Open Access Article
Open Access ArticleActivation and characterization of a cryptic gene cluster reveals a cyclization cascade for polycyclic tetramate macrolactams†
Subhasish
Saha‡
a,
Wenjun
Zhang‡
a,
Guangtao
Zhang‡
a,
Yiguang
Zhu
a,
Yuchan
Chen
b,
Wei
Liu
ac,
Chengshan
Yuan
a,
Qingbo
Zhang
a,
Haibo
Zhang
a,
Liping
Zhang
a,
Weimin
Zhang
b and
Changsheng
Zhang
 ac
ac
aCAS Key Laboratory of Tropical Marine Bio-resources and Ecology, Guangdong Key Laboratory of Marine Materia Medica, South China Sea Institute of Oceanology, Chinese Academy of Sciences, 164 West Xingang Road, Guangzhou 510301, China. E-mail: czhang2006@gmail.com; czhang@scsio.ac.cn
bState Key Laboratory of Applied Microbiology Southern China, Guangdong Institute of Microbiology, 100 Central Xianlie Road, Guangzhou 510070, China
cState Key Laboratory of Applied Microbiology Southern China, South China Sea Resource Exploitation and Protection Collaborative Innovation Center (SCS-REPIC), China
First published on 28th October 2016
Abstract
Polycyclic tetramate macrolactams (PTMs) are a growing class of natural products and are derived from a hybrid polyketide synthase (PKS)/non-ribosomal peptide synthetase (NRPS) pathway. PTM biosynthetic gene clusters are conserved and widely distributed in bacteria, however, most of them remain silent. Herein we report the activation of a PTM gene cluster in marine-derived Streptomyces pactum SCSIO 02999 by promoter engineering and heterologous expression, leading to the discovery of six new PTMs, pactamides A–F (11–16), with potent cytotoxic activity upon several human cancer cell lines. In vivo gene disruption experiments and in vitro biochemical assays reveal a reductive cyclization cascade for polycycle formation, with reactions sequentially generating the 5, 5/5 and 5/5/6 carbocyclic ring systems, catalysed by the phytoene dehydrogenase PtmB2, the oxidoreductase PtmB1, and the alcohol dehydrogenase PtmC, respectively. Furthermore, PtmC was demonstrated as a bifunctional cyclase for catalyzing the formation of the inner five-membered ring in ikarugamycin. This study suggests the possibility of finding more bioactive PTMs by genome mining and discloses a general mechanism for the formation of 5/5/6-type carbocyclic rings in PTMs.
Introduction
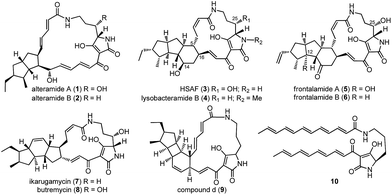
Polycyclic tetramate macrolactams (PTMs) feature a tetramate-embedding macrocyclic lactam ring that is fused to a subset of diverse carbocyclic ring systems (Fig. 1),1 such as the 5/5 ring system in alteramides (1, 2),2 5/5/6 ring system in HSAF (heat-stable antifungal factor, 3), lysobacteramide B (4),3,4 and frontalamides (5, 6),5 5/6/5 ring system in ikarugamycin (7)6 and butremycin (8),7 and 5/4/6 ring system in 9.8 PTMs exhibit a wide range of antifungal, antibacterial, antiprotozoal, antiulcer, antiviral, cytotoxic, and apoptosis-inducing biological activities.1 As such, PTMs have shown significant potential in both agricultural and medicinal applications.1The structural complexity of PTMs has greatly hampered efforts towards their total synthesis.9–12 In sharp contrast, nature has developed a conserved and compact biosynthetic pathway to construct the complex structures of PTMs.1,13 PTMs were revealed to have originated from a conserved hybrid polyketide synthase (PKS)/non-ribosomal peptide synthetase (NRPS) pathway.5 The PKS was iteratively used to generate two separate polyketide chains that were respectively condensed to the α- and δ-amino groups of an L-ornithine tethered in the NRPS, leading to a common tetramate-containing polyene precursor 10 (Fig. 1).8,14–17 The extraordinary simplicity of PTM biosynthesis was exemplified by a three-gene cassette (ikaABC) mediated production of ikarugamycin (7) in a heterologous host.15,16 However, the enzymatic machinery that dictates the manner of the divergent cyclization cascades that convert 10 into diverse ring systems in PTMs remains mysterious.1 The only well studied example was the formation of the inner five-membered ring in 7, which is catalyzed by the alcohol dehydrogenase IkaC in the manner of a Michael addition reaction.15
A genomic survey has revealed the wide distribution of conserved PTM biosynthetic gene clusters in phylogenetically diverse bacteria.5 However, most of them are silent.1,5 Recently, different strategies were employed to successfully activate silent PTM gene clusters for the production of new PTMs, including promoter engineering18 and the heterologous expression of a reconstituted PTM gene cluster via synthetic biology.8 The success of these strategies highlights the potential for the discovery of new PTMs by the genome mining-based activation of PTM gene clusters. Herein we report (i) the activation of a cryptic PTM biosynthetic gene cluster in the marine-derived Streptomyces pactum SCSIO 02999 via promoter engineering; (ii) the isolation, characterization and cytotoxicity determination of six new PTMs, the pactamides A–F; (iii) the discovery of a reductive cyclization cascade to sequentially generate the 5, 5/5 and 5/5/6 ring systems in pactamides by the inactivation of three oxidoreductase-encoding genes (ptmB2, ptmB1 and ptmC) and analysis of the corresponding intermediates; (iv) the biochemical demonstration of the alcohol dehydrogenase PtmC as a bifunctional cyclase capable of mediating the formation of both six- and five-membered inner rings in PTMs.
Results and discussion
Identification of a putative PTM gene cluster by genome mining
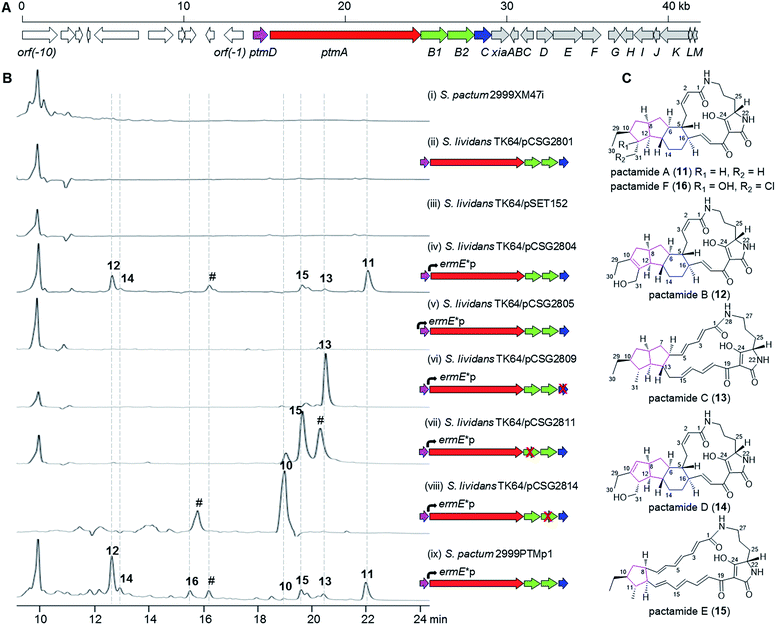
Deep-sea derived S. pactum SCSIO 02999 has been shown to produce xiamycin-related indolosesquiterpenes as major products, and the xiamycin biosynthetic gene cluster (xia) was identified by genome sequencing.19–22 Further genomic analysis reveals the presence of a 14.8 kb biosynthetic gene cluster (GenBank accession number KU569222) composed of five genes (ptmD, ptmA, ptmB1, ptmB2, and ptmC) that are highly homologous to the PTM biosynthetic genes, immediately neighbouring the xia gene cluster (Fig. 2A, Table S1†). The PtmD protein has over 50% identity with those hydroxylases (previously annotated as sterol desaturases3) which were established as introducing the hydroxy group at the ornithine moiety of PTMs.5,23,24 PtmA is predicted to be a typical hybrid PKS/NRPS protein, and shows a high similarity to PTM PKS/NRPS proteins such as IkaA,15,16 FtdB,5 and HSAF PKS/NRPS.3 PtmB1 is highly homologous to FAD-dependent oxidoreductases in other PTM pathways, while PtmB2 is highly similar to a number of phytoene dehydrogenases putatively involved in PTM biosynthesis (Table S1†).1 PtmC bears a striking resemblance to alcohol dehydrogenases involved in PTM biosynthesis, such as FtdE,5 OX4,14 and IkaC.15Activation and characterization of the cryptic PTM gene cluster
The presence of a conserved PTM biosynthetic gene cluster in S. pactum SCSIO 02999 suggests its potential for PTM production. However, no PTM-like compounds were detected either in the wild type S. pactum SCSIO 02999 (data not shown), or in the mutant 2999XM47i (Table S2†) where the xiaP gene (encoding an essential polyprenyl synthetase for xiamycin biosynthesis19) was in-frame deleted to abolish the production of xiamycin-type indolosesquiterpenes and hence facilitate the detection of secondary metabolites other than xiamycins (Fig. 2B, trace i). The cosmid pCSG2404 (Table S2†), obtained in our previous work in screening the xiamycin gene cluster,19 was found to contain the intact PTM gene cluster. However, introduction of the cosmid pCSG2801 (Table S2†), a derivative of pCSG2404 in which the neo gene (encoding kanamycin resistance) was replaced by a gene cassette containing aac(3)IV (encoding apramycin resistance), oriT (for conjugation), and IntϕC31 (for specific integration into Streptomyces species) from pSET152A′B25 (Table S2†) into Streptomyces lividans TK64, resulted in no observed production of PTM-like compounds (Fig. 2B, trace ii), similar to the control strain S. lividans TK64/pSET152 (Fig. 2B, trace iii).In attempts to activate the PTM gene cluster, the constitutive promoter ermE*p from pPWW50,26 was inserted independently in front of ptmA and ptmD for heterologous expression in S. lividans TK64 (Tables S2 and S3; Fig. S1 and S2†). Interestingly, a number of products with UV spectra similar to PTMs, such as 11–15 (Fig. 2B, trace iv), were detected when the ermE*p promoter was inserted in front of ptmA. Surprisingly, no PTMs were observed upon inserting the ermE*p promoter in front of ptmD (Fig. 2B, trace v). For in-frame gene deletion experiments, we subsequently constructed three cosmids, pCSG2809 (ΔptmC), pCSG2811 (ΔptmB1), and pCSG2814 (ΔptmB2), in which the ermE*p promoter was inserted in front of ptmA (Fig. S3–S5†). Compound 13 was found to be the major product upon inactivating ptmC (Fig. 2B, trace vi), whereas 15 became the major product when ptmB1 was inactivated (Fig. 2B, trace vii). When ptmB2 was in-frame deleted, compound 10 was the major product (Fig. 2B, trace viii). The identity of compound 10 (Fig. 1) was deduced from its UV spectrum and mass (m/z 475.4 [M + H]+, 473.6 [M − H]−) which were almost identical to those of 10 from the ΔikaB mutant in the biosynthesis of 7 (Fig. S6†).15
Finally, an attempt was made to in situ activate the ptm-gene cluster in S. pactum 2999XM47i to afford a corresponding mutant 2999PTMp1 (Table S2†), in which the ermE*p promoter was inserted in front of ptmA. The fermentation of 2999PTMp1 also led to the production of a number of PTMs, including 10–16 (Fig. 2B, trace ix).
Isolation, structure elucidation and cytotoxicity determination of new PTMs
Pactamides A–F (11–16, Fig. 2C) were isolated from the fermentation extracts of S. pactum 2999PTMp1, S. lividans TK64/pCSG2809, or S. lividans TK64/pCSG2811 (ESI†), and their structures were elucidated by extensive interpretation of the spectroscopic/spectrometric data (Tables S4–S7, Fig. S7–S13†) including high resolution electrospray ionization mass spectrometry (HRESIMS), 1D and 2D NMR, and electronic circular dichroism spectroscopy (ECD). The molecular formula of pactamide A (11) was established as C29H40N2O4 by HRESIMS. The planar structure of 11 was elucidated by comparing the NMR spectroscopic data of 11 (Tables S4 and S5, Fig. S7†) and 4 (Fig. 1).4 The relative configuration of 11 was deduced by the proton coupling constants (ZΔ2,3J2,3 11.5 Hz; EΔ17,18J17,18 15.0 Hz, Table S4†) and NOESY correlations (Fig. S7†). The assignment of H-5/H-11/H-13 on the same side of the 5/5/6-tricyclic ring system in 11, and H-6/H-8/H-10/H-12/H-16/H3-31 on another side, was supported by the observed NOESY correlations (Fig. S7†). On the basis of the biosynthetic origin of the L-ornithine of PTMs,1,4 the S-configuration was assignable to C-23. Given that 11 and 4 exhibited almost identical ECD spectra (Fig. S8†),4 the absolute configuration of 11 was deduced as 5R, 6S, 8S, 10S, 11R, 12R, 13S, 16R and 23S. Similarly, the structures of pactamides B–F (12–16) were determined (see the ESI† for the structure elucidation). The trans-orientation of H-10/H-11 in 11, 13 and 15 is similar to that in aburatubolactam A.27 In contrast, a cis-orientation of H-10/H-11 was found in other PTMs with determined relative configurations (Fig. 1 and S14†).1 Pactamide D (14) contains a Δ9,10 double bond, similar to that in the marine sponge metabolites cylindramide28 and geodin A29 (Fig. S14†), whereas 12 contains a unique Δ10,11 double bond. Pactamide F (16) represents the first example of a halogenated PTM. Pactamides A–F (11–16) exhibited in vitro antiproliferative activity against four cancer cell lines (SF-268, MCF-7, NCI-H460 and Hep-G2) with IC50 values ranging from 0.24–26 μM (Table 1). Pactamide A (11) displayed the best cytotoxic activity with IC50 values of 0.24–0.51 μM, and these were better than those of lysobacteramides and HSAF.4 Notably, the presence of a double bond in 12 and 14 greatly decreased their cytotoxicities.| IC50 (μM) | ||||
|---|---|---|---|---|
| SF-268 | MCF-7 | NCI-H460 | Hep-G2 | |
| a Cisplatin, positive control. | ||||
| 11 | 0.51 ± 0.01 | 0.26 ± 0.02 | 0.24 ± 0.02 | 0.37 ± 0.03 |
| 12 | 25.47 ± 1.09 | 24.45 ± 0.53 | 21.93 ± 0.58 | 26.15 ± 1.37 |
| 13 | 2.42 ± 0.15 | 0.71 ± 0.02 | 0.74 ± 0.02 | 1.71 ± 1.71 |
| 14 | 19.26 ± 0.69 | 14.50 ± 1.56 | 17.41 ± 0.92 | 17.23 ± 1.18 |
| 15 | 8.70 ± 0.25 | 5.10 ± 0.43 | 5.19 ± 0.11 | 6.88 ± 0.10 |
| 16 | 2.65 ± 0.29 | 2.66 ± 0.04 | 2.85 ± 0.22 | 2.66 ± 0.07 |
| CP | 3.99 ± 0.49 | 9.23 ± 0.41 | 1.53 ± 0.12 | 1.39 ± 0.18 |
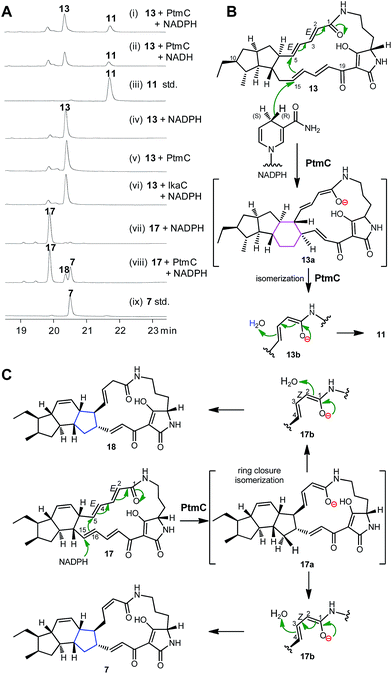
Biochemical characterization of PtmC
The alcohol dehydrogenase IkaC has been previously shown to catalyse the formation of the inner five-membered ring in ikarugamycin (7, Fig. 1). Given the high similarity of PtmC and IkaC, we expected that PtmC should have a similar function to IkaC. To probe its function in vitro, PtmC was overproduced in E. coli BL21(DE3) as a soluble N-His6-tagged protein (Fig. S15†). In vitro assays showed that 13 was converted to 11 by PtmC in the presence of NADPH or NADH (Fig. 3A, traces i–iii), while no conversion of 13 to 11 was observed in the absence of PtmC or NAD(P)H (Fig. 3A, traces iv & v), or upon replacing PtmC with IkaC (Fig. 3A, trace vi). These data unequivocally established that PtmC was able to catalyse the conversion of 13 to 11.We have previously established that the inner five-membered ring in ikarugamycin (7, Fig. 1) was formed through an IkaC-catalyzed 1,6-Michael addition reaction,15 by deuterium incorporation experiments in coupling assays with glucose dehydrogenases (GDH), providing either (S)-[4-2H]NADPH by BmGDH from Bacillus megaterium DSM 2894,30 or (R)-[4-2H]NADPH by TaGDH from Thermoplasma acidophilum ATCC 25905.31
The same strategy was employed to probe the reaction mechanism of PtmC. The LC-HRESIMS analyses of the reaction product (11) in the PtmC/GDH coupling assays, in buffers either prepared with H2O or 2H2O (Fig. S16†), revealed the incorporation of two protons, one from (R)-[4-H]NADPH and another from H2O, into the product 11. These observations revealed a similar 1,6-Michael addition reaction for the PtmC-catalysed formation of the six-membered ring in 11 from 13 (Fig. 3B). Similarly to IkaC catalysis, an isomerization from species 13a to 13b (Fig. 3B) was proposed to change ZΔ2,3 in the substrate 13 to EΔ2,3 in the product 11 during PtmC catalysis (Fig. 3B). Interestingly, PtmC could catalyse the conversion of 17 (the native substrate of IkaC15) into two products (Fig. 3A, traces vii & viii), one of which was identical to 7 by comparison with authentic 7 (Fig. 3A, trace ix). Another product 18 (1.7 mg) was isolated from a preparative PtmC enzyme reaction and was structurally elucidated as isoikarugamycin by comparing its 1H and 13C NMR data (Table S8, Fig. S17†) with the reported data.32 In contrast, IkaC was unable to convert 13 to 11 (Fig. 3B, trace vi), although IkaC has 65% identity with and 76% similarity to PtmC. The formation of 7 and 18 by PtmC catalysis was also consistent with a 1,6-Michael addition reaction (Fig. 3C).32 These data revealed that PtmC was a bifunctional enzyme capable of catalysing the formation of both six- and five-membered rings via a common Michael addition mechanism.
Implications of a reductive cyclization cascade in constructing a 5/5/6 ring system in PTMs
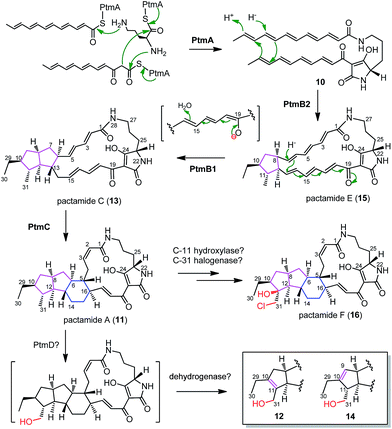
A substantial understanding of PTM biosynthesis has been gained for HSAF (3),3,14,17,23,33 frontalamides (5, 6),5 and ikarugamycin (7).1,15,16 The simplicity of PTM biosynthesis lies mainly in the beauty of synthesizing the polyene tetramate precursor 10 by a single PTM PKS/NRPS enzyme due to the functional iteration of PTM PKSs (Fig. 4).1 This study provides further support with 10 (generated by PtmA alone, Fig. 4) as a common biosynthetic intermediate in the PTM pathway. The in-frame deletion of ptmB2, ptmB1, and ptmC led to the accumulation of 10, 15, and 13, respectively, providing convincing evidence for a cascade of three reductive cyclization reactions to form the 5/5/6 ring system in 11 (Fig. 4). The phytoene dehydrogenase PtmB2 catalyses the first cyclization reaction, converting 10 to 15, in a not yet understood manner. Next, the oxidoreductase PtmB1 performs the second reductive cyclization to convert 15 to 13, probably undergoing a 1,8-Michael addition like reaction. Finally, the third reductive cyclization, mediated by the dehydrogenase PtmC, transforms 13 into 11 to furnish the 5/5/6 ring system (Fig. 4).In accordance with these hypotheses, feeding 15 into S. lividans TK64/pCSG2814 (where ptmB2 is in-frame deleted and ptmB1 and ptmC are intact) clearly led to the detection of compounds 13 and 11 (Fig. S18†), confirming that 15 is a biosynthetic intermediate en route to 11. PtmD displays high similarity to the hydroxylases involved in the hydroxylation at C-25 of the ornithine moiety (Fig. S19†),5,8,23,24 however, the absence of a C-25 hydroxy group in 11–16 suggests that PtmD might function as a C-31 hydroxylase in the biosynthesis of 12 and 14 (Fig. 4). The enzymes to generate the double bonds in 12 and 14 are still unknown, but should be encoded by genes shown in Fig. 2A, since 12 and 14 were also produced in S. lividans/pCSG2804 (Fig. 2B, trace v). Pactamide F (16) was only detected in S. pactum 2999PTMp1, but not in S. lividans TK64/pCSG2804 (Fig. 2B), indicating that the required tailoring enzymes, a halogenase and a C-11 hydroxylase, might be encoded by genes outside of the ptm-gene cluster.
Conclusions
Six new PTMs, pactamides A–F (11–16), have been discovered by the successful activation of a cryptic PTM gene cluster via promoter engineering in marine-derived S. pactum SCSIO 02999. The PTM gene clusters in S. pactum SCSIO 02999, S. albus J1074 (activated by promoter engineering18) and S. griseus IFO 13350 (activated by synthetic biology8) share the same genetic organization and have high sequence similarity in their encoded proteins, however, the activation of each cluster led to structurally distinct PTMs (Fig. S20†). This indicates a great potential to discover more new bioactive PTMs by activating the hundreds of conserved and silent PTM gene clusters in phylogenetically diverse bacteria.1,5 Furthermore, the discovery of the major products of the ΔptmB2 mutant (10), the ΔptmB1 mutant (15), and the ΔptmC mutant (13), provided solid evidence for three consecutive reductive cyclizations, in a stepwise manner, to generate the 5 (in 15 by PtmB2), 5/5 (in 13 by PtmB1) and 5/5/6 (in 11 by PtmC) carbocyclic ring systems in pactamides. This discovery highlights a general strategy for the formation of a subset of 5/5/6 ring systems in other PTMs, such as in 3 and 5. In contrast to the single enzyme IkaB-catalysed formation of the 5/6 ring system in 17,15 two oxidoreductases (PtmB2/B1) are required to generate the 5/5 ring system in 13. This discrepancy may serve as a general predictor for divergence dictating the 5/5/6 or 5/6/5 ring systems in PTM biosynthesis, and serve to guide the genome mining approach for predicting the anticipated PTM structures. Finally, PtmC was demonstrated herein as a bifunctional cyclase to mediate the formation of both six- and five-membered rings, via a common Michael addition reaction mechanism .Acknowledgements
This work is supported in part by the National Natural Science Foundation of China (31630004, 31290233, 31125001), the Chinese Academy of Sciences (XDA11030403, QYZDJ-SSW-DQC004), and the Administration of Ocean and Fisheries of Guangdong Province (A201601C03, GD2012-D01-002). Q. Z. is a “Pearl New Star” scholar and is supported by the Science and Technology Program of Guangzhou. We thank Prof. Hanjie Ying in Nanjing University of Technology for providing the plasmid pRSF-BmGDH and Prof. Lichuan Gu in Shandong University for providing genomic DNA of the strain Thermoplasma acidophilum ATCC 25905. We are grateful for the use of the analytical facilities at the SCSIO.Notes and references
- G. Zhang, W. Zhang, S. Saha and C. Zhang, Curr. Top. Med. Chem., 2016, 16, 1727–1739 CrossRef CAS PubMed.
- H. Shigemori, M. A. Bae, K. Yazawa, T. Sasaki and J. Kobayashi, J. Org. Chem., 1992, 57, 4317–4320 CrossRef CAS.
- F. Yu, K. Zaleta-Rivera, X. Zhu, J. Huffman, J. C. Millet, S. D. Harris, G. Yuen, X.-C. Li and L. Du, Antimicrob. Agents Chemother., 2007, 51, 64–72 CrossRef CAS PubMed.
- L. Xu, P. Wu, S. J. Wright, L. Du and X. Wei, J. Nat. Prod., 2015, 78, 1841–1847 CrossRef CAS PubMed.
- J. A. V. Blodgett, D. C. Oh, S. G. Cao, C. R. Currie, R. Kolter and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 11692–11697 CrossRef CAS PubMed.
- K. Jomon, M. Ajisaka, H. Sakai and Y. Kuroda, J. Antibiot., 1972, 25, 271–280 CrossRef CAS PubMed.
- K. Kyeremeh, K. S. Acquah, A. Sazak, W. Houssen, J. Tabudravu, H. Deng and M. Jaspars, Mar. Drugs, 2014, 12, 999–1012 CrossRef PubMed.
- Y. Luo, H. Huang, J. Liang, M. Wang, L. Lu, Z. Shao, R. E. Cobb and H. Zhao, Nat. Commun., 2013, 4 DOI:10.1038/ncomms3894.
- R. K. Boeckman, C. H. Weidner, R. B. Perni and J. J. Napier, J. Am. Chem. Soc., 1989, 111, 8036–8037 CrossRef CAS.
- L. A. Paquette, D. Macdonald, L. G. Anderson and J. Wright, J. Am. Chem. Soc., 1989, 111, 8037–8039 CrossRef CAS.
- N. Cramer, S. Laschat, A. Baro, H. Schwalbe and C. Richter, Angew. Chem., Int. Ed., 2005, 44, 820–822 CrossRef CAS PubMed.
- J. A. Henderson and A. J. Phillips, Angew. Chem., Int. Ed., 2008, 47, 8499–8501 CrossRef CAS PubMed.
- Y. X. Xie, S. Wright, Y. M. Shen and L. C. Du, Nat. Prod. Rep., 2012, 29, 1277–1287 RSC.
- L. L. Lou, G. L. Qian, Y. X. Xie, J. L. Hang, H. T. Chen, K. Zaleta-Riyera, Y. Y. Li, Y. M. Shen, P. H. Dussault, F. Q. Liu and L. C. Du, J. Am. Chem. Soc., 2011, 133, 643–645 CrossRef CAS PubMed.
- G. T. Zhang, W. J. Zhang, Q. B. Zhang, T. Shi, L. Ma, Y. G. Zhu, S. M. Li, H. B. Zhang, Y. L. Zhao, R. Shi and C. S. Zhang, Angew. Chem., Int. Ed., 2014, 53, 4840–4844 CrossRef CAS PubMed.
- J. Antosch, F. Schaefers and T. A. M. Gulder, Angew. Chem., Int. Ed., 2014, 53, 3011–3014 CrossRef CAS PubMed.
- Y. Y. Li, H. T. Chen, Y. J. Ding, Y. X. Xie, H. X. Wang, R. L. Cerny, Y. M. Shen and L. C. Du, Angew. Chem., Int. Ed., 2014, 53, 7524–7530 CrossRef CAS PubMed.
- C. Olano, I. Garcia, A. Gonzalez, M. Rodriguez, D. Rozas, J. Rubio, M. Sanchez-Hidalgo, A. F. Brana, C. Mendez and J. A. Salas, Microb. Biotechnol., 2014, 7, 242–256 CrossRef CAS PubMed.
- H. Li, Q. Zhang, S. Li, Y. Zhu, G. Zhang, H. Zhang, X. Tian, S. Zhang, J. Ju and C. Zhang, J. Am. Chem. Soc., 2012, 134, 8996–9005 CrossRef CAS PubMed.
- Q. Zhang, A. Mándi, S. Li, Y. Chen, W. Zhang, X. Tian, H. Zhang, H. Li, W. Zhang, S. Zhang, J. Ju, T. Kurtán and C. Zhang, Eur. J. Org. Chem., 2012, 5256–5262 CrossRef CAS.
- Q. Zhang, H. Li, S. Li, Y. Zhu, G. Zhang, H. Zhang, W. Zhang, R. Shi and C. Zhang, Org. Lett., 2012, 14, 6142–6145 CrossRef CAS PubMed.
- H. Li, Y. Sun, Q. Zhang, Y. Zhu, S. M. Li, A. Li and C. Zhang, Org. Lett., 2015, 17, 306–309 CrossRef CAS PubMed.
- Y. Y. Li, J. Huffman, Y. Li, L. C. Du and Y. M. Shen, Med. Chem. Commun., 2012, 3, 982–986 RSC.
- C. Greunke, J. Antosch and T. A. Gulder, Chem. Commun., 2015, 51, 5334–5336 RSC.
- Y. Zhang, H. Huang, Q. Chen, M. Luo, A. Sun, Y. Song, J. Ma and J. Ju, Org. Lett., 2013, 15, 3254–3257 CrossRef CAS PubMed.
- M. Doumith, P. Weingarten, U. F. Wehmeier, K. Salah-Bey, B. Benhamou, C. Capdevila, J. M. Michel, W. Piepersberg and M. C. Raynal, Mol. Gen. Genet., 2000, 264, 477–485 CrossRef CAS PubMed.
- M. A. Bae, K. Yamada, Y. Ijuin, T. Tsuji, K. Yazawa, Y. Tomono and D. Uemura, Heterocycl. Commun., 1996, 2, 315–318 CrossRef CAS.
- S. Kanazawa, N. Fusetani and S. Matsunaga, Tetrahedron Lett., 1993, 34, 1065–1068 CrossRef CAS.
- R. J. Capon, C. Skene, E. Lacey, J. H. Gill, D. Wadsworth and T. Friedel, J. Nat. Prod., 1999, 62, 1256–1259 CrossRef CAS.
- Q. Ye, H. Cao, M. Yan, F. Cao, Y. Zhang, X. Li, L. Xu, Y. Chen, J. Xiong, P. Ouyang and H. Ying, Bioresour. Technol., 2010, 101, 6761–6767 CrossRef CAS PubMed.
- J. Su, Q. Wang, J. Feng, C. Zhang, D. Zhu, T. Wei, W. Xu and L. Gu, Bioorg. Med. Chem., 2011, 19, 2991–2996 CrossRef CAS PubMed.
- R. Lacret, D. Oves-Costales, C. Gomez, C. Diaz, M. de la Cruz, I. Perez-Victoria, F. Vicente, O. Genilloud and F. Reyes, Mar. Drugs, 2014, 13, 128–140 CrossRef PubMed.
- L. Lou, H. Chen, R. L. Cerny, Y. Li, Y. Shen and L. Du, Biochemistry, 2012, 51, 4–6 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The experimental procedures, materials, and characterization of the compounds. See DOI: 10.1039/c6sc03875a |
| ‡ S. Saha, W. Zhang and G. Zhang contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |