Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Complementary oligonucleotides regulate induced fit ligand binding in duplexed aptamers†
Jeffrey D.
Munzar
 ab,
Andy
Ng
ab,
Andy
Ng
 ab,
Mario
Corrado
ab and
David
Juncker
ab,
Mario
Corrado
ab and
David
Juncker
 *abc
*abc
aMcGill University and Genome Quebec Innovation Centre, 740 Dr. Penfield Avenue, Montreal, Quebec H3A 0G1, Canada. E-mail: david.juncker@mcgill.ca
bDepartment of Biomedical Engineering, McGill University, 3775 Rue University, Montreal, Quebec H3A 2B4, Canada
cDepartment of Neurology and Neurosurgery, McGill University, 3801 Rue University, Montreal, Quebec H3A 2B4, Canada
First published on 8th December 2016
Abstract
Duplexed aptamers (DAs) are engineered by hybridizing an aptamer-complementary element (ACE, e.g. a DNA oligonucleotide) to an aptamer; to date, ACEs have been presumed to sequester the aptamer into a non-binding duplex state, in line with a conformational selection-based model of ligand binding. Here, we uncover that DAs can actively bind a ligand from the duplex state through an ACE-regulated induced fit mechanism. Using a widely-studied ATP DNA aptamer and a solution-based equilibrium assay, DAs were found to exhibit affinities up to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold higher than predicted by conformational selection alone, with different ACEs regulating the level of induced fit binding, as well as the cooperative allostery of the DA (Hill slope of 1.8 to 0.7). To validate these unexpected findings, we developed a non-equilibrium surface-based assay that only signals induced fit binding, and corroborated the results from the solution-based assay. Our findings indicate that ACEs regulate ATP DA ligand binding dynamics, opening new avenues for the study and design of ligand-responsive nucleic acids.
000-fold higher than predicted by conformational selection alone, with different ACEs regulating the level of induced fit binding, as well as the cooperative allostery of the DA (Hill slope of 1.8 to 0.7). To validate these unexpected findings, we developed a non-equilibrium surface-based assay that only signals induced fit binding, and corroborated the results from the solution-based assay. Our findings indicate that ACEs regulate ATP DA ligand binding dynamics, opening new avenues for the study and design of ligand-responsive nucleic acids.
Introduction
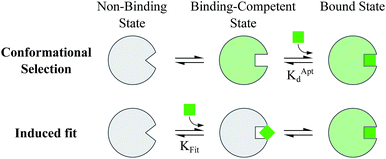
Proteins and functional nucleic acids often couple specific ligand-binding events with distinct structural changes central to biochemical regulation.1,2 Likewise, synthetic nucleic acid aptamers undergo distinct structural changes upon ligand binding, which can be leveraged for biosensing and applications in synthetic biology.3–9 The binding-related structural changes that occur in these ligand-binder systems can be described as proceeding via either conformational selection (MWC model10) or induced fit (KNF model11) (Fig. 1). In conformational selection, a binder exists at equilibrium between non-binding and binding-competent states, and is only capable of binding the ligand in the latter. Alternatively, induced fit describes a binding pathway from the non-binding to bound state actively catalyzed by the ligand.Interestingly, aptamers can be engineered with enhanced switching activity by hybridizing an aptamer-complementary element (ACE, such as a short DNA oligo) to a desired aptamer sequence, forming a duplexed aptamer (DA) that acts as a synthetic switch.12–15 The ease of engineering DAs from known aptamer sequences has led DAs to find numerous applications based on e.g. FRET,12 electrochemistry,16–22 colorimetry,23 SPR,24 fluorescence,25,26 and signaling cascades.27
However, to date, ligand binding in DAs has only been modeled based on conformational selection, in which the ACE acts as an inhibitor, sequestering the aptamer into a non-ligand-binding, passive duplex state. In this model, the observed affinity of a DA (KObsd) is a function of (i) the intrinsic affinity of the native aptamer (KAptd) and (ii) the hybridization free energy of the ACE-aptamer duplex (KHyb).28–36 In qualitative agreement with such a model (for details, see Fig. S1†), Porchetta et al. used a native cocaine DNA aptamer and varying length ACEs (10–15 bases) to engineer and tune the relative affinity of cocaine DAs over three orders of magnitude.32 To our knowledge, although concepts of 3-body side reactions and misfolded sensor states have been used to model DAs,28 induced fit ligand binding in DAs, in which a DA might actively sense and catalytically bind a ligand directly from the duplex state, has not been studied. In this regard, we note that small modifications to the length and location of ACEs have been documented to impact DA biosensors in ways that cannot be accounted for by differences in ACE-aptamer hybridization free energies.37–39
Here, we evaluate the possibility of induced fit ligand binding in DAs. We focused on DAs engineered from the Huizenga and Szostak ATP DNA aptamer introduced in 1995,40,41 as this aptamer is the most widely studied, is well characterized, and was the first to be implemented as a DA.12 The native aptamer binds ATP (6 μM KAptd) first through folding of the stem and loop regions, followed by the cooperative binding of two ATP molecules within the binding pocket (sites I and II, Fig. 2).40–43 Using DAs engineered from the ATP aptamer, we first performed equilibrium solution-based assays and uncovered (i) the existence of induced fit ligand binding in ATP DAs, and (ii) that ACEs allosterically regulate induced fit ligand binding, thereby modulating the affinity of ATP DAs in an unexpected manner. To confirm these findings, we performed a second set of experiments using a non-equilibrium surface-based assay that we developed.
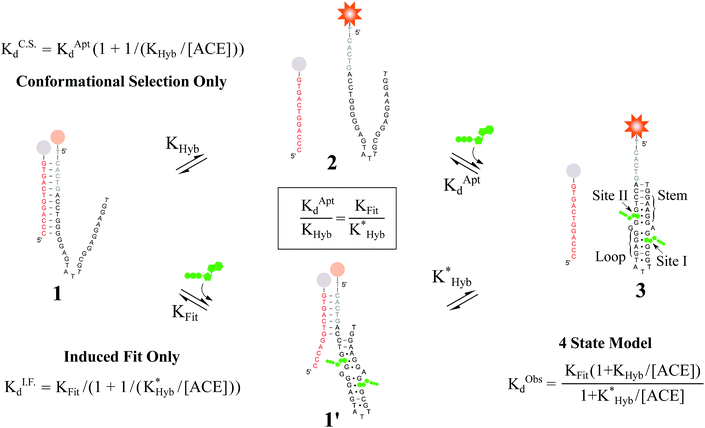 | ||
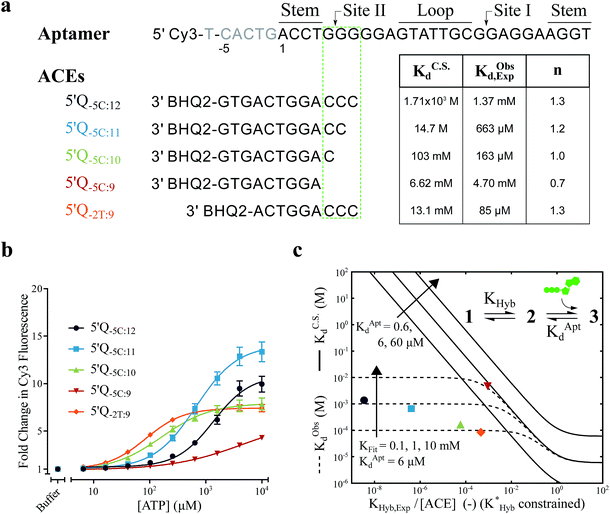
Fig. 2 Overview of ligand binding in duplexed aptamers. The 5′Q-5C:12 DA can be described by an equilibrium of four representative states and two binding pathways. The consensus aptamer sequence (black) and 5′Q-5C:12 ACE (red) are shown as an example, with canonical (dashes) and non-canonical (dots) base pairs included. The ATP binding sites and stem and loop regions of the native aptamer are labeled for state 3. The binding affinity of a DA is a function of the conformational selection pathway, governed by the duplex hybridization free energy (KHyb) and the apparent aptamer affinity (KAptd), as well as the proposed ATP-dependent induced fit binding pathway, which is governed by the induced fit binding affinity of the DA (KFit) and the hybridization free energy of the ligand-disrupted duplex 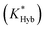 . Derivations of the analytical models for these two pathways (KC.S.d, KI.F.d) and the four-state model are provided in the ESI.† . Derivations of the analytical models for these two pathways (KC.S.d, KI.F.d) and the four-state model are provided in the ESI.† | ||
Results and discussion
For the solution-based assay, ATP DA constructs were designed starting with a 32-mer Cy3-labeled variant of the ATP aptamer, together with a previously reported 12-mer ACE, 3′-labeled with a BHQ-2 quencher.12 This ACE hybridizes to 12 bases at the 5′ end of the aptamer, beginning five bases outside the consensus aptamer sequence at the -5C nucleotide, and is termed 5′Q-5C:12. Representative states in the candidate binding pathways for the 5′Q-5C:12 DA are shown in Fig. 2. To assess any impact of ACE length and location on DAs, we engineered DAs using successively 5′-truncated 5′Q-5C:12 ACEs, as well as a three-base 3′-truncated ACE (Fig. 3a). After confirming duplex formation for all five ACEs (Fig. S2†), ATP DAs were tested using an equilibrium solution-based FRET assay29,32 (see ESI methods†), with ACE:aptamer (Q:F) ratios of 1:1 (Fig. 3b) and 3:1 (Fig. S3†).For a 1:1 Q:F ratio, the experimentally observed affinities of the 5 DAs are poorly predicted using a conformational selection-based analytical model of DAs (Fig. 3a and c). Based on experimentally measured hybridization DA free energies (obtained using FRET melting,44 see ESI methods, Table S1 and Fig. S5†), only the 5′Q-5C:9 data is consistent with a conformational selection model (Fig. 3c; KC.S.d, KAptd = 6 μM solid isoline), while the four other DAs tested are not. The analytical model underestimates 5′Q-5C:12 affinity by more than 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold, and predicts a 15-fold increase in apparent affinity for 5′Q-5C:9 over 5′Q-5C:10, whereas a 28-fold decrease in affinity was observed (Fig. 3a and c).
000-fold, and predicts a 15-fold increase in apparent affinity for 5′Q-5C:9 over 5′Q-5C:10, whereas a 28-fold decrease in affinity was observed (Fig. 3a and c).
An analytical model incorporating an induced fit binding pathway alongside conformational selection (and in which each DA has an ACE-dependent induced fit binding affinity (KFit)) can account for the unexpectedly high affinity of these DAs (Fig. 3c; see ESI derivation and Fig. S4† for additional modeling). This model suggests that induced fit binding is limited in the 5′Q-5C:9 DA (KFit > 10 mM, Fig. 3c), whereas the four other DAs exhibit KObsd,Exp in agreement with KFit values of 100–1000 μM.
Additionally, the ACEs tested gave rise to DAs with differing ATP binding cooperativities, as determined based on the Hill slope, n, suggesting that ACEs also modulate DA allostery. Here, reduced interaction of ACEs with site II promoted a shift from cooperative to anti-cooperative binding for Q:F ratios of 1:1 (n = 1.3 to 0.7) and 3:1 (n = 1.8 to 0.9) (Fig. 3a, b and S3†). Interestingly, positive cooperativity of the DA was restored using the site II-hybridizing 5′Q-2T:9 ACE (n = 1.3 and 1.7 for 1:1 and 3:1 Q:F ratios), suggesting that ACEs hybridized to site II yield DAs capable of cooperative ligand binding, as present in the native aptamer (n = 2.0 (ref. 42)).
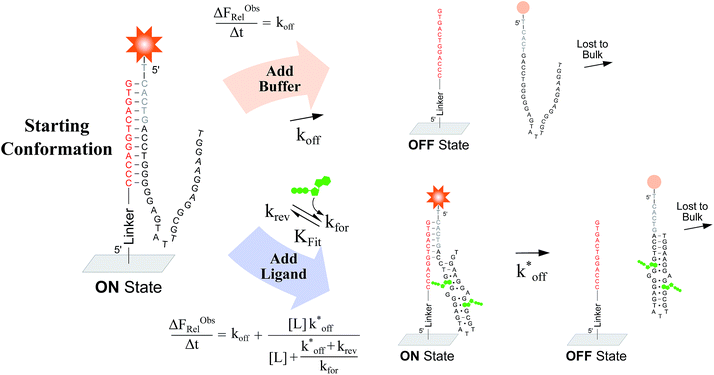
To verify the unexpected findings obtained from the solution-based assay, we developed a surface-based assay that signals only when a DA binds a ligand via induced fit, and in which conformational selection plays no role (Fig. 4). In contrast with the solution-based FRET assay, the surface-based fluorescence assay does not operate at equilibrium. Here, fluorophore-conjugated aptamers are first hybridized to ACEs covalently coupled on a slide surface, followed by washing off of non-hybridized aptamers, yielding surface-immobilized DAs (Fig. 4). After incubation with buffer (or buffer and ligand) for a specified time (Δt), DA dehybridization is measured as a loss of surface fluorescence (ΔFObsRel); owing to the low concentration of released aptamers, DA dissociation is effectively non-reversible. By assaying varying ligand concentrations, the surface-based fluorescence assay can be used to derive the induced fit affinity (KFit) of a DA.
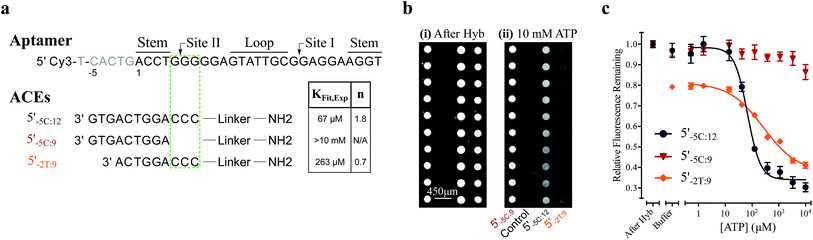
We used this surface-based assay to study ATP DAs constructed from three ACEs, corresponding to three ACEs used in the solution-based FRET assay (but missing quenchers). These ACEs varied in length and degree of site II hybridization to the ATP aptamer, termed 5′-5C:12, 5′-5C:9, and 5′-2T:9 (Fig. 5a). A 3-plex microarray was constructed with the 3 ACEs, incubated with Cy3-labeled aptamer, briefly washed, and imaged with a fluorescence scanner (ESI methods†) to establish a baseline. Next, sub-arrays on the microarray were incubated for 1 h each with ATP in buffer, or with buffer only, followed by a second fluorescence scan (Fig. 5b). Given the high concentrations of ATP assayed, the low dissociation rate of modified duplexes 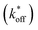 expected for the ACEs tested here (Table S1†), and assuming a steady state of intermediate duplexes on the surface, this assay can be modeled by Briggs-Haldane kinetics. By also assuming
expected for the ACEs tested here (Table S1†), and assuming a steady state of intermediate duplexes on the surface, this assay can be modeled by Briggs-Haldane kinetics. By also assuming 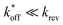 (Fig. 4), the experimental induced fit binding affinity of a DA (KFit,Exp) is equal to the Michaelis constant derived from ΔFObsRel with ATP titration (Fig. 5c).
(Fig. 4), the experimental induced fit binding affinity of a DA (KFit,Exp) is equal to the Michaelis constant derived from ΔFObsRel with ATP titration (Fig. 5c).
The surface-based fluorescence assay yielded a high induced fit binding affinity for the site-II hybridizing 5′-5C:12 ATP DA (KFit,Exp of 67 μM) (Fig. 5a and c). However, the 5′-5C:9 DA, which shares a footprint with 5′-5C:12 but with site II left unhybridized, displayed no induced fit binding (KFit,Exp > 10 mM, Fig. 5a and c), consistent with the solution-based FRET assay findings (Fig. 3a, c and S4†). Thus, despite having a much lower hybridization free energy than the 12-mer ACE, the 9-mer 5′-5C:9 ACE does not promote induced fit. Meanwhile, DAs engineered with the site-II hybridizing 5′-2T:9 ACE displayed a KFit,Exp of 263 μM (Fig. 5a and c). This value is in good agreement with an analytical model of the solution-based assay including both conformational selection and induced fit binding pathways (Fig. 3c and S4†). As a negative control ligand, we also assayed microarrays with GTP, and no induced fit binding was observed (Fig. S6†).
As observed in the solution-based FRET assay for site-II hybridizing ACEs, the 5′-5C:12 ACE also formed a DA with positive cooperativity for ATP (n = 1.8) (Fig. 5a and c). Interestingly, the 5′-2T:9 DA displayed no cooperativity (n = 0.7); given the unstable duplex expected for an ATP-disrupted 5′-2T:9 DA (Table S1†), this result may indicate that a single ATP is sufficient to displace the 5′-2T:9 ACE in the surface-based assay. Overall, the surface-based assay results corroborate the solution-based assay findings, supporting an ACE-dependent induced fit binding mechanism in ATP DAs. These findings also suggest that the ACE-based allosteric regulation of ATP DAs is relatively independent of biosensor design.
Conclusions
Taken together, our results indicate that induced fit ligand binding can be a dominating pathway in DAs, and that single nucleotide changes to ACEs can significantly modify ATP DA induced fit binding dynamics. These findings point towards ACEs as underappreciated functional regulators of the binding affinity, binding cooperativity, and allostery of DAs. This work opens new avenues for tuning aptamer-based systems, with particular relevance to the use of aptamers in biosensing and synthetic biology. However, it is not yet clear if induced-fit ligand binding in DAs is rare, with the family of ATP DAs studied here being an exception to the rule, or if induced fit is a general binding pathway common to duplexed ligand-responsive nucleic acids. In this regard, the non-equilibrium surface-based fluorescence assay introduced here could be expanded to investigate additional ACEs, to test other ACE-aptamer combinations, to investigate the effect of changes to structural regions of an aptamer on DAs (such as modifying the ligand binding pocket or tertiary structure-stabilizing bases), or to investigate other functionalities played by ACEs in DAs, such as perturbing ligand specificity.45,46Finally, this work also highlights the ACE-specific regulation of DA ligand binding as a novel model of the thermodynamic and structural determinants that govern transitions between ligand binding pathways. In this sense, we note that DAs may offer researchers a uniquely tractable and configurable nucleic acid-based alternative to existing protein-based models of allosteric regulation47–49 and collective motion in biopolymers.50–52
Acknowledgements
We thank Profs R. Sladek, M. Park and M. Tabrizian for equipment use, and Profs R. Sladek, A. Mittermaier, and members of our research group for helpful discussions. Funding was provided by the National Sciences and Engineering Research Council of Canada (NSERC) and the India-Canada Centre for Innovative Multidisciplinary Partnerships to Accelerate Community Transformation and Sustainability (IC-IMPACTS). J. D. M. acknowledges an NSERC CGS-D fellowship and D. J. acknowledges a Canada Research Chair.References
- D. D. Boehr, R. Nussinov and P. E. Wright, Nat. Chem. Biol., 2009, 5, 789–796 CrossRef CAS PubMed.
- A. Serganov and E. Nudler, Cell, 2013, 152, 17–24 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CAS.
- L. Gold, B. Polisky, O. Uhlenbeck and M. Yarus, Annu. Rev. Biochem., 1995, 64, 763–797 CrossRef CAS PubMed.
- D. J. Patel, et al. , J. Mol. Biol., 1997, 272, 645–664 CrossRef CAS PubMed.
- T. Hermann and D. J. Patel, Science, 2000, 287, 820–825 CrossRef CAS PubMed.
- J. Liu, Z. Cao and Y. Lu, Chem. Rev., 2009, 109, 1948–1998 CrossRef CAS PubMed.
- M. McKeague and M. C. DeRosa, J. Nucleic Acids, 2012, 2012, 20 Search PubMed.
- J. Monod, J. Wyman and J.-P. Changeux, J. Mol. Biol., 1965, 12, 88–118 CrossRef CAS PubMed.
- D. E. Koshland, G. Némethy and D. Filmer, Biochemistry, 1966, 5, 365–385 CrossRef CAS PubMed.
- R. Nutiu and Y. Li, J. Am. Chem. Soc., 2003, 125, 4771–4778 CrossRef CAS PubMed.
- Z. Tang, et al. , J. Am. Chem. Soc., 2008, 130, 11268–11269 CrossRef CAS PubMed.
- K. Sefah, et al. , Analyst, 2009, 134, 1765–1775 RSC.
- P. S. Lau, B. K. Coombes and Y. Li, Angew. Chem., Int. Ed., 2010, 49, 7938–7942 ( Angew. Chem. , 2010 , 122 , 8110–8114 ) CrossRef CAS PubMed.
- Y. Xiao, B. D. Piorek, K. W. Plaxco and A. J. Heeger, J. Am. Chem. Soc., 2005, 127, 17990–17991 CrossRef CAS PubMed.
- M. Zayats, Y. Huang, R. Gill, C.-a. Ma and I. Willner, J. Am. Chem. Soc., 2006, 128, 13666–13667 CrossRef CAS PubMed.
- X. Zuo, et al. , J. Am. Chem. Soc., 2007, 129, 1042–1043 CrossRef CAS PubMed.
- Z.-S. Wu, et al. , Anal. Chem., 2007, 79, 2933–2939 CrossRef CAS PubMed.
- J. Yoshizumi, S. Kumamoto, M. Nakamura and K. Yamana, Analyst, 2008, 133, 323–325 RSC.
- Y. Lu, et al. , Anal. Chem., 2008, 80, 1883–1890 CrossRef CAS PubMed.
- S. Zhang, J. Xia and X. Li, Anal. Chem., 2008, 80, 8382–8388 CrossRef CAS PubMed.
- J. Liu and Y. Lu, Angew. Chem., Int. Ed., 2006, 45, 90–94 ( Angew. Chem. , 2006 , 118 , 96–100 ) CrossRef CAS PubMed.
- J. Wang and H. S. Zhou, Anal. Chem., 2008, 80, 7174–7178 CrossRef CAS PubMed.
- D. Zheng, D. S. Seferos, D. A. Giljohann, P. C. Patel and C. A. Mirkin, Nano Lett., 2009, 9, 3258–3261 CrossRef CAS PubMed.
- A. Heilkenbrinker, et al. , Anal. Chem., 2015, 87, 677–685 CrossRef CAS PubMed.
- B. Shlyahovsky, et al. , J. Am. Chem. Soc., 2007, 129, 3814–3815 CrossRef CAS PubMed.
- I. Nakamura, A.-C. Shi, R. Nutiu, J. M. Y. Yu and Y. Li, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2009, 79, 031906 CrossRef PubMed.
- A. Vallée-Bélisle, F. Ricci and K. W. Plaxco, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13802–13807 CrossRef PubMed.
- S. G. Rouleau, R. Jodoin, M. Bisaillon and J.-P. Perreault, ACS Chem. Biol., 2012, 7, 1802–1806 CrossRef CAS PubMed.
- F. Ricci, A. Vallée-Bélisle, A. Porchetta and K. W. Plaxco, J. Am. Chem. Soc., 2012, 134, 15177–15180 CrossRef CAS PubMed.
- A. Porchetta, A. Vallée-Bélisle, K. W. Plaxco and F. Ricci, J. Am. Chem. Soc., 2012, 134, 20601–20604 CrossRef CAS PubMed.
- A. Porchetta, A. Vallée-Bélisle, K. W. Plaxco and F. Ricci, J. Am. Chem. Soc., 2013, 135, 13238–13241 CrossRef CAS PubMed.
- A. Porchetta, A. Idili, A. Vallée-Bélisle and F. Ricci, Nano Lett., 2015, 15, 4467–4471 CrossRef CAS PubMed.
- J. Pang, Z. Zhang and H. Jin, Biosens. Bioelectron., 2016, 77, 174–181 CrossRef CAS PubMed.
- Y. Du, et al. , Anal. Chem., 2016, 88, 2250–2257 CrossRef CAS PubMed.
- J. A. Cruz-Aguado and G. Penner, Anal. Chem., 2008, 80, 8853–8855 CrossRef CAS PubMed.
- G. Contreras Jiménez, et al. , Anal. Chem., 2015, 87, 1075–1082 CrossRef PubMed.
- C. Gu, T. Lan, H. Shi and Y. Lu, Anal. Chem., 2015, 87, 7676–7682 CrossRef CAS PubMed.
- D. E. Huizenga and J. W. Szostak, Biochemistry, 1995, 34, 656–665 CrossRef CAS PubMed.
- C. H. Lin and D. J. Patel, Chem. Biol., 1997, 4, 817–832 CrossRef CAS PubMed.
- R. E. Armstrong and G. F. Strouse, Bioconjugate Chem., 2014, 25, 1769–1776 CrossRef CAS PubMed.
- T. Xia, J. Yuan and X. Fang, J. Phys. Chem. B, 2013, 117, 14994–15003 CAS.
- R. A. J. Darby, et al. , Nucleic Acids Res., 2002, 30, 39e CrossRef.
- R. Kierzek, D. H. Turner and E. Kierzek, Nucleic Acids Res., 2015, 43, 1–12 CrossRef CAS PubMed.
- P. Yu, X. He, L. Zhang and L. Mao, Anal. Chem., 2015, 87, 1373–1380 CrossRef CAS PubMed.
- S.-R. Tzeng and C. G. Kalodimos, Nature, 2009, 462, 368–372 CrossRef CAS PubMed.
- G. G. Hammes, Y.-C. Chang and T. G. Oas, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13737–13741 CrossRef CAS PubMed.
- H. N. Motlagh, J. O. Wrabl, J. Li and V. J. Hilser, Nature, 2014, 508, 331–339 CrossRef CAS PubMed.
- K.-C. Chou, Biopolymers, 1987, 26, 285–295 CrossRef CAS PubMed.
- K.-C. Chou and B. Mao, Biopolymers, 1988, 27, 1795–1815 CrossRef CAS PubMed.
- K.-C. Chou, Biopolymers, 1988, 30, 3–48 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6sc03993f |
| This journal is © The Royal Society of Chemistry 2017 |