Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Direct cryopreservation of adherent cells on an elastic nanofiber sheet featuring a low glass-transition temperature
Onon Batnyama,
Shin-ichiro Suyeab and
Satoshi Fujita *ab
*ab
aDepartment of Frontier Fiber Technology and Science, Graduate School of Engineering, University of Fukui, 3-9-1 Bunkyo, Fukui-city, Fukui 910-8507, Japan. E-mail: fujitas@u-fukui.ac.jp
bLife Science Innovation Center, University of Fukui, 3-9-1 Bunkyo, Fukui-city, Fukui 910-8507, Japan
First published on 3rd November 2017
Abstract
Cryopreservation of ready-made cell-biomaterial composites is an essential aspect of modern regenerative medicine and tissue engineering. However, in the freeze–thawing of adherent cells, a problem encountered is the detachment of cells due to the intracellular tensile stress caused by dehydration. To reduce the cell detachment, a scaffold that retains elasticity at freezing temperatures was investigated here for use in direct cryopreservation of adherent cells. We focused on electrospun polyurethane (PU) nanofiber sheets, which featured a lower glass-transition temperature than the freezing temperature used and presented a loose mesh-like structure of nanofibers. Consequently, the recovery of cells cultured on the PU nanofiber sheets and then freeze–thawed was higher than the recovery of cells cultured on polystyrene films and fibers. Furthermore, higher cell recovery was obtained with randomly oriented PU nanofibers than with highly aligned PU nanofibers. These results suggest that the elasticity of the polymer and the looseness of the fiber mesh are key factors that enhance the recovery of adherent cells after freeze–thawing. This is the first report demonstrating that the elastic nanofiber scaffold is an available material that enables the cryopreservation of adherent cells; the use of this scaffold could substantially improve the cryopreservation outcome of cell-biomaterial composites.
Introduction
Recent advances in the research and development of numerous ready-made cell-biomaterial composites hold the potential to provide not only standardized research tools for laboratories, but also safe and convenient treatments for patients.1 For the practical application of these composites, reliable cryopreservation of adherent cells is required to ensure long shelf-life and safe stocks. One of the problems encountered in cryopreservation is the injury caused by the ice crystals that are formed intracellularly and extracellularly during the freeze–thawing process.2 For preventing the formation of ice crystals, two widely recognized conventional methods are vitrification and slow freezing.3 In vitrification, rapid freezing is used to cryopreserve embryonic stem cells, induced pluripotent stem cells, and oocytes while maintaining high viability of the cells;4–6 however, this method cannot be readily applied in the case of adherent cells because the presence of the substrate triggers uneven cooling and ice-crystal formation.7 Conversely, slow freezing, which is the “gold standard” in the cryopreservation of mammalian cells, dehydrates cells gradually by enhancing the permeability of the cell membrane in the presence of dimethyl sulfoxide (DMSO) without the formation of intracellular ice crystals.8 Some of DMSO-free cryoprotectant solutions were also commercialized for slow freezing, such as CryoSOfree™ (Sigma-Aldrich, USA), CryoScarless® (BioVerde, Japan), and Stem-Cellbanker® (Zenoaq, Japan). However, even with slow freezing, the cryopreservation of adherent cells is considerably more challenging than that of single-cell suspensions; this is because of the shrinkage and deformation of cells caused by the exclusion of water,9 which results in the detachment and breakage of cells. To overcome these hurdles and successfully cryopreserve adherent cells, several approaches have been reported, including enhancement of cell-substrate interaction,10,11 supportive use of hydrogels,12–14 augmentation of cell–cell interaction,15 and improvement of freezing conditions.16,17 Notably, the use of high concentrations of the cryoprotectant DMSO, which enhances the dehydration effect and inhibits of ice-crystal formation, was reported to allow direct cryopreservation of adherent cells cultured on polydimethylsiloxane substrate.18 However, the development of a systematic approach for the cryopreservation of adherent cells remains under investigation.The attachment of cells to any substrate requires the recruitment of the transmembrane receptor integrin and the formation of focal adhesions, where cells sense and adapt themselves to their microenvironment,19 and the maintenance of cell adhesion after the freeze–thawing process is critical for cell viability and function. However, during the freeze–thawing process, cells adhered onto a stiff substrate detach and break due to the differential thermal contraction of the cells and the substrate.20 Therefore, we focused here on the elasticity of the scaffold for maintaining tight adhesion with cells after freeze–thawing. We hypothesized that a substrate that retains its elasticity at a freezing temperature would inhibit the exfoliation of adherent cells from the surface that results from the contraction caused by the dehydration occurring in a slow-freezing procedure. To test this possibility, we designed a substrate based on considering two viewpoints: first, we focused on an electrospun nanofiber sheet as an elastic scaffold; this nanofiber sheet, which is one of the promising forms of scaffolds suitable for cell-biomaterial composites, mimics the natural structure of the extracellular matrix and thus controls cell proliferation, migration, and differentiation.21–24 The loose mesh-like structure of an electrospun nanofiber sheet would mitigate the detachment and breakage of adherent cells even if the cells deform and shrink. Second, we focused on a polymer featuring a glass-transition temperature (Tg) that is lower than the freezing temperature. Here, we selected polyurethane, the Tg of which is −40 °C.25 Thus, for the cryopreservation of adherent cells, we investigated the suitability of a scaffold that retains its elasticity at freeze–thawing temperatures.
Experimental
Materials
The following materials were from commercial sources: thermoplastic polyether-polyurethane-elastomer (PU; Mw 146![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Elastollan® 1180A10), BASF (Ludwigshafen, Germany); polystyrene (PS; Mw 350
000, Elastollan® 1180A10), BASF (Ludwigshafen, Germany); polystyrene (PS; Mw 350![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), Sigma-Aldrich (St. Louis, MO, USA); and tetrahydrofuran (THF), N,N-dimethylformamide (DMF), and DMSO, Wako Pure Chemical Industries (Osaka, Japan). All other chemicals and reagents were of analytical grade and were used without further purification.
000), Sigma-Aldrich (St. Louis, MO, USA); and tetrahydrofuran (THF), N,N-dimethylformamide (DMF), and DMSO, Wako Pure Chemical Industries (Osaka, Japan). All other chemicals and reagents were of analytical grade and were used without further purification.
Fabrication of substrates
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) THF at 1
THF at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
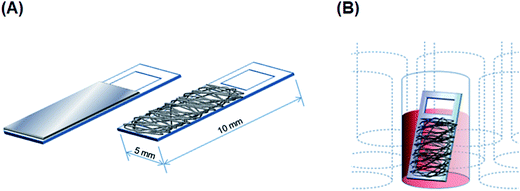
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 ratio, respectively, and gently mixed overnight on a rotator (BC-710, BIO-CRAFT, Tokyo, Japan) at room temperature. PU and PS fiber scaffolds were electrospun on acrylic frames (Scheme 1A) by using a commercial electrospinning setup (NANON, MECC, Fukuoka, Japan), which consisted of a 23 G-needle nozzle, a syringe pump, a high-voltage power supply, and a rotating collector in a closed chamber. The electrospinning conditions were the following: applied electric field: 1.67 kV cm−1; infusion flow rate: 0.6 mL h−1; collector rotation speed: 1500 rpm (linear velocity 15 m s−1) for aligned fibers and 50 rpm (0.5 m s−1) for random fibers. Sterilization and surface-hydrophilicity enhancement of nanofibrous scaffolds were once again achieved through oxygen-plasma treatment (as mentioned above).
9 ratio, respectively, and gently mixed overnight on a rotator (BC-710, BIO-CRAFT, Tokyo, Japan) at room temperature. PU and PS fiber scaffolds were electrospun on acrylic frames (Scheme 1A) by using a commercial electrospinning setup (NANON, MECC, Fukuoka, Japan), which consisted of a 23 G-needle nozzle, a syringe pump, a high-voltage power supply, and a rotating collector in a closed chamber. The electrospinning conditions were the following: applied electric field: 1.67 kV cm−1; infusion flow rate: 0.6 mL h−1; collector rotation speed: 1500 rpm (linear velocity 15 m s−1) for aligned fibers and 50 rpm (0.5 m s−1) for random fibers. Sterilization and surface-hydrophilicity enhancement of nanofibrous scaffolds were once again achieved through oxygen-plasma treatment (as mentioned above).Characterization of fabricated substrates
S = 〈3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos2 cos2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ − 1〉/2 θ − 1〉/2
| (1) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ.
δ.Cryopreservation of myoblast cells
Statistical analysis
All experiments were conducted at least thrice, and all values are reported as means ± standard deviation (SD). For statistical analysis, nonparametric Kruskal–Wallis tests were performed using R (Version 2.1.7); p < 0.05 was considered statistically significant.Results
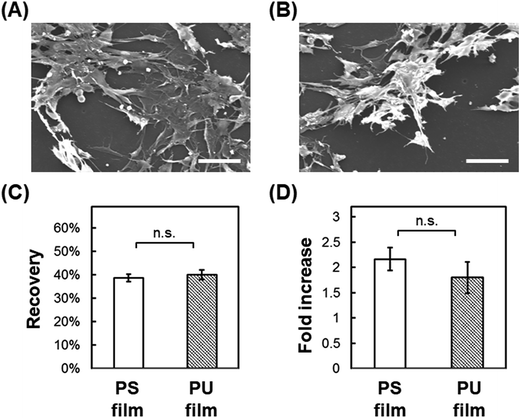
Direct cryopreservation on films
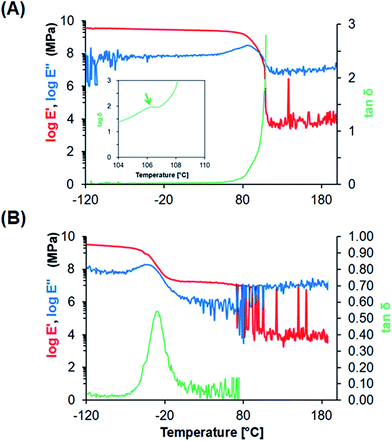
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ, which corresponds to its glass-transition temperature (Tg), PU films showed a peak at −30 °C in tan
δ, which corresponds to its glass-transition temperature (Tg), PU films showed a peak at −30 °C in tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ, which reflects the micro-Brownian segmental motion of amorphous PU at a freezing temperature.
δ, which reflects the micro-Brownian segmental motion of amorphous PU at a freezing temperature.
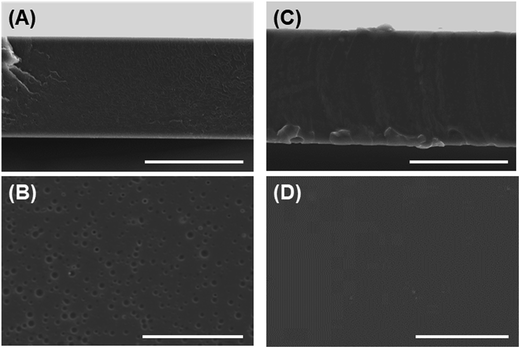 | ||
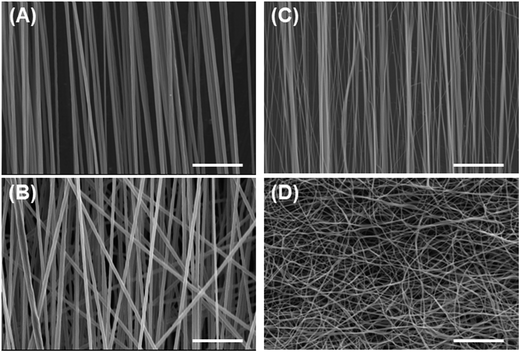
| Fig. 1 Morphological observation of fabricated films. (A, B) PS film, (C, D) PU film; (A, C) cross-sectional views, (B, D) top views. Bars = 50 μm. | ||
Direct cryopreservation on nanofiber sheets
Discussion
Effect of polymer elasticity
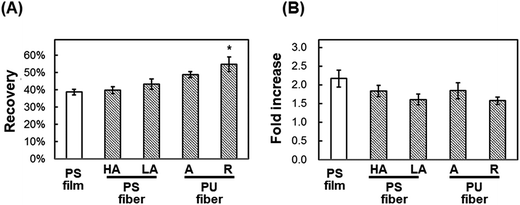
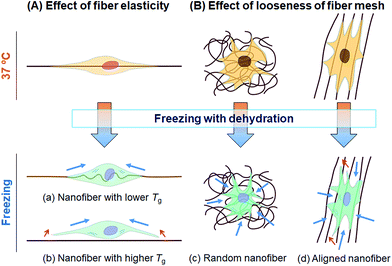
In this study, we tested whether a polymer scaffold that exhibits elasticity at the freezing temperature would enable cells to maintain their adhesion even if the adherent cells contract because of dehydration during the freezing process, and thus increase the viability of freeze–thawed adherent cells. The elasticity of a polymer at the freezing temperature depends on its Tg. If the polymer Tg is lower than the melting temperature of a medium containing 10% DMSO, approximately −20 °C,29 the polymer can deform by creeping during the freezing process (Scheme 2A). To investigate whether a polymer featuring a low Tg can be used for developing an effective cryopreservation scaffold, we compared the recovery of cells cultured on PS and PU scaffolds, fabricated in the form of films and fibers, after direct freeze–thawing of the cells on the scaffolds. We found that whereas 60% of the cells cultured on both films and on the PS fiber exfoliated after freeze–thawing, cell recovery was high with the PU fiber. We can attribute these results to the stiffness of the scaffold: at the melting temperature of the medium, PS was in a stiff glassy state, but PU was in a rubbery state. The elasticity of the PU used in this study was approximately 8 MPa at −10 °C, as determined through DMA measurement, and SEM analysis revealed that the diameter of each PU fiber was roughly 1 μm. Therefore, a force of 6 × 106 pN would be required to bend a single PU fiber. Dembo and Wang reported that the contraction force of cells in tight adhesions is 2000 pN μm−2.30 When cells extend to 5000 μm2, the force required to detach the cells from the substrate is 10 × 106 pN, which is larger than the force necessary to bend the fiber.Effect of geometry of fiber mesh
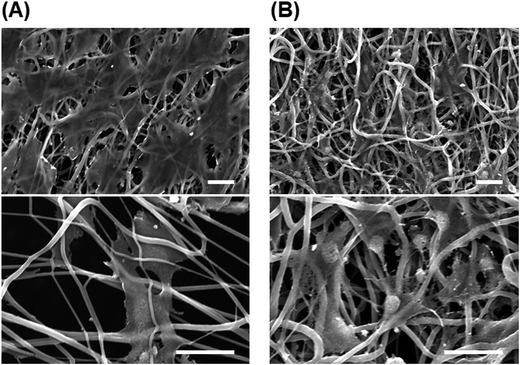
We also investigated the effect of the fiber mesh structure on cell recovery: the recovery was low with highly oriented fibers, which indicated that the tight and tensioned mesh structure caused cell detachment, particularly in the direction of fiber alignment; by contrast, random PU fibers exhibited a looseness in their mesh structure in the fiber sheets, which resulted in the maintenance of cell adhesions (Scheme 2B). Here, the loosenes was indexed by the randomness of fiber geometry, S. Moreover, the diameter and the pore sizes of the fibers were very small relative to the dimension of cells, and thus the cells attached to the substrate at multiple focal-adhesion sites. SEM analysis revealed that these multiple adhesions were retained after the freeze–thawing process, and this resulted in the high cell recovery observed with random PU fibers.Intriguingly, cell proliferation after culture for 24 h on aligned PU fibers was higher than that after culture on random PU nanofibers. Cells have been previously reported to be sensitive to the topographical arrangement of the substrate. Cell stretching can induce the accumulation of vinculin at talin-containing focal adhesions by exposing vinculin-binding sites in the talin rod domain, and can thereby allow the reinforcement of the focal adhesions with vinculin.31 The extension of cells triggers the development of strong adhesion and pronounced stress fibers.32–35 Furthermore, several studies have provided evidence that cells prefer random arrangements, which can regulate cellular functions such as attachment, proliferation, and survival, over perfect grooves, pits, and pillars.36–38 Thus, random geometry is favorable for mitigating the tension that arises during freeze–thawing, whereas high orientation is preferable for cell proliferation after thawing. Overall, these findings suggest the requirement of an optimal geometry of the fiber mesh structure, which should be further investigated to maximize the yield of cells after freeze–thawing and culturing the cells.
Conclusions
To meet the increasing demand for tissue allografts, the development of artificial tissue-engineering constructs has been widely investigated; however, the maintenance of the shelf-life and post-thaw biofunctionality of the constructs is a challenge that remains poorly addressed. Here, we first found that nanofiber materials, which present a loose mesh structure and exhibit elasticity at freezing temperatures due to the use of polymers featuring a low Tg, can be used for direct cryopreservation of adherent cells. Our study can potentially contribute to the commercial application and transportation of therapeutic cells for regenerative medicine by providing a standard procedure for the cryopreservation of adherent cells, and the study should also facilitate the further development of this research field. We expect that our strategy is also applicable to other emerging cryoprotectants for slow freezing from the proposed mechanism of cryoprotection. Moreover, the results of cell-culture assays indicated that highly efficient cryoprotectant materials can be developed by modifying the surface cell adhesion.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by JSPS KAKENHI Grant Number 16K05908.References
- R. H. Harrison, J. P. St-Pierre and M. M. Stevens, Tissue Eng., Part B, 2014, 20, 1–16 CrossRef PubMed.
- G. J. Morris and E. Acton, Cryobiology, 2013, 66, 85–92 CrossRef PubMed.
- D. Balci and A. Can, Curr. Stem Cell Res. Ther., 2013, 8, 60–72 CrossRef CAS PubMed.
- T. Miyazaki and H. Suemori, Methods Mol. Biol., 2015, 1235, 97–104 CAS.
- C. J. Hunt, Transfus. Med. Hemother., 2011, 38, 107–123 CrossRef PubMed.
- A. Cobo and C. Diaz, Fertil. Steril., 2011, 96, 277–285 CrossRef PubMed.
- L. L. Kuleshova, S. S. Gouk and D. W. Hutmacher, Biomaterials, 2007, 28, 1585–1596 CrossRef CAS PubMed.
- J. O. Karlsson, E. G. Cravalho, I. H. Borel Rinkes, R. G. Tompkins, M. L. Yarmush and M. Toner, Biophys. J., 1993, 65, 2524–2536 CrossRef CAS PubMed.
- P. Mazur, Am. J. Physiol.: Cell Physiol., 1984, 247, C125–C142 CAS.
- L. Ji, J. J. de Pablo and S. P. Palecek, Biotechnol. Bioeng., 2004, 88, 299–312 CrossRef CAS PubMed.
- X. Xu, Y. Liu and Z. F. Cui, J. Tissue Eng. Regener. Med., 2014, 8, 664–672 CrossRef CAS PubMed.
- R. Malpique, F. Ehrhart, A. Katsen-Globa, H. Zimmermann and P. M. Alves, Tissue Eng., Part C, 2009, 15, 373–386 CrossRef CAS PubMed.
- A. Katsen-Globa, I. Meiser, Y. A. Petrenko, R. V. Ivanov, V. I. Lozinsky, H. Zimmermann and A. Y. Petrenko, J. Mater. Sci.: Mater. Med., 2014, 25, 857–871 CrossRef CAS PubMed.
- H. F. Ahmad and A. Sambanis, Acta Biomater., 2013, 9, 6814–6822 CrossRef CAS PubMed.
- J. P. Acker, A. Larese, H. Yang, A. Petrenko and L. E. McGann, Cryobiology, 1999, 38, 363–371 CrossRef CAS PubMed.
- T. L. Bailey, M. Wang, J. Solocinski, B. P. Nathan, N. Chakraborty and M. A. Menze, Cryobiology, 2015, 71, 472–480 CrossRef CAS PubMed.
- X. Xu, Y. Liu, Z. Cui, Y. Wei and L. Zhang, J. Biotechnol., 2012, 162, 224–231 CrossRef CAS PubMed.
- Y. Nagahara, H. Sekine, M. Otaki, M. Hayashi and N. Murase, Cryobiology, 2016, 72, 53–59 CrossRef CAS PubMed.
- J. D. Humphrey, E. R. Dufresne and M. A. Schwartz, Nat. Rev. Mol. Cell Biol., 2014, 15, 802–812 CrossRef CAS PubMed.
- B. L. Liu, J. McGrath, L. McCabe and M. Baumann, Afr. J. Biotechnol., 2006, 5, 2014–2019 CAS.
- C. Huang and R. Ogawa, FASEB J., 2010, 24, 3625–3632 CrossRef CAS PubMed.
- J. Holst, S. Watson, M. S. Lord, S. S. Eamegdool, D. V. Bax, L. B. Nivison-Smith, A. Kondyurin, L. Ma, A. F. Oberhauser, A. S. Weiss and J. E. Rasko, Nat. Biotechnol., 2010, 28, 1123–1128 CrossRef CAS PubMed.
- S. Fujita, H. Shimizu and S. Suye, J. Nanotechnol., 2012, 2012, 429890 Search PubMed.
- O. Batnyam, H. Shimizu, K. Saito, T. Ishida, S. Suye and S. Fujita, RSC Adv., 2015, 5, 80357–80364 RSC.
- V. Pistor, D. de Conto, F. G. Ornaghi and A. J. Zattera, J. Nanomater., 2012, 2012, 283031 CrossRef.
- R. S. Stein and F. H. Norris, J. Polym. Sci., Part A: Polym. Chem., 1956, 21, 381–396 CAS.
- S. Nomura, H. Kawai, I. Kimura and M. Kagiyama, J. Polym. Sci., Part B: Polym. Phys., 1970, 8, 383–400 CrossRef CAS.
- S. Kurita, US. Pat., US5105627 A, 1992.
- T. M. Gurina, A. V. Pakhomov, A. L. Polyakova, E. I. Legach and G. A. Bozhok, Cell Tissue Banking, 2016, 17, 303–316 CrossRef PubMed.
- M. Dembo and Y. L. Wang, Biophys. J., 1999, 76, 2307–2316 CrossRef CAS PubMed.
- H. Hirata, H. Tatsumi, C. T. Lim and M. Sokabe, Am. J. Physiol.: Cell Physiol., 2014, 306, C607–C620 CrossRef CAS PubMed.
- S. Pellegrin and H. Mellor, J. Cell Sci., 2007, 120, 3491–3499 CrossRef CAS PubMed.
- E. G. Rens and R. M. H. Merks, Biophys. J., 2017, 112, 755–766 CrossRef CAS PubMed.
- C. H. Seo, H. Jeong, Y. Feng, K. Montagne, T. Ushida, Y. Suzuki and K. S. Furukawa, Biomaterials, 2014, 35, 2245–2252 CrossRef CAS PubMed.
- S. Y. Tee, J. Fu, C. S. Chen and P. A. Janmey, Biophys. J., 2011, 100, L25–L27 CrossRef CAS PubMed.
- D. J. Kyle, A. Oikonomou, E. Hill and A. Bayat, Biomaterials, 2015, 52, 88–102 CrossRef CAS PubMed.
- E. A. Cavalcanti-Adam, T. Volberg, A. Micoulet, H. Kessler, B. Geiger and J. P. Spatz, Biophys. J., 2007, 92, 2964–2974 CrossRef CAS PubMed.
- J. Huang, S. V. Grater, F. Corbellini, S. Rinck, E. Bock, R. Kemkemer, H. Kessler, J. Ding and J. P. Spatz, Nano Lett., 2009, 9, 1111–1116 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |