Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bio-conjugation of graphene quantum dots for targeting imaging†
Fei Jia *,
Shuyu Lv and
Sha Xu
*,
Shuyu Lv and
Sha Xu
School of Chemical Engineering, Changchun University of Technology, Changchun, 130012, China. E-mail: jiafei19861112@163.com
First published on 21st November 2017
Abstract
Graphene quantum dots (GQDs) with strong photoluminescence (PL) and chemical modification have attracted tremendous interest for fundamental and applied research owing to their special low-cost production and low toxicity. However, most of the research focused on the GQDs without much targeted design, which limited their applications in specific bio-based fields. Inspired by this fascinating conundrum, we demonstrate GQD-based bio-conjugation. Relying on sedimentation coefficient differences for the density gradient ultracentrifuge (DGU) separation of conjugates with different proteins, ultra-purified conjugates were obtained. We demonstrate the first targeted imaging in both cells and tissue models with single or multi color staining, which shows universality for different kinds of biomolecules.
Fluorescent materials play an important role in investigating biological issues as a “visible” marker.1,2 In the evolution of fluorescent materials, increasing efforts have been devoted to the development of heavy-metal-free and low-toxicity photoluminescence (PL) nano-probes. Carbon dots (CDs: including graphene quantum dot (GQDs),3–7 carbon nanodots (CNDs),8–11 polymer dots (PDs)12–15 and polymer carbon dots (PCDs)16–20), one class of these nano-probes, have drawn much attention during recent years.10,21 GQDs possess high photo-stability, tunable emission, chemical inertness, good biocompatibility, low toxicity, photoelectric/optical properties22–26 and convenient surface modification, and can be achieved by low-cost production. Therefore, GQDs could find use in biomedical applications, such as bioimaging,27,28 drug delivery,29,30 thermal therapy,31,32 DNA cleavage,33 sensors,34 etc.
Bioimaging is an emerging research field aimed at using sophisticated bioimaging probes to visualize specific molecular pathways in vivo or in vitro.35,36 Currently, the majority of bioimaging probes utilized in clinical practice are small-molecule compounds that tend to be unstable, toxic, nonspecific, and rapidly cleared. GQDs have the potential to be remarkably successful in the field of bioimaging due in part to their excellent optical properties and extremely low cytotoxicity, but also due to their suitable size and surface chemistry.37,38 The size of GQDs is always ca. 3–10 nm and lots of oxygen or amino based groups, which are beneficial for bio-labeling process (e.g. endocytose), are found on the surface of GQDs.7,39 However, there are few work focusing on the targeting imaging through the bio-conjugation of GQDs. It is very important to use the targeting GQDs for specific imaging position, which can afford efficient imaging speed, high signal-to-noise ratio (SNR) as well as multi-colors issue imaging.40–42
Although there are kinds of bio-conjugation methods developing with high reaction activity, all the reaction mixture generally contains unbound fluorophores (small molecules or nanomaterials), which cause the background problem in the staining process.43–45 Therefore, the efficient separation of fluorophore bioconjugates from unbound fluorophore is necessary. The density gradient ultracentrifuge (DGU) is a reliable and flexible approach to purify these kinds of conjugates. DGU works by exploiting subtle differences in buoyant density. The species of interest are loaded into an aqueous solution with a known density gradient. Under the centripetal force of an ultracentrifuge, the species sediment toward their respective isopycnic points (i.e. the position where their density matches that of the gradient).46 As a result, it is very important to purify the GQDs bio-conjugation for further targeting staining.
Herein, graphene quantum dots with green PL emission and carboxyl groups were used for the bio-conjugation with amino groups of proteins, and the conjugates of streptavidin (SA)@GQDs, anti-mouse@GQDs and Erbitux@GQDs were successfully prepared and purified by the density gradient ultracentrifuge (DGU) route. The conjugates between GQDs and different proteins possess controllable targeting capacity to label the specific position of biological systems. For example, the anti mouse@GQDs/mouse anti-Her 2 was used to stain SKOV-3 (human ovarian carcinoma) cell; cluster of differentiation 31 (CD 31)@biotin and SA@GQDs were used to stain vessel; Erbitux@GQDs were used to stain squamous cell carcinoma (SCC) cell.
GQDs were prepared by previously reported “nano-cutting” and solvothermal method from graphite powder and HNO3/H2SO4, developed by Yang group,47,48 as shown in Scheme S1.† The average diameter of the prepared GQDs was ca. 4.5 nm with crystal lattice ca. 0.21 nm (Fig. 1a and S2†). The MALDI-TOF MS spectra of GQDs showed the molecular weights were mainly between 1250–2750 with an regular interval (58 Da). It indicated that the neighbouring GQDs differed by “C2O2H2” groups (Fig. 1b), and the actual molecule weight will be larger than this value because big GQDs are hard to be ionized. In the FTIR analysis of GQDs, stretching vibrations of C–OH at 3430 cm−1, and a vibrational absorption band of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at 1635 cm−1 were all observed (Fig. 1c).8,47 In addition, surface groups were also investigated by XPS analysis (Fig. S3†). The –OH was mainly produced by the cutting position through epoxy, while the –COOH and carbonyl was the stable oxidation state. The carboxyl groups were further used for bio-conjugation. GQDs possess optimal excitation and emission wavelengths at ca. 468 nm and 525 nm, respectively. We have tested the absolute quantum yield of present GQDs, and the value is 4.7%. The PL center of GQDs was in surface state which was induced by the hybridization structure of edge groups and connected carbon core, and the participant edge groups for green emission mainly resulted from carboxyl and amide.49,50
O at 1635 cm−1 were all observed (Fig. 1c).8,47 In addition, surface groups were also investigated by XPS analysis (Fig. S3†). The –OH was mainly produced by the cutting position through epoxy, while the –COOH and carbonyl was the stable oxidation state. The carboxyl groups were further used for bio-conjugation. GQDs possess optimal excitation and emission wavelengths at ca. 468 nm and 525 nm, respectively. We have tested the absolute quantum yield of present GQDs, and the value is 4.7%. The PL center of GQDs was in surface state which was induced by the hybridization structure of edge groups and connected carbon core, and the participant edge groups for green emission mainly resulted from carboxyl and amide.49,50
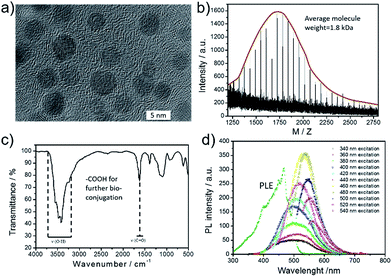 | ||
| Fig. 1 The used GQDs for bio-conjugation. (a) TEM image of GQDs. (b) MALDI-TOF MS spectrum of GQDs. (c) FT-IR spectrum of GQDs. (d) The excitation and emission spectra of GQDs. | ||
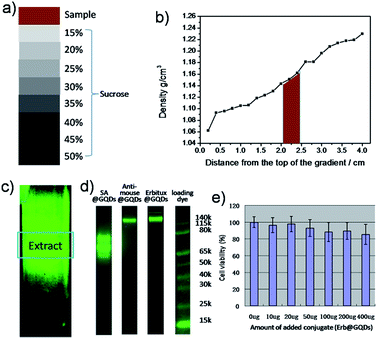
The specific binding used in this work was based on the bio-conjugation, e.g., streptavidin (SA) and biotin.51 First of all, the covalent SA@GQDs and bovine serum albumin (BSA)@biotin were prepared by standard 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) method and biotinylated reagent, respectively (EDC was used to react with the carboxyl groups on the surface of GQDs, forming an unstable reactive O-acylisourea intermediate. Subsequently, N-hydroxysulfosuccinimide (Sulfo-NHS) was added to convert it to a semi-stable amine-reactive NHS-ester.52,53 The amine groups of proteins then covalently conjugated with the NHS-ester modified GQDs to produce protein@GQDs).51 For purifying the BSA@biotin, due to the biotinylated molecules are relatively small, the centrifuge filter with molecule weight 30 kDa can be used to remove the un-conjugated biotin. For purifying the SA@GQDs, the density gradient ultracentrifuge (DGU) was applied to separate the SA@GQDs from the mixtures of free SA, free GQDs. The used gradient sucrose was from 15 to 50% (5% interval, the final column will be a homogeneous linear gradient after tilting 20° for 45 minutes), then we tested the DGU column after ultracentrifuge (Fig. 2a–c), and the desired SA@GQDs conjugates were concentrated at the gradient positions of 20–25% (Fig. 2b). On the top of the fluorescent column section, there are free GQDs and SA; in the middle of the fluorescent section, there should be the desired conjugates between single SA and GQDs, or few SA and GQDs; at the bottom of the fluorescent section, there are possible the multimer with several SA and GQDs conjugates.
To achieve multiple targeting tissue staining with the bio-conjugates, GQDs were further covalently bonded to another two typical proteins (anti-mouse secondary antibodies and Erbitux). Similar DGU purification was used to obtain the selected conjugation with high purifications. For testing the conjugation between GQDs and different proteins, the Bolt 4–12% Bis–Tris gel electrophoresis was used to confirm the conjugate formation of proteins and GQDs via fluorescence imaging, since GQDs have strong fluorescence signals under blue light illumination. As shown in Fig. 2d, SA@GQDs possess three PL bands between 50 kDa and 70 kDa, resulting from the dimer and trimer conjugates; anti-mouse@GQDs show the molecule weight at 115–140 kDa; for Erbitux@GQDs, there is a strong band at 150 kDa, and also two weak bands at larger molecule section, which are due to the dimer and trimer conjugations. Fortunately, the multimer of the protein, which is also powerful for targeting conjugations, don't affect the biological activity.54,55 More importantly, the GQDs show very cell toxicity which is ideal for in vivo staining.
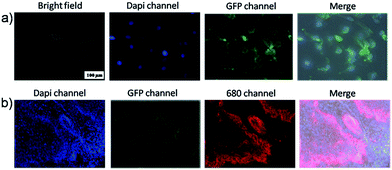
To test the performance of the targeting system, first of all, the targeting cell imaging was tested by SKOV-3 cell model. The cultured SKOV-3 stained by anti-mouse@GQDs/mouse anti-Her 2 and 4′,6-diamidino-2-phenylindole dihydrochloride (Dapi).56 There are Her 2 proteins receptors located on the surface of SKOV-3 cell membrane, which can be conjugated by mouse anti-Her 2 followed by anti-mouse@GQDs. From bright field image of Fig. 3a, the cells kept the normal morphology, and the blue fluorescence (Dapi channel) can be seen in the nucleus while the green fluorescence (GFP channel) was located on the cell membrane of SKOV-3. The overlay images (Fig. 3a and S4†) indicated perfect staining of the nucleus and Her 2 proteins receptors of cell membrane, respectively. Furthermore, due to the universality of the conjugation with bio-molecules, other targeting staining could be achieved.
The targeting imaging provides the possibility to stain different positions of the tissue or living body at the same time. We tested the multi-color imaging by using the SKOV 3 tumor tissue as a model case. The Dapi with blue emission was used for staining the nucleus; the anti mouse@IR-680 and mouse anti-Her 2 were used to stain the SKOV 3 cells of the tumor; the anti CD31@biotin and SA@GQDs were used to stain the vessels of the tumor. Fig. 3b shows the three color imaging of the SKOV 3 tumor. The blue nucleus was encircled by the red cell membrane, and the green vessel can also be recognized spreading the whole tumor slide.
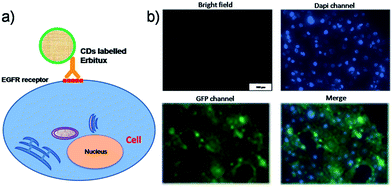
The GQDs can also be used for monoclonal antibody conjugation, which will be “one-step” binding to the cell or organ efficiently.57 Erbitux was used as a proof-of-concept model. Erbitux (cetuximab) is a recombinant, human/mouse chimeric monoclonal antibody that binds specifically to the extracellular domain of the EGFR. The Erbitux@GQDs conjugation possesses a molecule weight slightly more than 150 kDa, and bright fluorescence (Fig. 2d). Fig. 4a shows the scheme of biological fixation between the Erbitux@GQDs and EGFR of the SCC cells. After suitable cultivating, the targeted imaging could be observed on the membrane of the SCC cells (Fig. 4b and S5†). The bright field, Dapi staining of the nucleus and the merger imaging indicated the perfect staining of the EGFR of the SCC cells. The in vivo targeting imaging was also conducted on SCC tumor model (Fig. S6†), showing great tumor targeting capacity also with the non-specific liver uptake. H&E staining study of GQDs@conjugate was added in Fig. S7,† and it show very low in vivo toxicity of the present conjugate.
Conclusions
In conclusion, high-performance GQDs based bio-conjugates were developed. Relying on sedimentation coefficient differences for the density gradient ultracentrifuge (DGU) separation of conjugate with different proteins, ultra-purified conjugates were obtained. By using gel electrophoresis to quickly screen conjugation efficiency, we demonstrate that DGU separated conjugates display the highest selectivity out of any carbon based imaging agent to date. At last, the targeting cell imaging and tissue staining were achieved by the conjugates of SA@GQDs, Erbitux@GQDs and anti mouse@GQDs with very high efficiency, demonstrating the first histological imaging with GQDs based conjugates. The present work endows the fluorescent carbon based materials with promising targeting property, which is of great significance to practical bio-based applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the NSFC 20160323MA1.Notes and references
- C. Vinegoni, I. Botnaru, E. Aikawa, M. A. Calfon, Y. Iwamoto, E. J. Folco, V. Ntziachristos, R. Weissleder, P. Libby and F. A. Jaffer, Sci. Transl. Med., 2011, 3, 84ra45 CrossRef PubMed.
- H. Zhang, Y. Liu, D. Yao and B. Yang, Chem. Soc. Rev., 2012, 41, 6066–6088 RSC.
- Y. Li, Y. Hu, Y. Zhao, G. Shi, L. Deng, Y. Hou and L. Qu, Adv. Mater., 2011, 23, 776–780 CrossRef CAS PubMed.
- S. Zhu, J. Zhang, C. Qiao, S. Tang, Y. Li, W. Yuan, B. Li, L. Tian, F. Liu, R. Hu, H. Gao, H. Wei, H. Zhang, H. Sun and B. Yang, Chem. Commun., 2011, 47, 6858–6860 RSC.
- S. Zhu, J. Zhang, X. Liu, B. Li, X. Wang, S. Tang, Q. Meng, Y. Li, C. Shi, R. Hu and B. Yang, RSC Adv., 2012, 2, 2717 RSC.
- S. Zhu, X. Zhao, Y. Song, J. Zhang and B. Yang, Graphene Science Handbook, 2016, vol. 10, p. 163 Search PubMed.
- S. Zhu, S. Tang, J. Zhang and B. Yang, Chem. Commun., 2012, 48, 4527–4539 RSC.
- B. Kong, A. Zhu, C. Ding, X. Zhao, B. Li and Y. Tian, Adv. Mater., 2012, 24, 5844–5848 CrossRef CAS PubMed.
- S. Zhu, X. Zhao, Y. Song, S. Lu and B. Yang, Nano Today, 2016, 11, 128–132 CrossRef CAS.
- Y. Song, S. Zhu and B. Yang, RSC Adv., 2014, 4, 27184 RSC.
- S. Zhu, Q. Meng, L. Wang, J. Zhang, Y. Song, H. Jin, K. Zhang, H. Sun, H. Wang and B. Yang, Angew. Chem., Int. Ed., 2013, 52, 3953–3957 CrossRef CAS PubMed.
- S. Zhu, J. Zhang, L. Wang, Y. Song, G. Zhang, H. Wang and B. Yang, Chem. Commun., 2012, 48, 10889–10891 RSC.
- S. Zhu, L. Wang, N. Zhou, X. Zhao, Y. Song, S. Maharjan, J. Zhang, L. Lu, H. Wang and B. Yang, Chem. Commun., 2014, 50, 13845–13848 RSC.
- S. Zhu, Y. Song, J. Shao, X. Zhao and B. Yang, Angew. Chem., Int. Ed., 2015, 54, 14626–14637 CrossRef CAS PubMed.
- S. Zhu, J. Zhang, Y. Song, G. Zhang, H. Zhang and B. Yang, Acta Chim. Sin., 2012, 70, 2311 CrossRef CAS.
- J. Shao, S. Zhu, H. Liu, Y. Song, S. Tao and B. Yang, Adv. Sci., 2017, 1700395 CrossRef.
- Y. Song, S. Zhu, J. Shao and B. Yang, J. Polym. Sci., Part A: Polym. Chem., 2017, 55, 610–615 CrossRef CAS.
- S. Tao, S. Zhu, T. Feng, C. Xia, Y. Song and B. Yang, Mater. Today Chem., 2017, 6, 13–25 CrossRef.
- S. Tao, Y. Song, S. Zhu, J. Shao and B. Yang, Polymer, 2017, 116, 472–478 CrossRef CAS.
- S. Lu, L. Sui, J. Liu, S. Zhu, A. Chen, M. Jin and B. Yang, Adv. Mater., 2017, 29, 1603443 CrossRef PubMed.
- S. Zhu, Y. Song, X. Zhao, J. Shao, J. Zhang and B. Yang, Nano Res., 2015, 8, 355–381 CrossRef CAS.
- S. Y. Lim, W. Shen and Z. Gao, Chem. Soc. Rev., 2015, 44, 362–381 RSC.
- C. Yu, T. Xuan, Y. Chen, Z. Zhao, X. Liu, G. Lian and H. Li, J. Alloys Compd., 2016, 688, 611–619 CrossRef CAS.
- C. Yu, T. Xuan, D. Yan, S. Lou, X. Hou, Y. Chen, J. Wang and H. Li, Sens. Actuators, B, 2017, 253, 900–910 CrossRef CAS.
- Q. Qu, A. Zhu, X. Shao, G. Shi and Y. Tian, Chem. Commun., 2012, 48, 5473–5475 RSC.
- X. Li, S. Zhu, B. Xu, K. Ma, J. Zhang, B. Yang and W. Tian, Nanoscale, 2013, 5, 7776–7779 RSC.
- S. Zhu, J. Zhang, S. Tang, C. Qiao, L. Wang, H. Wang, X. Liu, B. Li, Y. Li, W. Yu, X. Wang, H. Sun and B. Yang, Adv. Funct. Mater., 2012, 22, 4732–4740 CrossRef CAS.
- L. Wang, Y. Wang, T. Xu, H. Liao, C. Yao, Y. Liu, Z. Li, Z. Chen, D. Pan, L. Sun and M. Wu, Nat. Commun., 2014, 5, 5357 CrossRef CAS PubMed.
- L. Lin, M. Rong, F. Luo, D. Chen, Y. Wang and X. Chen, TrAC, Trends Anal. Chem., 2014, 54, 83–102 CrossRef CAS.
- L. Li, G. Wu, G. Yang, J. Peng, J. Zhao and J. J. Zhu, Nanoscale, 2013, 5, 4015–4039 RSC.
- Z. M. Markovic, B. Z. Ristic, K. M. Arsikin, D. G. Klisic, L. M. Harhaji-Trajkovic, B. M. Todorovic-Markovic, D. P. Kepic, T. K. Kravic-Stevovic, S. P. Jovanovic, M. M. Milenkovic, D. D. Milivojevic, V. Z. Bumbasirevic, M. D. Dramicanin and V. S. Trajkovic, Biomaterials, 2012, 33, 7084–7092 CrossRef CAS PubMed.
- J. Ge, M. Lan, B. Zhou, W. Liu, L. Guo, H. Wang, Q. Jia, G. Niu, X. Huang, H. Zhou, X. Meng, P. Wang, C. S. Lee, W. Zhang and X. Han, Nat. Commun., 2014, 5, 4596 CAS.
- X. Zhou, Y. Zhang, C. Wang, X. Wu, Y. Yang, B. Zheng, H. Wu, S. Guo and J. Zhang, ACS Nano, 2012, 6, 6592–6599 CrossRef CAS PubMed.
- Y. Mao, Y. Bao, L. Yan, G. Li, F. Li, D. Han, X. Zhang and L. Niu, RSC Adv., 2013, 3, 5475 RSC.
- P. G. Luo, F. Yang, S.-T. Yang, S. K. Sonkar, L. Yang, J. J. Broglie, Y. Liu and Y.-P. Sun, RSC Adv., 2014, 4, 10791 RSC.
- J. Zhang and S.-H. Yu, Mater. Today, 2016, 19, 382–393 CrossRef CAS.
- C. Wu, C. Wang, T. Han, X. Zhou, S. Guo and J. Zhang, Adv. Healthcare Mater., 2013, 2, 1613–1619 CrossRef CAS PubMed.
- M. Nurunnabi, Z. Khatun, K. M. Huh, S. Y. Park, D. Y. Lee, K. J. Cho and Y. K. Lee, ACS Nano, 2013, 7, 6858–6867 CrossRef CAS PubMed.
- J. Shen, Y. Zhu, X. Yang and C. Li, Chem. Commun., 2012, 48, 3686–3699 RSC.
- A. Zhu, C. Ding and Y. Tian, Sci. Rep., 2013, 3, 2933 CrossRef PubMed.
- Q. Liu, B. Guo, Z. Rao, B. Zhang and J. R. Gong, Nano Lett., 2013, 13, 2436–2441 CrossRef CAS PubMed.
- M. Zheng, S. Liu, J. Li, D. Qu, H. Zhao, X. Guan, X. Hu, Z. Xie, X. Jing and Z. Sun, Adv. Mater., 2014, 26, 3554–3560 CrossRef CAS PubMed.
- X. T. Zheng, H. L. He and C. M. Li, RSC Adv., 2013, 3, 24853 RSC.
- X. Wang, X. Sun, J. Lao, H. He, T. Cheng, M. Wang, S. Wang and F. Huang, Colloids Surf., B, 2014, 122, 638–644 CrossRef CAS PubMed.
- P. Nigam, S. Waghmode, M. Louis, S. Wangnoo, P. Chavan and D. Sarkar, J. Mater. Chem. B, 2014, 2, 3190 RSC.
- M. Grossi, M. Morgunova, S. Cheung, D. Scholz, E. Conroy, M. Terrile, A. Panarella, J. C. Simpson, W. M. Gallagher and D. F. O'Shea, Nat. Commun., 2016, 7, 10855 CrossRef CAS PubMed.
- S. Zhu, J. Shao, Y. Song, X. Zhao, J. Du, L. Wang, H. Wang, K. Zhang, J. Zhang and B. Yang, Nanoscale, 2015, 7, 7927–7933 RSC.
- S. Zhu, N. Zhou, Z. Hao, S. Maharjan, X. Zhao, Y. Song, B. Sun, K. Zhang, J. Zhang, H. Sun, L. Lu and B. Yang, RSC Adv., 2015, 5, 39399–39403 RSC.
- L. Wang, S. J. Zhu, H. Y. Wang, S. N. Qu, Y. L. Zhang, J. H. Zhang, Q. D. Chen, H. L. Xu, W. Han, B. Yang and H. B. Sun, ACS Nano, 2014, 8, 2541–2547 CrossRef CAS PubMed.
- S. Zhu, Y. Song, J. Wang, H. Wan, Y. Zhang, Y. Ning and B. Yang, Nano Today, 2017, 13, 10–14 CrossRef CAS.
- M. Kano, A. E. Sosulski, L. Zhang, H. D. Saatcioglu, D. Wang, N. Nagykery, M. E. Sabatini, G. Gao, P. K. Donahoe and D. Pepin, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E1688–E1697 CrossRef CAS PubMed.
- N. Zhou, Z. Hao, X. Zhao, S. Maharjan, S. Zhu, Y. Song, B. Yang and L. Lu, Nanoscale, 2015, 7, 15635–15642 RSC.
- B. Sun, B. Zhao, D. Wang, Y. Wang, Q. Tang, S. Zhu, B. Yang and H. Sun, Nanoscale, 2016, 8, 9837–9841 RSC.
- E. L. Rosenthal, J. M. Warram, E. de Boer, T. K. Chung, M. L. Korb, M. Brandwein-Gensler, T. V. Strong, C. E. Schmalbach, A. B. Morlandt, G. Agarwal, Y. E. Hartman, W. R. Carroll, J. S. Richman, L. K. Clemons, L. M. Nabell and K. R. Zinn, Clin. Cancer Res., 2015, 21, 3658–3666 CrossRef CAS PubMed.
- S. Zhu, Y. Li, J. Zhang, C. Lu, X. Dai, F. Jia, H. Gao and B. Yang, J. Colloid Interface Sci., 2010, 344, 541–546 CrossRef CAS PubMed.
- H. S. Choi, S. L. Gibbs, J. H. Lee, S. H. Kim, Y. Ashitate, F. Liu, H. Hyun, G. Park, Y. Xie, S. Bae, M. Henary and J. V. Frangioni, Nat. Biotechnol., 2013, 31, 148–153 CrossRef CAS PubMed.
- C. Ding, A. Zhu and Y. Tian, Acc. Chem. Res., 2014, 47, 20–30 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Characterization of GQDs, more cell imaging and in vivo data. See DOI: 10.1039/c7ra11963a |
| This journal is © The Royal Society of Chemistry 2017 |