Open Access Article
Open Access ArticleCharacterizing aliphatic moieties in hydrocarbons with atomic force microscopy†
Bruno
Schuler
 *a,
Yunlong
Zhang
*a,
Yunlong
Zhang
 *b,
Sara
Collazos
c,
Shadi
Fatayer
a,
Gerhard
Meyer
a,
Dolores
Pérez
c,
Enrique
Guitián
c,
Michael R.
Harper
b,
J. Douglas
Kushnerick
b,
Diego
Peña
*b,
Sara
Collazos
c,
Shadi
Fatayer
a,
Gerhard
Meyer
a,
Dolores
Pérez
c,
Enrique
Guitián
c,
Michael R.
Harper
b,
J. Douglas
Kushnerick
b,
Diego
Peña
 *c and
Leo
Gross
*c and
Leo
Gross
 a
a
aIBM Research – Zurich, Säumerstrasse 4, 8803 Rüschlikon, Switzerland. E-mail: bschuler@lbl.gov
bExxonMobil Research and Engineering Company, Annandale, NJ 08801, USA. E-mail: yunlong.zhang@exxonmobil.com
cCentro de Investigación en Química Biolóxica e Materiais Moleculares (CIQUS), Departamento de Química Orgánica, Universidade de Santiago de Compostela, Santiago de Compostela 15782, Spain. E-mail: diego.pena@usc.es
First published on 12th December 2016
Abstract
We designed and studied hydrocarbon model compounds by high-resolution noncontact atomic force microscopy. In addition to planar polycyclic aromatic moieties, these novel model compounds feature linear alkyl and cycloaliphatic motifs that exist in most hydrocarbon resources – particularly in petroleum asphaltenes and other petroleum fractions – or in lipids in biological samples. We demonstrate successful intact deposition by sublimation of the alkyl-aromatics, and differentiate aliphatic moieties from their aromatic counterparts which were generated from the former by atomic manipulation. The characterization by AFM in combination with atomic manipulation provides clear fingerprints of the aromatic and aliphatic moieties that will facilitate their assignment in a priori unknown samples.
Introduction
High resolution atomic force microscopy (AFM) with functionalized tips1 is evolving towards a valuable tool for molecular structure identification of synthetic2–5 and natural6–8 compounds, and products formed by on-surface synthesis9–16 and atomic manipulation.2,10,17,18 Structure identification by AFM has the strategic advantage that one can address individual molecules, i.e. one can isolate different constituents in a mixture and look at them one by one.8,9 If enough molecules are imaged, a statistically significant analysis is possible13 and even rare side products can be identified.3,19 Moreover, AFM analysis requires only a small amount of material (about 0.1–1 mg)20 and does not demand the compound to be soluble. Especially the single molecule sensitivity renders AFM an interesting tool for the investigation of complex molecular mixtures like petroleum, which contains a tremendous diversity and number of molecular species,21,22 of which asphaltenes are one fraction.8 The compositional complexity of these mixtures is a great scientific challenge with economic relevance in oil production and processing.23–29 Besides the tremendous diversity of chemical structures in such complex asphaltene mixtures, the poor solubility and difficult volatilization and ionization have considerably impeded their structure elucidation.AFM is most powerful on planar molecules and much more challenging on non-planar ones. For molecules exhibiting a planar aliphatic hydrocarbon backbone it was demonstrated that a sp3 and sp2 hybridized carbon can be distinguished.2,10 In the present study we investigate molecules with several neighboring tetrahedrally coordinated carbons, resulting in a non-planar carbon backbone and multiple possible adsorption geometries. The low abundance of alkyl and cycloaliphatic moieties and archipelago-type motifs found in a preceding AFM study on asphaltenes8 has triggered questions whether such molecules can sustain the thermal sublimation step during preparation and be recognized by AFM. This work resolves that issue.
In this work we identified linear and cyclic aliphatic groups in different adsorption geometries and distinguished them from their aromatic planar counterparts that were generated by tip-induced dehydrogenation reactions. The model compounds introduced here have characteristics typical for aliphatic moieties and archipelago-type molecules found in asphaltenes. The alkyl moieties selected span a great enough range to be representative of petroleum. It is demonstrated that the compounds sustain the deposition and can be unambiguously identified by AFM. Identification was based on the AFM contrast on different substrates and by employing atomic manipulation, to: (i) switch the non-planar geometry of aliphatic compounds, and (ii) cleave C–H bonds. We thus show that such molecules can be detected by AFM, and we obtained fingerprints for the identification of aliphatic hydrocarbon moieties, facilitating and improving their chemical identification for future determination of a priori unknown molecules by AFM.
Results and discussion
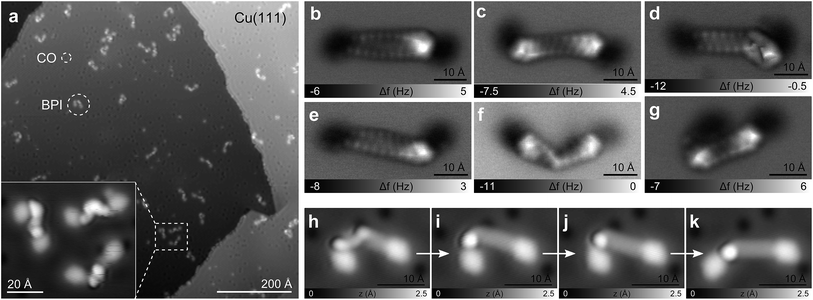
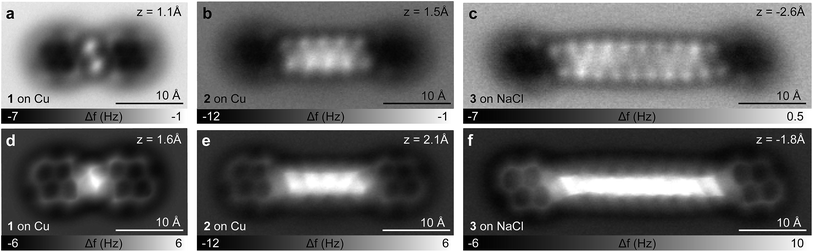
We investigated a set of five synthetic model compounds featuring aliphatic moieties (see Fig. 1): 1,2-di(pyren-1-yl)ethane (BPE, 1), 1,10-di(pyren-1-yl)decane (BPD, 2), 1,20-di(pyren-1-yl)icosane (BPI, 3), 11,12-dihydro-10H-cyclopenta[6,7]naphtho[1,2,3,4-ghi]perylene (CPNP, 4) and 10,11,12,13-tetrahydroanthra [1,2,3,4-ghi]perylene (CHNP, 5). BPE, BPD and BPI were particularly designed to address the linkage between polycyclic aromatic hydrocarbon (PAH) cores, which is the main distinguishing feature between the archipelago and island models of asphaltene structure. CPNP and CHNP comprise cycloaliphatic units. The model molecules introduced here are consistent with asphaltene molecular properties regarding molecular weight, boiling point, H/C ratio and double bond equivalent as reported previously.30–32In Fig. 2a, a scanning tunneling microscopy (STM) overview image of the BPI sample preparation is shown. Even for this comparatively large molecule with a molecular weight of 683 Da and a C20H40 aliphatic chain as a linker, the vast majority of the molecules does not dissociate or form aggregates. The two pyrene units of BPI adsorb flat on the substrate and the alkyl chain is typically buckled or twisted. The number of possible conformations of the aliphatic linker increases rapidly with chain length. In Fig. 2b–g AFM images of BPI in a few conformations are shown. Often the typical zig–zag pattern of the alkyl chain lying (partially) flat on the surface can be recognized. Non-planar conformations, that is, sections and twists where the C–C bonds of the alkyl chain are not in a plane parallel to the surface, are more difficult to assign. By providing external stimuli through tip-induced inelastic excitations33 (Fig. 2h–k) or thermal annealing of the sample to 300 K (Fig. S5†), however, the molecules unbend and preferentially adopt a planar adsorption geometry with an overall straight alkyl chain, suggesting this geometry to be energetically favored on both the Cu and NaCl substrate.
Similar to BPI, also its smaller analogues BPD and BPE could be deposited intact by fast thermal evaporation, despite the presence of one of the weakest C–C bonds in BPE's aliphatic linker (65.2 kcal mol−1).34 Because of the generally increasing sublimation temperature with increasing molecular weight, fragmentation will become more problematic for larger molecules. Experiments show that at masses of about 1000 Da, even for completely aromatic PAHs, fragmentation can dominate over intact sublimation,3 providing an approximate upper bound for molecule deposition by sublimation.
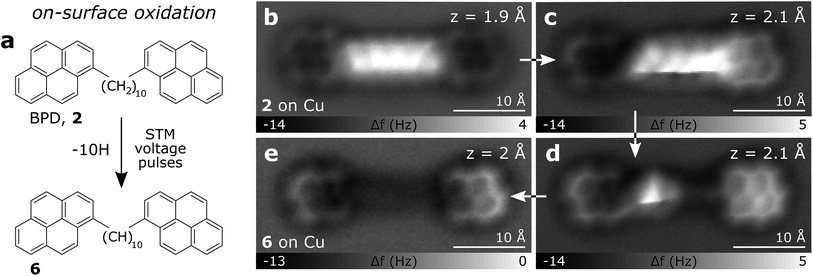
In Fig. 3a–f AFM measurements of BPE, BPD and BPI in the planar conformation at different scan heights are shown. All the methylene units can be clearly identified and differentiated from the sp2 carbon atoms in pyrene. Because the atoms are not all in the same plane, different scan heights are used for identifying the alkyl chain and the PAH units, respectively (compare upper and lower panels of Fig. 3). The brighter appearance of the alkyl chains compared to the pyrene units indicates the larger adsorption height of the former.2 In fact, tip induced dehydrogenation35 to form alkenyl chains significantly reduced the adsorption height and thus the brightness of the linker chain (see Fig. 4). Moreover, also the qualitative contrast of alkyl and alkenyl chains is different. In case of the alkyl chains the brightest features are at the rim of the chain corresponding to the apparent location of the protruding hydrogen atoms. Note that there is an apparent increase in width due to the well known effect of CO tip tilting towards the molecule.36,37 For alkenyl chains the brightest features are in the central part of the chain, above apparent carbon atom positions and carbon–carbon bonds (see Fig. 4). The tip-induced oxidation of the alkyl chain to an alkenyl chain demonstrates (i) the capability to clearly distinguish the two different chains and (ii) the initial integrity of the alkyl chain after deposition. For the more extended alkyl chains even small brightness modulations of the methylene units can be observed, which are probably related to minute differences in adsorption height. For straight alkyl chains the different brightness of methylene units seems to be related to their non-equivalent location with respect to the underlying substrate atoms (see Fig. S2 in the ESI†).
 | ||
| Fig. 3 BPE, BPD and BPI in straight conformation. (a–f) CO tip AFM images of BPE (a, d) and BPD (b, e) on Cu(111) and BPI on NaCl(2ML)/Cu(111) (c, f) at different tip heights. | ||
In the BPD and BPI samples we find a low number (<10%) of additional molecules on the surface. Some examples of such molecules are shown in Fig. S9 in the ESI.† The abundance of additional molecules observed is comparable to the impurity content of the material of about 4 wt%, indicating that a significant amount of the deposited additional molecules originate from impurities of the source compound. Although we can exclude fragmentation or chemical transformation of the vast majority of molecules, it cannot be ruled out for a few percent of the BPD and BPI molecules. For the other samples, which were of higher purity, we did not observe additional molecules by AFM.
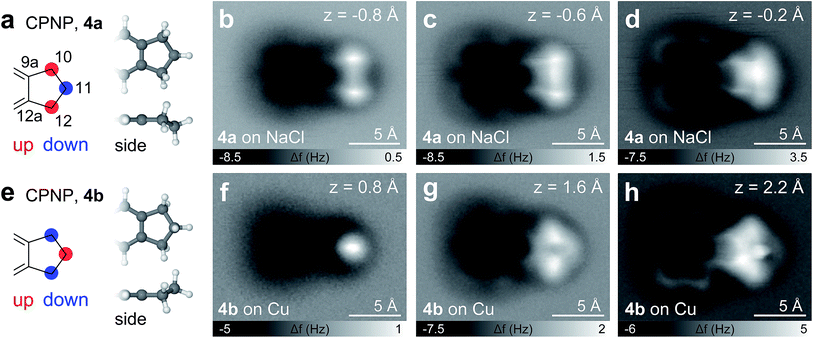
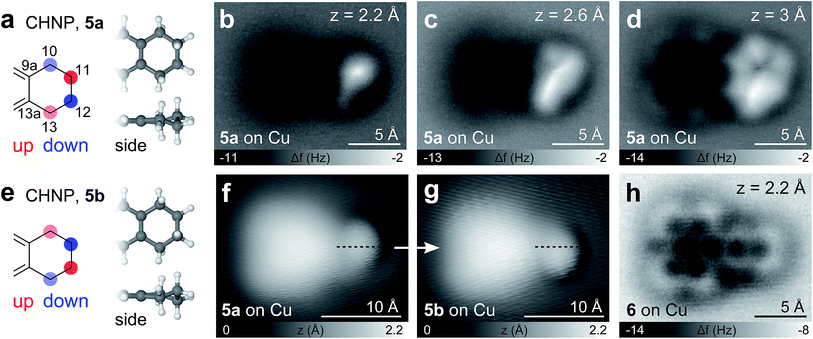
Besides PAHs and aliphatic chains, other important structural elements in hydrocarbons in general and in asphaltenes in particular are cycloaliphatic moieties. As models for such compounds we synthesized CPNP and CHNP that feature fused cyclopentene and cyclohexene rings, respectively. The CPNP molecule adsorbs differently on Cu and NaCl, showing two adsorption geometries of the same envelope-type conformation (4a or 4b, see Fig. 5). On NaCl, the 5-membered ring adopts an alternating ‘up-down-up’ geometry (4a), meaning that the carbon atoms at the 10 and 12 positions are buckled upwards (increased adsorption height) and the C at the 11 position downwards (see Fig. 5e). The assignments of the adsorption geometries are supported by AFM simulations based on the probe particle model by Hapala et al.37 (see Fig. S1†). Note that the apparent bond between the opposing methylene groups in Fig. 5b and c stems from a proximity effect as observed previously.37–40 On Cu on the other hand, the C in the 11 position buckles upwards to form a ‘down-up-down’ sequence in the aliphatic ring (4b). At close distance (Fig. 5d and h), however, it is unclear whether the native geometry can be fully maintained or whether repulsive tip forces might deflect the atoms from their intrinsic positions. Nonetheless, the cyclopentene moiety can be identified in either adsorption geometry, for both of which we obtained characteristic fingerprints.
Also for CHNP, two different molecular geometries were observed (see Fig. 6 and see Fig. S1† for simulated AFM images). This time, however, they represent the two enantiomers of a chiral half-chair conformation and were found on both, the Cu and NaCl substrate. The cyclohexene moiety adopts an alternating ‘down–UP–DOWN–up’ (5a) or ‘up–DOWN–UP–down’ (5b) geometry, where the degree of deflection is higher for the two carbons at the front of the molecule (positions 11, 12). Again an apparent bond-like feature arises connecting the positions of the two most protruding hydrogen atoms at positions 11 and 13 of 5a or positions 10 and 12 of 5b (most pronounced in Fig. 6c). On both substrates, reversible chiral switching between the two enantiomers41 could be induced by applying a voltage pulse of |V| > 0.4 V in either polarity as shown in Fig. 6f and g.
Like for BPE, BPD and BPI it is also possible to abstract hydrogen atoms from CPNP and CHNP by voltage pulses of about 3.5 V on Cu(111), demonstrating an on-surface oxidation process. In the case of CHNP, the removal of 4H from the cyclohexene group leads to a fully aromatic molecule (see Fig. 6h), reinforcing that the hydrogens did not dissociate during evaporation.
Conclusions
We synthesized and investigated model compounds featuring aliphatic moieties and unambiguously identified them by AFM. All molecules were deposited intact by fast thermal evaporation demonstrating that alkyl chain linkages between multiple aromatic moieties are stable to the preparation process applied for high-resolution AFM. This corroborates the significance of AFM for the structural investigation of complex organic mixtures, and reinforces the higher abundance of island-type asphaltene molecules found in a previous AFM study.8 By atomic manipulation we accessed different adsorption geometries and oxidized the aliphatic moieties. Using both AFM imaging and atomic manipulation we established fingerprints of alkyl chains and non-planar aliphatic rings in different adsorption geometries for their identification in a priori unknown samples.Methods
STM/AFM experiments
The experiments were performed with a combined STM and AFM in ultrahigh vacuum (p ≈ 10−10 mbar) and low-temperature (T ≈ 5 K) using a qPlus sensor.42 AFM measurements were acquired in the frequency-modulation mode43 (f0 ≈ 30 kHz and A = 0.5 Å) at constant-height and zero sample bias. If not stated otherwise, STM measurements were recorded at I = 2 pA and V = 0.2 V. Tip heights z relate to this tunneling set-point, with z > 0 being closer to the substrate. The tip was functionalized with a single CO molecule (referred to as CO tip).1,44 As a sample we used a Cu(111) single crystal partially covered by islands of bilayer NaCl,45 denoted as NaCl(2ML)/Cu(111) or simply NaCl. To evaporate the molecules we apply a small grain of the solid material ex situ on a (oxidized) Si wafer. In the ultrahigh vacuum this Si wafer is resistively heated to about 900 K within 2 s such that the molecules are evaporated onto the sample, which is held at T ≈ 10 K. The rapid heating typically enhances the volatility of fragile species.46,47Synthesis
Details on the synthesis of the model compounds and additional characterization can be found in the ESI.†Author contributions
All authors contributed to the manuscript. B. S., S. F., G. M. and L. G. performed the scanning probe measurements. S. C., D. P., E. G. and D. P. synthesized the CPNP and CHNP molecules. Y. Z. M. R. H. and J. D. K. designed the BPE, BPD and BPI molecules. L. G., B. S., Di. P., Y. Z. and J. D. K. conceived the experiments.Acknowledgements
We thank Z. Majzik, R. Allenspach and S. P. Rucker for discussions. We acknowledge financial support from the ERC Grants CEMAS (agreement no. 291194) and AMSEL (682144), the EU project PAMS (610446) and the Spanish Ministry of Science and Competitiveness for financial support (MAT2013-46593-C6-6-P).References
- L. Gross, F. Mohn, N. Moll, P. Liljeroth and G. Meyer, Science, 2009, 325, 1110–1114 CrossRef CAS PubMed.
- B. Schuler, W. Liu, A. Tkatchenko, N. Moll, G. Meyer, A. Mistry, D. Fox and L. Gross, Phys. Rev. Lett., 2013, 111, 106103 CrossRef PubMed.
- B. Schuler, S. Collazos, L. Gross, G. Meyer, D. Pérez, E. Guitián and D. Peña, Angew. Chem., Int. Ed., 2014, 53, 9004–9006 CrossRef CAS PubMed.
- Z. Majzik, A. B. Cuenca, N. Pavliček, N. Miralles, G. Meyer, L. Gross and E. Fernández, ACS Nano, 2016, 10, 5340–5345 CrossRef CAS PubMed.
- N. J. van der Heijden, P. Hapala, J. A. Rombouts, J. van der Lit, D. Smith, P. Mutombo, M. Švec, P. Jelinek and I. Swart, ACS Nano, 2016, 10, 8517–8525 CrossRef CAS PubMed.
- L. Gross, F. Mohn, N. Moll, G. Meyer, R. Ebel, W. M. Abdel-Mageed and M. Jaspars, Nat. Chem., 2010, 2, 821–825 CrossRef CAS PubMed.
- K. Ø. Hanssen, B. Schuler, A. J. Williams, T. B. Demissie, E. Hansen, J. H. Andersen, J. Svenson, K. Blinov, M. Repisky, F. Mohn, G. Meyer, J.-S. Svendsen, K. Ruud, M. Elyashberg, L. Gross, M. Jaspars and J. Isaksson, Angew. Chem., Int. Ed., 2012, 51, 12238–12241 CrossRef CAS PubMed.
- B. Schuler, G. Meyer, D. Peña, O. C. Mullins and L. Gross, J. Am. Chem. Soc., 2015, 137, 9870–9876 CrossRef CAS PubMed.
- D. G. d. Oteyza, P. Gorman, Y.-C. Chen, S. Wickenburg, A. Riss, D. J. Mowbray, G. Etkin, Z. Pedramrazi, H.-Z. Tsai, A. Rubio, M. F. Crommie and F. R. Fischer, Science, 2013, 340, 1434–1437 CrossRef PubMed.
- J. van der Lit, M. P. Boneschanscher, D. Vanmaekelbergh, M. Ijäs, A. Uppstu, M. Ervasti, A. Harju, P. Liljeroth and I. Swart, Nat. Commun., 2013, 4, 2023 Search PubMed.
- T. Dienel, S. Kawai, H. Söde, X. Feng, K. Müllen, P. Ruffieux, R. Fasel and O. Gröning, Nano Lett., 2015, 15, 5185–5190 CrossRef PubMed.
- C. Rogers, C. Chen, Z. Pedramrazi, A. A. Omrani, H.-Z. Tsai, H. S. Jung, S. Lin, M. F. Crommie and F. R. Fischer, Angew. Chem., Int. Ed., 2015, 54, 15143–15146 CrossRef CAS PubMed.
- A. Riss, A. P. Paz, S. Wickenburg, H.-Z. Tsai, D. G. De Oteyza, A. J. Bradley, M. M. Ugeda, P. Gorman, H. S. Jung, M. F. Crommie, A. Rubio and F. R. Fischer, Nat. Chem., 2016, 8, 678–683 CrossRef CAS PubMed.
- P. Ruffieux, S. Wang, B. Yang, C. Sánchez-Sánchez, J. Liu, T. Dienel, L. Talirz, P. Shinde, C. A. Pignedoli and D. Passerone, et al. , Nature, 2016, 531, 489–492 CrossRef CAS PubMed.
- Y. He, M. Garnica, F. Bischoff, J. Ducke, M.-L. Bocquet, M. Batzill, W. Auwärter and J. V. Barth, Nat. Chem., 2016 DOI:10.1038/nchem.2600 Search PubMed.
- S. Kawai, V. Haapasilta, B. D. Lindner, K. Tahara, P. Spijker, J. A. Buitendijk, R. Pawlak, T. Meier, Y. Tobe and A. S. Foster, et al. , Nat. Commun., 2016, 7, 12711 CrossRef CAS PubMed.
- N. Pavliček, B. Schuler, S. Collazos, N. Moll, D. Pérez, E. Guitián, G. Meyer, D. Peña and L. Gross, Nat. Chem., 2015, 7, 623–628 CrossRef PubMed.
- B. Schuler, S. Fatayer, F. Mohn, N. Moll, N. Pavliček, G. Meyer, D. Peña and L. Gross, Nat. Chem., 2016, 8, 220–224 CrossRef CAS PubMed.
- A. Mistry, B. Moreton, B. Schuler, F. Mohn, G. Meyer, L. Gross, A. Williams, P. Scott, G. Costantini and D. J. Fox, Chem.–Eur. J., 2015, 21, 2011–2018 CrossRef CAS PubMed.
- B. Schuler, F. Mohn, L. Gross, G. Meyer and M. Jaspars, in Modern NMR Approaches to the Structure Elucidation of Natural Products: Volume 1: Instrumentation and Software, The Royal Society of Chemistry, 2015, vol. 1, pp. 306–320 Search PubMed.
- K. Qian, R. P. Rodgers, C. L. Hendrickson, M. R. Emmett and A. G. Marshall, Energy Fuels, 2001, 15, 492–498 CrossRef CAS.
- K. Qian, W. K. Robbins, C. A. Hughey, H. J. Cooper, R. P. Rodgers and A. G. Marshall, Energy Fuels, 2001, 15, 1505–1511 CrossRef CAS.
- O. C. Mullins, E. Y. Sheu, A. Hammami and A. G. Marshall, Asphaltenes, Heavy Oils, and Petroleomics, Springer, New York, 2007, vol. 1 Search PubMed.
- S. Larter, B. Bowler, M. Li, M. Chen, D. Brincat, B. Bennett, K. Noke, P. Donohoe, D. Simmons, M. Kohnen, J. Allan, N. Telnaes and I. Horstad, Nature, 1996, 383, 593–597 CrossRef CAS.
- R. Quann and S. Jaffe, Chem. Eng. Sci., 1996, 51, 1615–1635 CrossRef CAS.
- I. M. Head, D. M. Jones and S. R. Larter, Nature, 2003, 426, 344–352 CrossRef CAS PubMed.
- C. M. Aitken, D. Jones and S. Larter, Nature, 2004, 431, 291–294 CrossRef CAS PubMed.
- J. Ancheyta, F. Trejo and M. S. Rana, Asphaltenes: Chemical Transformation during Hydroprocessing of Heavy Oils, CRC Press, Boca Raton, FL, 2010 Search PubMed.
- J. Sheng, Modern Chemical Enhanced Oil Recovery: Theory and Practice, Gulf Professional Publishing, Burlington, MA, 2010 Search PubMed.
- A. E. Pomerantz, M. R. Hammond, A. L. Morrow, O. C. Mullins and R. N. Zare, J. Am. Chem. Soc., 2008, 130, 7216–7217 CrossRef CAS PubMed.
- A. G. Marshall and R. P. Rodgers, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18090–18095 CrossRef CAS PubMed.
- D. C. Podgorski, Y. E. Corilo, L. Nyadong, V. V. Lobodin, B. J. Bythell, W. K. Robbins, A. M. McKenna, A. G. Marshall and R. P. Rodgers, Energy Fuels, 2013, 27, 1268–1276 CrossRef CAS.
- I. Swart, T. Sonnleitner, J. Niedenführ and J. Repp, Nano Lett., 2012, 12, 1070–1074 CrossRef CAS PubMed.
- S. J. Blanksby and G. B. Ellison, Acc. Chem. Res., 2003, 36, 255–263 CrossRef CAS PubMed.
- Y. Kim, T. Komeda and M. Kawai, Phys. Rev. Lett., 2002, 89, 126104 CrossRef PubMed.
- L. Gross, F. Mohn, N. Moll, B. Schuler, A. Criado, E. Guitián, D. Peña, A. Gourdon and G. Meyer, Science, 2012, 337, 1326–1329 CrossRef CAS PubMed.
- P. Hapala, G. Kichin, C. Wagner, F. S. Tautz, R. Temirov and P. Jelinek, Phys. Rev. B: Condens. Matter Mater. Phys., 2014, 90, 085421 CrossRef.
- N. Pavliček, B. Fleury, M. Neu, J. Niedenführ, C. Herranz-Lancho, M. Ruben and J. Repp, Phys. Rev. Lett., 2012, 108, 086101 CrossRef PubMed.
- A. Sweetman, S. P. Jarvis, H. Sang, I. Lekkas, P. Rahe, Y. Wang, J. Wang, N. R. Champness, L. Kantorovich and P. Moriarty, Nat. Commun., 2014, 5, 3931 CAS.
- S. K. Hämäläinen, N. van der Heijden, J. van der Lit, S. den Hartog, P. Liljeroth and I. Swart, Phys. Rev. Lett., 2014, 113, 186102 CrossRef PubMed.
- Y.-S. Fu, J. Schwöbel, S.-W. Hla, A. Dilullo, G. Hoffmann, S. Klyatskaya, M. Ruben and R. Wiesendanger, Nano Lett., 2012, 12, 3931–3935 CrossRef CAS PubMed.
- F. J. Giessibl, Appl. Phys. Lett., 1998, 73, 3956 CrossRef CAS.
- T. R. Albrecht, P. Grütter, D. Horne and D. Rugar, J. Appl. Phys., 1991, 69, 668 CrossRef.
- F. Mohn, B. Schuler, L. Gross and G. Meyer, Appl. Phys. Lett., 2013, 102, 073109 CrossRef.
- R. Bennewitz, V. Barwich, M. Bammerlin, C. Loppacher, M. Guggisberg, A. Baratoff, E. Meyer and H.-J. Güntherodt, Surf. Sci., 1999, 438, 289–296 CrossRef CAS.
- R. Beuhler, E. Flanigan, L. Greene and L. Friedman, J. Am. Chem. Soc., 1974, 96, 3990–3999 CrossRef CAS PubMed.
- L. Grill, J. Phys.: Condens. Matter, 2010, 22, 084023 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional scanning tunneling microscopy (STM) and atomic force microscopy (AFM) measurements as well as NMR, optical spectroscopy and gas chromatography (GC) characterization of the five model compounds are provided. For CPNP and CHNP the synthetic route is described. See DOI: 10.1039/c6sc04698c |
| This journal is © The Royal Society of Chemistry 2017 |