengfei Tua,
Yitao Wang*c and
Yuelin Song *a
*a
aModern Research Center for Traditional Chinese Medicine, School of Chinese Materia Medica, Beijing University of Chinese Medicine, Beijing 100029, China. E-mail: syltwc2005@163.com
bInstitute of Chinese Minority Traditional Medicine, Minzu University of China, Beijing 100081, China
cState Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Taipa 999078, Macao, China. E-mail: ytwang@umac.mo
First published on 15th November 2017
Abstract
The medicinal values of bear bile are believed to rely on both hydrophilic and hydrophobic components regardless of their abundant or minor distributions. However, there is usually a technical bottleneck to simultaneously monitor components regardless of polarity and concentration due to the retention threshold of single column liquid chromatography (LC) as well as the relatively narrow linear dynamic range of tandem mass spectrometry (MS/MS). A new integrated method was developed and demonstrated to be reliable via diverse validation assays towards simultaneous determination of four nucleosides together with ten bile acids in bear bile (Chinese name: Xiongdan) as well as its analogues, although these compounds-of-interest spanned not only a large polarity range but also a great content scale. Improvements were made in both LC and MS/MS domains. In the LC domain, reversed phase LC and hydrophilic interaction liquid chromatography were coupled in series (RPLC-HILIC) to afford satisfactory chromatographic behaviors for bile acids along with nucleosides. In the MS domain, to ensure the locations of all abundant bile acids, such as TUDCA, TCA, GHDCA, CA, UDCA, HDCA, GDCA, CDCA, and DCA, in their linear dynamic ranges, response tailoring was carried out through the employment of inferior collision energies in multiple reaction monitoring (MRM) mode. The validated approach was then successfully applied for quantitative characterization of bear bile and some other medicinal bile samples. Significant differences occurred between bear bile and its analogues originating from other species as well as among bear bile samples from different manufacturers. The distributions of TUDCA, GUDCA, UDCA and CDCA, particularly TUDCA, might enable reflection of, to some extent, the quality of bear bile. Consequently, RPLC-HILIC-tailored MRM provides a qualified tool for simultaneous determination of multiple components with large polarity and content spans in bear bile as well as other bile-derived medicines.
1. Introduction
Bile plays a vital role in regular physiological metabolism of most vertebrates, in particular the digestion of lipids. Because of their inherent beneficial nature, bile or gall bladders originating from various animals are widely utilized for the treatment of many kinds of bile metabolism disorders, such as gallstones and cholestasis,1,2 being exactly consistent with certain traditional Chinese medical principles. Among diverse bile-derived traditional Chinese medicines (TCMs), Ursi Fellis Pulvis (folk name: bear bile, Chinese name: Xiongdan) always occupies the hot spot attributed to not only the high price, but also the convincing therapeutic outcomes.3–6 As a complicated matrix, bear bile consists of a vast array of hydrophobic and hydrophilic components that also exhibit a great content range, and the therapeutic benefits are believed as the synergic outcomes of those ingredients regardless of polarity and content.7,8 Except for bile acids, attention has seldom, however, been paid to the quantitative characterization of those hydrophilic and/or minor compounds, indicating an urgent task to search for a fit-for-purpose approach able to completely fulfill the requirements of the in-depth quality assessment of bear bile.Liquid chromatography (LC) coupled with tandem mass spectrometry (MS/MS), equipped with a single column in most cases, currently serves as the work horse for quantitative analysis of TCMs.9–13 However, technical obstacles still occur at the chromatographic behaviors as well as appropriate responses for all compounds-of-interest.14 Improvements are thereby called for both LC and MS/MS domains to afford desired chromatographic and spectrometric patterns.10 Recently, two dimensional LC, even multi-dimensional LC, is blooming in response to chemical diversity of complicated matrices according to orthogonally hyphenating separation mechanisms with online manner.15–22 Those schemes are usually attractive in term of peak capacity, and therefore provide superior choices for qualitative characterization; however, they always suffer from unsatisfactory precision attributing to numerous uncertainties accompanying with the sophisticated instrument setup. Moreover, several instrumentations have also been configured to achieve simultaneous measurements of both hydrophilic and hydrophobic substances via online coupling hydrophilic interaction liquid chromatography (HILIC) and reversed phase liquid chromatography (RPLC).23–25 Among those accessible schemes, serially coupling of RPLC and HILIC (RPLC-HILIC) is worldwide favored because it enables to yield polarity-extended chromatographic separation without sacrificing the instrumental facility.10,26–31 On the other side, the measurement of primary components, as well as minor compounds, is also quite annoying for MS/MS, owing to the mismatching between the relative linear dynamic range of mass spectrometer and the content coverage of complex matrices. The key issue dampening the reliable ion counting is the sensor saturation. Because multiple monitoring mode (MRM) is a unique function of triple-quadrupole (QqQ) mass spectrometer, and quantification is accomplished on the basis of the linear correlation between the content and the count of targeted product ion, a robust strategy being able to flexibly tailor mass response has therefore been proposed to decrease the amount of product ions through the deliberate employment of inferior compound-dependent parameters, e.g. MRM ion transitions and collision energies.10,13,32,33
The objective of current study is to develop a practical method possessing the potential for simultaneous determination of analytes with great polarity and content ranges, including ten bile acids along with four nucleosides, in bear bile as well as its analogues. The integration of RPLC-HILIC and tailored MRM might be feasible to exactly suit for the goal. The findings are envisioned: (1) to clarify the quantitative properties of bear bile; (2) to explore the differences between bear bile and its analogues; and (3) to offer an analytical tool for the in-depth quality assessment of bear bile as well as other bile-derived drugs.
2. Experimental
2.1 Chemicals and materials
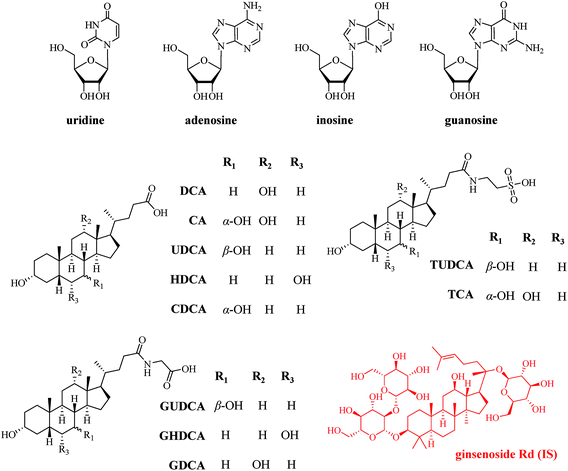
All methanol, acetonitrile (ACN), formic acid, and ammonium formate were of HPLC grade and purchased from Thermo-Fisher (Pittsburgh, PA, USA). HPLC grade dimethylsulfoxide (DMSO) was supplied by Merck (Darmstadt, Germany). De-ionized water was prepared in-lab by a Milli-Q Integral water purification system (Millipore, Bedford, MA, USA).Four nucleosides, including uridine, adenosine, inosine and guanosine, were supplied by Xinjingke Biotechnology Company (Beijing, China). Cholic acid (CA), ursodeoxycholic acid (UDCA), hyodeoxycholic acid (HDCA), chenodeoxycholic acid (CDCA), and deoxycholic acid (DCA) were purchased from National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China). Taurocholic acid (TCA), tauroursodeoxycholic acid (TUDCA), glycoursodeoxycholic acid (GUDCA), and glycohyodeoxycholic acid (GHDCA) were obtained from Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China). Glycodeoxycholic acid (GDCA) was gained from BioBioPha Co., Ltd. (Kunming, China). Ginsenoside Rd supplied by Shanghai Standard Biotech Co., Ltd. (Shanghai, China) was served as internal standard (IS). The chemical structures of all analytes together with IS are elucidated in Fig. 1.
Ten batches of Ursi Fellis Pulvis (folk name: bear bile, BB1–10) were collected from different manufacturers, including Yunnan Daweishan biological pharmaceutical co. Ltd. (BB1–2), Sichuan province Xinlu pharmaceutical co., Ltd. (BB3), Heilongjiang Heibao pharmaceutical co., Ltd. (BB4–7), Yanji Federal specialty co., Ltd. (BB8), and Yanji Baitoufeng bear garden (BB9–10). Bovis Fellis Pulvis (folk name: cattle bile), Bos Grunniens Fellis Pulvis (folk name: yak bile), Caprae Fellis Pulvis (folk name: goat bile), Gallus Domesticus Fellis Pulvis (folk name: chicken bile), and Anser Cygnoides Fellis Pulvis (folk name: geese bile) were collected from a local slaughter house (Beijing Langzhong Slaughter Factory, Beijing, China). Artificial and natural Bovis Calculus (folk name: artificial/natural cow-bezoar) were purchased from Beijing Tongrentang Co. Ltd. (Beijing, China). All voucher specimens are deposited in the herbarium of our institute, Beijing University of Chinese Medicine (Beijing, China).
2.2 Sample preparation
The stock solutions of all analytes and IS were individually prepared with DMSO at a concentration of 10 mM, and stored at 4 °C until use. The mixed stock solution was generated by pooling stock solutions to yield desired concentrations for all compounds-of-interest. A panel of mixed standard solutions was then obtained by diluting the mixed stock solution with DMSO. Then, an aliquot of each obtained solution was 20-fold diluted with 25% aqueous acetonitrile to yield serial working solutions (total volume of each: 150 μL); afterwards, 50 μL of 25% aqueous ACN containing IS was spiked into each diluted working solution. Subsequently, the mixture was vortex for 1 min and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min to remove any possible residues, and a set of calibration standard solutions were ultimately generated (final concentration as 93.75 μg mL−1 for IS in each solution).
000 rpm for 10 min to remove any possible residues, and a set of calibration standard solutions were ultimately generated (final concentration as 93.75 μg mL−1 for IS in each solution).
All batches of bile samples (approximately 20.0 mg for each) were suspended in 10 mL of 25% aqueous ACN and then extracted in an ultrasonication-manner for 30 min at 25 °C. Following centrifugation and filtration, each filtrate was 10-fold diluted with 25% aqueous ACN, and a 150 μL aliquot was further fortified with 50 μL of 25% aqueous ACN containing IS.
2.3 RPLC-HILIC configuration
RPLC-HILIC was configured on a Shimadzu modular LC system (Kyoto, Japan) as depicted in our previous article.10 Briefly, four LC-20ADXR pumps (pumps A–D), a SIL-20ACXR auto-sampler, a CTO-20AC column oven, a DGU-20A3R degasser, a SUS-mixer, a CBM-20A controller, as well as some polyether ether ketone (PEEK) tubings in charge of connecting different modules and columns, took part in the configuration of RPLC-HILIC. An HSS T3 column (2.1 × 100 mm, 1.8 μm, Waters, Milford, MA, USA) was responsible for RPLC separation, and HILIC chromatographic mechanism was performed on an Xbridge Amide column (4.6 × 150 mm, 3.5 μm, Waters) that was coupled to the outlet of T3 column through a SUS-mixer (Shimadzu, Kyoto, Japan). Pumps A and B were in charge of delivering 0.1% aqueous formic acid containing 5 mM ammonium formate (A) and ACN (B), respectively, into T3 column. The gradient elution was programmed as below: 0–15 min, 35% B; 15–20 min, 35–40% B; 20–24 min, 40–60% B; 24–27 min, 60–100% B; 27.1–33 min, 35% B; and flow rate, 0.15 mL min−1. Pumps C and D delivered 5 mM ammonium formate fortified with 0.1% aqueous formic acid (C) and ACN (D), respectively, at a total flow rate of 1.0 mL min−1 into the SUS-mixer following a gradient program, as follows: 0–15 min, 100–92% D; 15–20 min, 92–72% D; 20–24 min, 72–65% D; 24–27 min, 65% D; and 27.1–33 min, 100% D. Both columns were maintained thermal (40 °C) in the column oven and the injection volume was set at 2 μL.2.4 Tailored MRM detection
The MS/MS detection was carried out on a SCIEX 5500 Qtrap mass spectrometer (Foster City, CA, USA) equipped with a Turbo V™ electrospray ionization (ESI) interface that always took the load of receiving a fluid at a total flow rate of 1.15 mL min−1. The ion source parameters for negative polarity ionization were optimized as following conditions: GS1, GS2, and CUR, 55, 55, and 35 psi, respectively; ion spray needle voltage, −4500 V; heater gas temperature, 550 °C. Scheduled multiple reaction monitoring (sMRM) mode34 was employed to monitor all analytes, and detection window was set as 60 s. All optimized compound-dependent parameters of fourteen analytes as well as IS, such as MRM ion transitions, declustering potential (DP) and collision energy (CE), are summarized in Table 1. Analyst Software package (Version 1.6.2, SCIEX) was utilized to synchronize the entire system, as well as for data acquisition and processing. In addition, MRM mode also acted as the survey experiment to trigger two separate enhanced product ion (EPI) scans through an information dependent acquisition (IDA) algorithm with a threshold as 300 cps. CE and collision energy spread (CES) that were the key parameters for EPI scans were set at −45 eV and 35 eV, respectively.| Analyte | tR (min) | MRM transitions | DP (V) | CE (eV) | Calibration curve | r | Weighting | Linear range (ng mL−1) | LOD (ng mL−1) | LOQ (ng mL−1) |
|---|---|---|---|---|---|---|---|---|---|---|
| a The CE value in the bracket was the optimal value gained by manual tuning, while the value with * symbol was the modified value for extending linearity range. | ||||||||||
| Uridine | 6.17 | 243 > 110 | −90 | −20 | y = 0.0607x − 0.00226 | 0.997 | None | 9.6–3000 | 1.92 | 4.80 |
| Adeonosine | 6.54 | 266 > 134 | −100 | −30 | y = 0.0184x − 6.66 × 10−6 | 0.999 | None | 4.0–1500 | 2.00 | 4.00 |
| Inosine | 8.32 | 267 > 135 | −80 | −30 | y = 0.344x + 0.00242 | 0.995 | None | 2.0–1500 | 0.960 | 2.00 |
| TUDCA | 8.44 | 498.3 > 80 | −150 | −50 * (−120) | y = 0.00128x + 0.00542 | 0.993 | None | 749–78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
30.0 | 60.0 |
| TCA | 10.78 | 514.3 > 80 | −150 | −60 * (−120) | y = 0.00104x + 0.000123 | 0.991 | 1/x | 155–77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
62.0 | 155 |
| Guanosine | 11.83 | 282 > 150 | −150 | −25 | y = 0.0573x − 0.000131 | 0.998 | None | 4.8–3000 | 1.92 | 4.80 |
| GUDCA | 13.90 | 448.3 > 74 | −120 | −80 | y = 0.0251x + 0.00286 | 0.993 | 1/x | 36–2000 | 18.0 | 36.0 |
| GHDCA | 14.18 | 448.3 > 448.3 | −120 | −50 * (−15) | y = 0.00765x + 0.00144 | 0.991 | None | 21.0–33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
10.0 | 21.0 |
| CA | 22.12 | 407.3 > 407.3 | −120 | −45 * (−15) | y = 0.0176x + 0.000612 | 0.992 | 1/x | 6.00–15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
3.0 | 6.00 |
| UDCA | 22.76 | 391.3 > 391.3 | −120 | −45*(−15) | y = 0.0297x − 0.0061 | 0.992 | 1/x | 118–14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
24.0 | 47.0 |
| HDCA | 23.01 | 391.3 > 391.3 | −120 | −45 * (−15) | y = 0.0261x + 0.00193 | 0.991 | 1/x | 23.5–7350 | 18.0 | 23.5 |
| GDCA | 23.23 | 448.3 > 448.3 | −120 | −50 * (−15) | y = 0.00495x − 0.000322 | 0.992 | None | 26.0–16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 850 850 |
14.0 | 26.0 |
| CDCA | 26.12 | 391.3 > 391.3 | −120 | −45 * (−15) | y = 0.0252x − 0.000528 | 0.993 | 1/x | 94.1–29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
18.0 | 70.0 |
| DCA | 26.43 | 391.3 > 391.3 | −120 | −45 * (−15) | y = 0.059x − 0.0167 | 0.991 | None | 48–14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
9.00 | 24.0 |
| Ginsenoside Rd (IS) | 18.93 | 991 > 945 | −90 | −32 | ||||||
2.5 Method validation
Terms including linearity, limit of detection (LOD), limit of quantification (LOQ), precision, repeatability, stability, and recovery were assayed to validate the developed method. Linearity was evaluated over more than six different concentration levels of calibration standard solutions. Calibration curves were constructed by plotting the peak area ratio (y) of analyte and IS against the concentrations ratio (x). And 1/x weighting was utilized to advance the linear regression if necessary. The LOD and LOQ corresponding to a signal-noise ratios (S/N) of 3 and 10, respectively, were determined by serially diluting calibration standard solutions. The intra- and inter-day variations were employed to assess the precision of the method. For intra-day assays, three different concentration levels (low, medium, and high) of calibration samples, served as quality control (QC) samples, were measured for six times in a single day. For inter-day assays, QC samples were analyzed for three times per day within three consecutive days. Regarding the repeatability and stability assays, six replicates of a selected bile sample (BB3) was run continuously and one of the selected sample stored at 4 °C was chosen to be analyzed within 24 h (0 h, 2 h, 4 h, 6 h, 12 h, and 24 h). The RSD% (relative standard deviation%) of the concentration of each analytes was employed to assess the deviation of precision, repeatability, and stability. Recoveries were evaluated by spiking the mixed standard solutions to the selected sample (BB3) at three different concentrations. And the recovery was calculated with the following equation: recovery (%) = (amount found − original amount)/amount spiked × 100%.3. Results and discussions
3.1 Optimization of RPLC-HILIC conditions
Two complementary columns, being orthogonal regarding separation mechanism, however, compatible in term of mobile phase, RP and HILIC ones were carefully evaluated for promoting the chromatographic pattern. Because the dilution pumps (pumps C and D) supplied ACN-enriched solvent to facilitate the enrichment of those hydrophilic components that weren't retained by RP column at the head of HILIC column, a wide-bore HILIC column that permitted fluid at a high flow rate was required to receive the diluted effluent from a narrow-bore RP column. Besides, particle shelled with amide group was proved to be a suitable choice for the retention and separation of hydrophilic compounds in our previous study.10 Hence, an amide-type column (Xbridge Amide column, 4.6 × 150 mm, 3.5 μm, Waters, Milford, MA, USA) with internal diameter as 4.6 mm was employed as the exact role for HILIC separation being responsible for accepting those non-retained compounds in the eluate from the front RP column. Regarding the demand of a narrow-bore RP column as the front separation choice, several available candidates with 2.1 mm internal diameter were screened, such as Capcell core C18 column (2.1 × 150 mm, 2.7 μm, Shiseido, Tokyo, Japan), Waters HSS T3 column (2.1 × 100 mm, 1.8 μm), and Waters BEH Shield RP18 (2.1 × 100 mm, 1.7 μm). In particular, HSS T3 column and BEH C18 column were most popular stationary phases for bile acid profiling.35 Following careful evaluations, HSS T3 column was superior to other candidates attributing to better resolution for bile acid isomers, lower back pressure, and greater peak shape.To promote chromatographic performances as far as possible, except for columns and gradient elution program, mobile phase additives were carefully assayed as well. The addition of ammonium formate was found to be able to strengthen the mass response of taurine-conjugated bile acids, e.g. TUDCA, that were the primary family in bear bile (Fig. 2); however, the mass response decreased when more than 5 mM ammonium formate was fortified into the solvents corresponding to pumps A and C. Moreover, the further introduction of 0.1% formic acid into the solvent of pump A was found to possess the potential of ameliorating chromatographic behaviors in terms of both peak shape and peak width for those conjugated bile acids, e.g. TUDCA (Fig. 2C). Above all, the combination of 5 mM ammonium formate and 0.1% formic acid was introduced as the modifiers into the solvents of pump A and C, to achieve satisfactory retention as well as resolution for bile acids, particularly conjugated bile acids.
Moreover, the solvent of the sample was also optimized. Because the pulse injection of 2.0 μL solution with auto-sampler, corresponding to 1/75 of the total flow rate of pump A and B, might result in significant solvent effect, in particular the employment of organic solvents, e.g. methanol, poor chromatographic behaviors were observed for some compounds-of-interest. Notably, the signals of all four nucleosides were completely split into two peaks when organic solvent (methanol) acted as the solvent. Hence, aqueous ACN was employed as the solvent attributing to its potential for dissolving both hydrophilic and hydrophobic components, and 25% aqueous ACN was ultimately selected because it could afford superior peak shape and intensity for most analytes in comparison of 50% and 70% aqueous ACN as well as those aqueous methanol solvents.
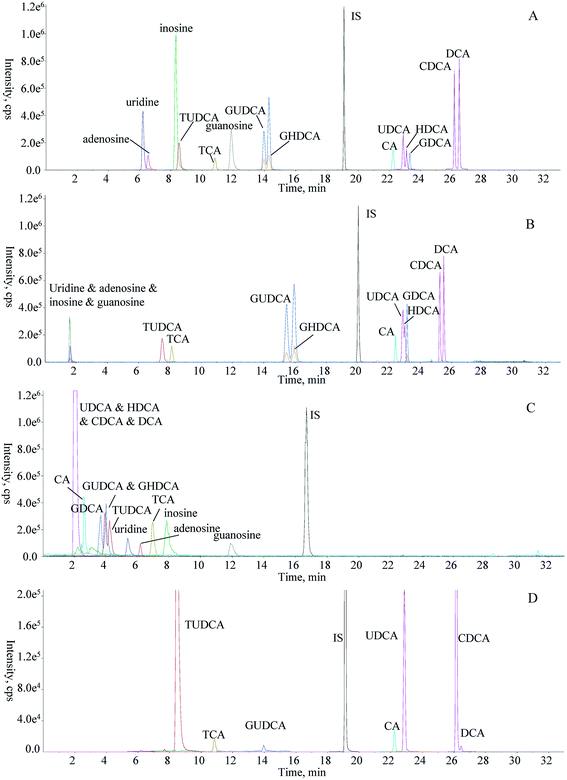
With the application of those optimized parameters, the representative chromatogram of mixed standard solution is shown in Fig. 3A. Obviously, satisfactory chromatographic profile was yielded for the chemical pool.
3.2 Comparison of RPLC-HILIC and single column LC
To highlight the necessity of the employment of RPLC-HILIC for simultaneous determination of four nucleosides along with ten bile acids in bear bile, comparisons among RPLC-HILIC and single column instrumentations were conducted. PEEK tubing with comparable length was implemented to substitute RP column or HILIC column for the configuration of single column LC to achieve parallel comparison, and identical elution program was scheduled for each run. Regarding RPLC equipped with a single T3 column, four nucleosides were co-eluted at the void time (tR < 2.0 min), and even worse, extensive overlaying occurred between UDCA (tR, 22.9 min) and HDCA (tR, 23.0 min) that shared identical ion transition (Fig. 3B). Meanwhile, the other analytes showed comparable retention behaviors between RPLC-HILIC and single RP column (Fig. 3B). When the sample was injected into single column HILIC system, the majority of analytes were eluted among 2–9 min, indicating the significant possibility for the matrix effect resulted from ionization competition among co-eluted substances. It was worthwhile to mention that the response of CA yielded from single column HILIC was significant greater than that generated from RPLC-HILIC and RPLC, and the possible reason was attributed to the matrix effect initiated by co-eluted components. Those four nucleosides were well retained and separated; however, those four unconjugated bile acids, such as UDCA, HDCA, CDCA, and DCA, were co-eluted at the void time on the HILIC column (Fig. 3C). In regard of resolution, satisfactory separation wasn't accomplished between GUDCA and GHDCA, the structural difference of which only occurred at the substitute position of the hydroxyl groups (3,7-dihydroxyl substitution for GUDCA and 3,6-dihydroxyl substitution for GHDCA), attributing to the insufficient separation capacity for the amide column. When RPLC-HILIC served as the separation combination, not only the hydrophilic compounds but bile acids were assigned satisfactory retention due to the hybrid retention mechanism (Fig. 3A). Overall, great resolution of fourteen analytes was observed, and particularly, acceptable separation was achieved between HDCA and UDCA. Above all, RPLC-HILIC, although facile instrumentation, was able to overcome the technical barriers in terms of comprehensive retention and resolution occurred for single column LC towards simultaneous determination of both polar and apolar components in bear bile.3.3 Method validation
The identities of all captured signals using MRM mode were firstly confirmed by matching retention times and MS2 spectra generated by EPI experiments between calibration samples and testing samples. In our preliminary assay, we found that the upper limits of quantitation for TUDCA, TCA, GHDCA, CA, UDCA, HDCA, GDCA, CDCA, and DCA could not completely cover their contents in those bile samples. It was definitely practicable to dilute the sample, nonetheless, sacrificing the sensitivity of those minor components such as those nucleosides. In theory, the maximum of the linear dynamic range was governed by the saturation of the sensor located at the back of the Q3 chamber for the QqQ mass spectrometer, and it was viable to increase the upper limit of linear range for MRM mode according to suppressing the generation of product ions.10,13 Consequently, response suppression was conducted through the employment of inferior CEs to extend the upper limits of quantitation for those abundant bile acids. All suitable CEs were marked with symbol “*” and utilized to replace those optimum ones that were exhibited in the brackets in the monitoring schedule (Table 1). Afterwards, satisfactory linearity with correlation coefficient (r) greater than 0.99 for each regressive calibration curve was achieved for each linear range (Table 1), and the upper limits of TUDCA, TCA, GHDCA, CA, UDCA, HDCA, GDCA, CDCA, and DCA were 78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800, 77
800, 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300, 33
300, 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700, 15
700, 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300, 14
300, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700, 7350, 16
700, 7350, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 850, 29
850, 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400, and 14
400, and 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 ng mL−1, respectively, all of which could fully fulfill their quantitative requirements.
700 ng mL−1, respectively, all of which could fully fulfill their quantitative requirements.
The LODs of all analytes were lower than 62 ng mL−1 and better sensitivity (each LOD < 2.4 ng mL−1) was observed for four nucleosides (Table 1). LOQs were in range of 2.40–155 ng mL−1 (Table 1). For repeatability assay, RSDs% of those detectable compounds in BB3, including TUDCA, TCA, GUDCA, CA, UDCA, CDCA, were less than 5.59% (Table 2). Besides, bile extract stored at 4 °C were found stable within 24 h, and RSDs% of six detectable analytes were less than 8.87% (Table 2). For intra- and inter-day assays, RSDs% of all analytes at three concentration levels of QC samples were among 1.72–11.04% and 3.87–13.26%, respectively (Table 2). Regarding the recovery, mean recoveries of quantifiable analytes in BB3 were among 74.12–124.83% with RSDs% lower than 10.94% (Table 2). Together, the newly developed method was proved to be sensitive, accurate and reproducible, and suitable for quantitative analysis of medicinal bile samples.
| Analyte | Repeatability | Stability | Intra-day RSD (%, n = 6) | Inter-day RSD (%, n = 9) | Recovery (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (%, n = 6) | (%, n = 6) | Low | Medium | High | Low | Medium | High | Low (n = 3) | Medium (n = 3) | High (n = 3) | ||||
| Mean | RSD | Mean | RSD | Mean | RSD | |||||||||
| a N.A.: not applicable. | ||||||||||||||
| Uridine | N.A. | N.A. | 5.86 | 3.16 | 4.34 | 7.55 | 7.41 | 5.76 | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. |
| Adeonosine | N.A. | N.A. | 3.46 | 6.85 | 5.11 | 8.99 | 6.95 | 6.79 | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. |
| Inosine | N.A. | N.A. | 5.25 | 3.67 | 5.78 | 6.12 | 5.65 | 6.59 | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. |
| TUDCA | 2.65 | 7.60 | 4.67 | 2.51 | 4.57 | 5.70 | 3.87 | 7.25 | 98.84 | 7.65 | 96.54 | 4.68 | 87.89 | 4.88 |
| TCA | 4.46 | 5.89 | 3.74 | 4.13 | 2.63 | 6.98 | 4.69 | 7.58 | 124.83 | 6.59 | 94.85 | 8.80 | 74.12 | 7.66 |
| Guanosine | N.A. | N.A. | 6.57 | 1.72 | 4.60 | 7.08 | 4.25 | 4.77 | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. |
| GUDCA | 3.99 | 8.87 | 12.65 | 9.21 | 4.85 | 11.56 | 12.14 | 8.58 | 83.89 | 10.94 | 94.16 | 1.85 | 86.11 | 1.52 |
| GHDCA | N.A. | N.A. | 5.65 | 7.40 | 11.04 | 8.37 | 6.00 | 7.29 | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. |
| CA | 4.70 | 6.96 | 5.36 | 2.20 | 4.19 | 4.80 | 4.90 | 7.72 | 91.23 | 2.32 | 87.15 | 0.89 | 100.76 | 3.63 |
| UDCA | 2.55 | 2.43 | 3.93 | 5.01 | 6.97 | 6.86 | 8.24 | 13.26 | 105.95 | 6.66 | 86.55 | 2.37 | 97.73 | 3.67 |
| HDCA | N.A. | N.A. | 7.48 | 5.26 | 4.89 | 14.55 | 5.38 | 11.60 | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. |
| GDCA | N.A. | N.A. | 4.61 | 1.98 | 2.95 | 4.73 | 4.98 | 8.20 | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. |
| CDCA | 5.59 | 6.53 | 2.00 | 3.52 | 5.86 | 5.08 | 8.31 | 13.19 | 97.51 | 2.66 | 84.35 | 2.17 | 106.01 | 9.83 |
| DCA | N.A. | N.A. | 4.51 | 6.70 | 7.91 | 4.70 | 8.80 | 11.16 | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. |
3.4 Determination of 14 analytes in bear bile and its analogues using RPLC-HILIC-tailored MRM
The verified method was subsequently applied for simultaneous determination of fourteen compounds, including four nucleosides (i.e. uridine, adenosine, inosine, guanosine), five unconjugated bile acids (i.e. CA, UDCA, HDCA, CDCA, DCA) and five conjugated bile acids (i.e. TUDCA, TCA, GUDCA, GHDCA, GDCA), in ten batches of bear bile as well as seven samples originated from other species. The representative chromatogram for simultaneous determination of fourteen analytes in bear bile sample (BB3) is shown in Fig. 3D, and the chromatograms of bile samples, seven in total, originated from other species are displayed in Fig. S1 (ESI).† The details of quantitative results are illustrated in Table 3.| Sample | Uridine | Adenosine | Inosine | TUDCA | TCA | Guanosine | GUDCA | GHDCA | CA | UDCA | HDCA | GDCA | CDCA | DCA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a N.D.: not detected; N.Q.: not quantified. | ||||||||||||||
| BB1 | N.D. | 0.02 | N.Q. | 250.33 | 7.33 | N.D. | 0.23 | 0.91 | 1.54 | 13.83 | N.D. | 0.13 | 31.03 | 0.43 |
| BB2 | N.D. | N.D. | N.D. | 217.00 | 8.60 | N.D. | 0.18 | N.D. | 1.30 | 5.17 | N.D. | N.D. | 12.37 | N.Q. |
| BB3 | N.D. | N.D. | N.D. | 190.33 | 7.40 | N.D. | 0.19 | N.D. | 0.51 | 4.23 | N.D. | N.D. | 10.50 | N.Q. |
| BB4 | N.D. | N.D. | N.D. | 136.00 | 7.00 | N.D. | N.Q. | N.D. | 0.28 | 2.36 | N.D. | N.D. | 7.77 | N.D. |
| BB5 | N.D. | N.D. | N.D. | 207.33 | 3.26 | N.D. | N.Q. | N.D. | 0.03 | 0.85 | N.D. | N.D. | 1.51 | N.D. |
| BB6 | N.D. | N.D. | N.D. | 219.33 | 2.87 | N.D. | 0.18 | N.D. | 0.04 | 1.14 | N.D. | N.D. | 2.98 | N.D. |
| BB7 | N.Q. | 0.03 | 0.03 | 236.33 | 3.93 | 0.03 | N.Q. | N.D. | N.Q. | 1.03 | N.D. | N.D. | 1.74 | N.D. |
| BB8 | N.D. | N.D. | N.D. | 6.10 | N.D. | N.D. | N.D. | 15.40 | 0.05 | N.D. | 6.37 | 0.09 | 11.20 | N.D. |
| BB9 | N.Q. | 0.03 | N.Q. | 108.33 | 45.67 | N.D. | N.Q. | 16.20 | 3.43 | 7.63 | N.D. | 17.07 | 20.30 | 1.18 |
| BB10 | N.Q. | N.D. | N.D. | 227.00 | 4.53 | N.D. | N.Q. | N.D. | 0.07 | 1.29 | N.D. | N.D. | 4.13 | N.D. |
| Artificial cow-bezoar | N.Q. | 0.03 | 0.03 | N.D. | 41.00 | 0.06 | N.D. | 0.34 | 59.33 | N.D. | 28.60 | 4.23 | 6.40 | 21.07 |
| Natural cow-bezoar | N.Q. | 0.03 | 0.01 | N.Q. | 30.13 | N.D. | N.D. | N.D. | 35.00 | N.D. | N.D. | 14.47 | 2.21 | 19.00 |
| Geese bile | 0.62 | 0.07 | 0.42 | N.Q. | 3.10 | 0.30 | N.D. | N.D. | 0.10 | N.D. | N.D. | 0.13 | 9.03 | N.D. |
| Goat bile | 0.51 | N.D. | 1.34 | N.D. | 235.33 | 0.09 | N.D. | N.Q. | 38.67 | N.D. | N.D. | 3.43 | N.Q. | N.Q. |
| Cattle bile | 0.17 | N.Q. | 0.01 | N.Q. | 84.00 | N.Q. | N.D. | 19.13 | 2.12 | N.D. | N.D. | 31.43 | N.Q. | N.Q. |
| Yak bile | 0.18 | N.Q. | N.D. | N.Q. | 120.33 | N.D. | N.D. | N.Q. | 1.68 | N.D. | N.D. | 49.33 | N.D. | N.D. |
| Chicken bile | 0.82 | 0.07 | 0.12 | N.Q. | 44.67 | 0.21 | N.D. | 0.20 | 0.74 | N.D. | N.D. | 0.31 | 1.85 | N.D. |
Overall, the contents of bile acids and nucleosides extensively varied among different medicinal bile samples. The content range of all analytes was as many as four orders of magnitude (from 0.01 μg mg−1 inosine in natural cow-bezoar/cattle bile to 250.33 μg mg−1 TUDCA in BB1). Minor distributions occurred for those nucleosides (<0.82 μg mg−1 for uridine, <0.07 μg mg−1 for adenosine, <1.34 μg mg−1 for inosine, and <0.30 μg mg−1 for adenosine) in most samples, especially in bear bile samples. It was obvious that TUDCA was the most abundant component in bear bile samples (108.33–250.33 μg mg−1), except for BB8 (6.10 μg mg−1). However, TUDCA was found trace occurrences in the other bile samples (Table 3). Therefore, it was possible for employing TUDCA as the diagnostic marker to differentiate bear from other medicinal bile samples. GUDCA (<0.23 μg mg−1) and UDCA (<0.23 μg mg−1), less abundant than TUDCA, were only detected in bear bile samples as well.36 TCA was the abundant component in goat bile (235.33 μg mg−1) and yak bile (120.33 μg mg−1), and also exhibited significant distributions (2.87–84.00 μg mg−1) in other samples, nonetheless, undetectable in BB8. HDCA was merely quantifiable in artificial cow-bezoar (28.60 μg mg−1) and BB8 (6.37 μg mg−1). Notably, GHDCA showed high contents in BB8 (15.40 μg mg−1) and BB9 (16.20 μg mg−1) which were collected from Yanji pharmaceuticals. It was worthy to note that significant differences occurred between artificial and natural cow-bezoar. Except for TUDCA, GUDCA and UDCA, the other bile acids exhibited extensive distributions in the artificial sample, whereas the occurrences of GHDCA and HDCA were not observed in natural one. Judging from the quantitative properties of these bile samples, BB8 that exhibited different quantitative profiles from other bear bile might be contaminated by bile originated from other species.
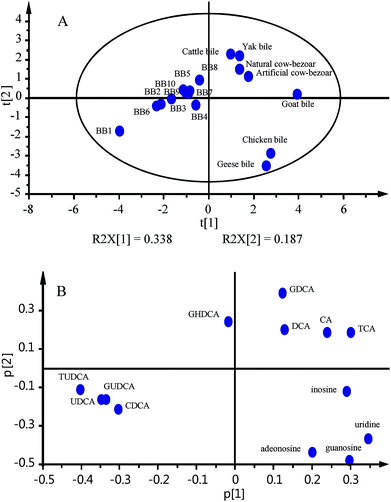
The quantitative results were subjected into multivariated statistical analysis software package (SIMCA-P v14.1, Umetrics, Umeå, Sweden) to carry out principal cluster analysis (PCA). A 5-component model explained 95.1% of the variance, with the first and second components explaining 33.8% and 18.7%, respectively. The score scattering plot together with loading plot are shown in Fig. 4A and B, respectively. Obviously, all bear bile samples located at the left region of the ellipse corresponding to 95% confidence interval, whereas the other samples were distributed in the right part. To obtain more intuitive information, the hierarchical clustering tree is also shown in Fig. S2 (ESI).† BB1–10 were grouped into a single class attributing to the significant differences regarding quantitative properties between bear bile and bile samples from other species. Great similarity occurred, as expected, between yak bile and cattle bile; and then, they were sorted into a single cluster with goat bile, attributing to their similar food spectrum as well as adjacent taxonomic positions. Interestingly, the cluster distance within bear bile samples was equal to that from goat bile to the cluster consisting of yak bile and cattle bile, indicating the variations among different habitats were comparable with the difference between subfamilies of Bovidae. Artificial and natural cow-bezoars were grouped into a single family. Geese and chicken bile samples belonging to poultry also showed similar chemical profiles. It is worthy to note that BB8 almost located at the edge of bear bile cluster, however, closing to cattle bile and yak bile dots; hence, it was possible that BB8 was an adulteration of bear bile via introducing bile from other species, such as cattle. Regarding loading plot, significant enrichment occurred for TUDCA, GUDCA, UDCA and CDCA in all bear bile samples, in particular BB1, indicating that this chemical cluster might be qualified to act as quality indicators for bear bile samples. Chicken bile and geese bile are rich of those nucleosides, such as uridine, adenosine, inosine and guanosine, whereas accumulation of GDCA, DCA, CA, and TCA were observed in cattle bile, yak bile, natural cow bezoar, and artificial cow bezoar. In particular, abundant distributions were observed for CA and TCA in goat bile.
4. Conclusions
In response to the large polarity and content spans of components in bear bile and its analogues, a versatile method namely RPLC-HILIC-tailored MRM was developed by conducting improvements for both LC and MS/MS domains. RP and HILIC column were serially connected to afford desired chromatographic performances for all fourteen analytes in a single run. Due to the employment of tailored MRM algorithm, inferior CEs were employed to enlarge the upper limits of linear ranges for those abundant compounds, including TUDCA, TCA, GHDCA, CA and GDCA. The method validation assays demonstrated the current approach to be reliable for quantitative analysis of four nucleosides together with ten bile acids in bear bile although those targeted compounds exhibited great polarity and content coverage. Quantitative properties regarding those fourteen compounds were raveled in ten batches of bear bile as well as seven bile samples originated from other animals. Significant variations in terms of the content and chemical type were observed for both nucleosides and bile acids among not only different batches of bear bile, but also different species. TUDCA, GUDCA, UDCA and CDCA, particularly TUDCA, might be qualified to serve as the quality markers for bear bile samples. The results proved that RPLC-HILIC-tailored MRM was a promising choice to achieve polarity- and content-extended determination of components and should be a versatile tool for quality control of bear bile, as well as other traditional Chinese medicines derived from animal bile.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was financially supported by National Science Fund of China (No. 81773875, 81403073, and 81530097), Quality guarantee system of Chinese Herbal Medicines (No. 201507002), foundation from Beijing University of Chinese Medicine (No. 2016-JYB-XJQ004), and the Macao Science and Technology Development Fund (007/2014/AMJ).References
- Y. Feng, K. Siu, N. Wang, K. M. Ng, S. W. Tsao, T. Nagamatsu and Y. Tong, J. Ethnobiol. Ethnomed., 2009, 5, 2 CrossRef PubMed.
- S. Li, H. Y. Tan, N. Wang, M. Hong, L. Li, F. Cheung and Y. Feng, Evid. Based Complement. Alternat. Med., 2016, 2016, 4305074 Search PubMed.
- Z. Liu, Z. Jiang, H. Fang, C. Li, A. Mi, J. Chen, X. Zhang, S. Cui, D. Chen, X. Ping, F. Li, C. Li, S. Tang, Z. Luo, Y. Zeng and Z. Meng, PLoS One, 2016, 11, e0145901 Search PubMed.
- D. Q. Wang and M. C. Carey, World J. Gastroenterol., 2014, 20, 9952–9975 CrossRef PubMed.
- Yerigui, X. H. Wu, X. J. Wang and C. M. Ma, Anal. Sci., 2016, 32, 499–503 CrossRef CAS PubMed.
- J. Y. Zhao, W. Lin, Q. C. Zhuang, X. Y. Zhong, J. Peng and Z. F. Hong, Chin. J. Integr. Med., 2015, 21, 369–375 CrossRef PubMed.
- X. Qiao, W. Song, X. H. Lin, Q. Wang, T. Bo, D. A. Guo, J. Liu and M. Ye, Anal. Methods, 2014, 6, 596–601 RSC.
- C. Feng, X. Li, C. Zhang, G. He, Y. Xu, W. Li, Z. Yu, H. Cai and D. Liu, Anal. Methods, 2015, 7, 7606–7617 RSC.
- J. Sun, Y. Song, H. Sun, W. Liu, Y. Zhang, J. Zheng, Q. Zhang, Y. Zhao, W. Xiao, P. Tu and J. Li, J. Pharm. Biomed. Anal., 2017, 145, 462–472 CrossRef CAS PubMed.
- Y. Yan, Q. Q. Song, X. J. Chen, J. Li, P. Li, Y. T. Wang, T. X. Liu, Y. L. Song and P. F. Tu, J. Chromatogr. A, 2017, 1501, 39–50 CrossRef CAS PubMed.
- Y. L. Song, Q. Q. Song, J. Li, N. Zhang, Y. F. Zhao, X. Liu, Y. Jiang and P. F. Tu, J. Chromatogr. A, 2016, 1429, 238–247 CrossRef CAS PubMed.
- X. L. Ma, X. Y. Guo, Y. L. Song, L. R. Qiao, W. G. Wang, M. B. Zhao, P. F. Tu and Y. Jiang, Sci. Rep., 2016, 6, 38379 CrossRef CAS PubMed.
- Y. L. Song, Q. Q. Song, Y. Liu, J. Li, J. B. Wan, Y. T. Wang, Y. Jiang and P. F. Tu, Anal. Chim. Acta, 2017, 953, 40–47 CrossRef CAS PubMed.
- Q. Q. Song, Y. F. Zhao, X. J. Chen, J. Li, P. Li, Y. Jiang, Y. T. Wang, Y. L. Song and P. F. Tu, RSC Adv., 2017, 7, 31838–31849 RSC.
- Y. Q. Wang, X. Tang, J. F. Li, Y. L. Wu, Y. Y. Sun, M. J. Fang, Z. Wu, X. M. Wang and Y. K. Qiu, J. Chromatogr. A, 2017, 1519, 145–151 CrossRef CAS PubMed.
- L. Montero, E. Ibanez, M. Russo, L. Rastrelli, A. Cifuentes and M. Herrero, Anal. Chim. Acta, 2017, 985, 202–212 CrossRef CAS PubMed.
- S. Ji, D. D. He, T. Y. Wang, J. Han, Z. Li, Y. Du, J. H. Zou, M. Z. Guo and D. Q. Tang, J. Pharm. Biomed. Anal., 2017, 146, 68–78 CrossRef CAS PubMed.
- N. Sheng, H. Zheng, Y. Xiao, Z. Wang, M. Li and J. Zhang, J. Chromatogr. A, 2017, 1517, 97–107 CrossRef CAS PubMed.
- L. I. Tsonev and A. G. Hirsh, J. Chromatogr. A, 2016, 1468, 173–182 CrossRef CAS PubMed.
- A. T. Hanke, P. D. Verhaert, L. A. van der Wielen, M. H. Eppink, E. J. van de Sandt and M. Ottens, J. Chromatogr. A, 2015, 1394, 54–61 CrossRef CAS PubMed.
- P. Schoenmakers and P. Aarnoutse, Anal. Chem., 2014, 86, 6172–6179 CrossRef CAS PubMed.
- S. Wang, X. Shi and G. Xu, Anal. Chem., 2017, 89, 1433–1438 CrossRef CAS PubMed.
- Y. L. Song, N. Zhang, S. P. Shi, J. Li, Q. Zhang, Y. F. Zhao, Y. Jiang and P. F. Tu, J. Chromatogr. A, 2015, 1407, 106–118 CrossRef CAS PubMed.
- S. Wang, J. Li, X. Shi, L. Qiao, X. Lu and G. Xu, J. Chromatogr. A, 2013, 1321, 65–72 CrossRef CAS PubMed.
- S. Wang, L. Qiao, X. Shi, C. Hu, H. Kong and G. Xu, Anal. Bioanal. Chem., 2015, 407, 331–341 CrossRef CAS PubMed.
- T. Alvarez-Segura, J. R. Torres-Lapasio, C. Ortiz-Bolsico and M. C. Garcia-Alvarez-Coque, Anal. Chim. Acta, 2016, 923, 1–23 CrossRef CAS PubMed.
- S. Bieber, G. Greco, S. Grosse and T. Letzel, Anal. Chem., 2017, 89, 7907–7914 CrossRef CAS PubMed.
- J. Chen, L. Gao, Z. Li, S. Wang, J. Li, W. Cao, C. Sun, L. Zheng and X. Wang, Anal. Chim. Acta, 2016, 914, 117–126 CrossRef CAS PubMed.
- J. Haggarty, M. Oppermann, M. J. Dalby, R. J. Burchmore, K. Cook, S. Weidt and K. E. Burgess, Metabolomics, 2015, 11, 1465–1470 CrossRef CAS PubMed.
- W. Y. Sun, Q. W. Lu, H. Gao, L. Tong, D. X. Li, Z. Q. Zhou, Z. J. Jiang, H. Sun and K. S. Bi, Sci. Rep., 2017, 7, 7061 CrossRef PubMed.
- Z. Yan and R. Yan, Anal. Chim. Acta, 2015, 894, 65–75 CrossRef CAS PubMed.
- H. Liu, L. Lam and P. K. Dasgupta, Talanta, 2011, 87, 307–310 CrossRef CAS PubMed.
- Z. Yan and R. Yan, J. Chromatogr. A, 2016, 1443, 101–110 CrossRef CAS PubMed.
- Q. Q. Song, Y. L. Song, N. Zhang, J. Li, Y. Jiang, K. R. Zhang, Q. Zhang and P. F. Tu, RSC Adv., 2015, 5, 57372–57382 RSC.
- S. Yin, M. Su, G. Xie, X. Li, R. Wei, C. Liu, K. Lan and W. Jia, Anal. Bioanal. Chem., 2017, 409, 5533–5545 CrossRef CAS PubMed.
- X. Qiao, M. Ye, D. L. Pan, W. J. Miao, C. Xiang, J. Han and D. A. Guo, J. Chromatogr. A, 2011, 1218, 107–117 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra10229a |
| This journal is © The Royal Society of Chemistry 2017 |