Open Access Article
Open Access ArticleOrange, red, yellow: biosynthesis of azaphilone pigments in Monascus fungi†
Wanping
Chen‡
 a,
Runfa
Chen‡
a,
Qingpei
Liu‡
ac,
Yi
He
a,
Kun
He
a,
Xiaoli
Ding
a,
Lijing
Kang
a,
Xiaoxiao
Guo
a,
Nana
Xie
a,
Youxiang
Zhou
*b,
Yuanyuan
Lu
cd,
Russell J.
Cox
a,
Runfa
Chen‡
a,
Qingpei
Liu‡
ac,
Yi
He
a,
Kun
He
a,
Xiaoli
Ding
a,
Lijing
Kang
a,
Xiaoxiao
Guo
a,
Nana
Xie
a,
Youxiang
Zhou
*b,
Yuanyuan
Lu
cd,
Russell J.
Cox
 e,
István
Molnár
e,
István
Molnár
 *c,
Mu
Li
a,
Yanchun
Shao
a and
Fusheng
Chen
*a
*c,
Mu
Li
a,
Yanchun
Shao
a and
Fusheng
Chen
*a
aKey Laboratory of Environment Correlative Dietology, College of Food Science and Technology, Huazhong Agricultural University, Wuhan, Hubei Province 430070, China. E-mail: chenfs@mail.hzau.edu.cn; Fax: +86-27-87282111; Tel: +86-27-87282111
bInstitute of Quality Standard and Testing Technology for Agro-Products, Hubei Academy of Agricultural Sciences, Wuhan, Hubei Province 430064, China. E-mail: zhouyouxiang@gmail.com
cNatural Products Center, The University of Arizona, 250 E. Valencia Rd., Tucson, Arizona 85706, USA. E-mail: imolnar@email.arizona.edu
dState Key Laboratory of Natural Medicines, School of Life Science and Technology, China Pharmaceutical University, 24 Tong Jia Xiang, Nanjing 210009, China
eInstitut fur Organische Chemie, BMWZ, Leibniz Universitat Hannover, Schneiderberg 1B, 30167 Hannover, Germany
First published on 24th April 2017
Abstract
Monascus azaphilone pigments (MonAzPs) are very widely used as food colorants, but their biosynthetic pathway has remained poorly characterized for more than half a century. In this study, the individual steps of MonAzPs biosynthesis in Monascus ruber M7 were elucidated by a combination of targeted gene knockouts, heterologous gene expression, and in vitro chemical and enzymatic reactions. This study describes the first rational engineering of MonAzPs biosynthesis and provides a roadmap for future pathway engineering efforts directed towards the selective production of the most valuable pigments and serves as a model for the biosynthesis of fungal azaphilones in general.
Introduction
The earliest historical document describing the production of Monascus azaphilone pigments (MonAzPs) as food colorants in the Orient dates to the 1st century CE.1 Since then, MonAzPs have gained widespread acceptance in the food industry due to their efficient production by fermentation using inexpensive and renewable substrates and because of their diverse colors, good solubility in water and ethanol, and high safety.2,3 Thus, in East Asian countries, particularly in China, Japan, the Philippines, Thailand and Indonesia, MonAzPs have traditionally been added to various foods such as sausages, fermented bean curd, red rice wine, kamaboko, and preserved dry meat and fish products.1,4 In Europe, MonAzPs have been considered as partial substitutes for nitrate and nitrite salts for the preservation and color enhancement of meat and poultry products, including sausages, pâté, ham and frankfurters.5–8In addition to serving as food coloring agents, MonAzPs have various promising applications in the pharmaceutical, cosmetics, textile, printing and dyeing industries.9–11 It is estimated that the annual production of MonAzPs is nearly 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 tons in China alone, and that more than one billion people eat MonAzPs-containing products during their daily life.12
000 tons in China alone, and that more than one billion people eat MonAzPs-containing products during their daily life.12
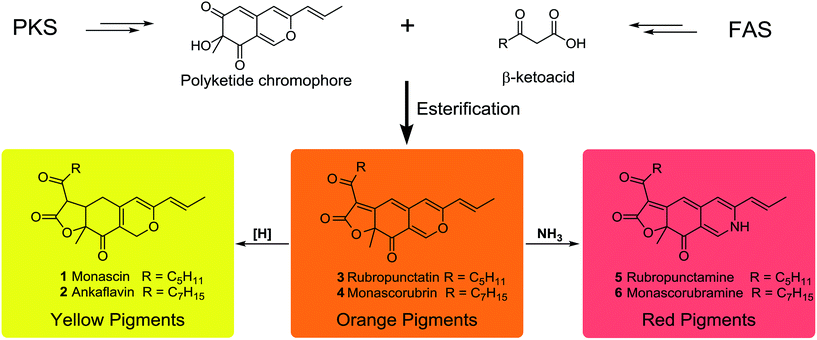
MonAzPs are a complex mixture of compounds with a common azaphilone skeleton. They are traditionally classified as red, orange, and yellow pigments based on their absorbance maxima at 490–530, 460–480 and 330–450 nm, respectively (Scheme 1).13 Between 1930 and 1980, six MonAzPs constituents were identified,13 including two yellow (monascin 1 and ankaflavin 2),14,15 two orange (rubropunctatin 3 and monascorubrin 4),15,16 and two red pigments (rubropunctamine 5 and monascorubramine 6).17 Since 1980, many more family members have been identified, and to the best of our knowledge, these include 44 yellow, 8 orange and 42 red pigments.18
In spite of their large scale commercial production and tremendous economic potential, the understanding of the biosynthesis of MonAzPs has remained incomplete. Investigations of the biosynthetic pathway began in as early as the 1960s,19–21 and several groups, including ours, continued their study in the same, leading to a partial understanding of the pathway.13,22–25 The generally recognized pathway proposes that the orange MonAzPs rubropunctatin 3 and monascorubrin 4 are formed by the esterification of a β-ketoacid (from the fatty acid synthase (FAS) pathway) to the chromophore (derived from the polyketide synthase (PKS) pathway). Then, reduction of the orange MonAzPs yields the yellow pigments monascin 1 and ankaflavin 2. In contrast, amination of the orange pigments with NH3 leads to the red MonAzPs rubropunctamine 5 and monascorubramine 6 (Scheme 1).26 However, these widely recognized steps remained assumptions based on chemical principles that may not accurately describe the in vivo biosynthetic steps. This situation started to change with the first few reports on the functional analysis of MonAzPs biosynthetic gene clusters in 2013.27,28
In addition to their economic importance and potential health benefits, MonAzPs are also widely regarded as ideal models for studying the biosynthesis of azaphilones due to their high productivity and ease of detection.29 Thus, the present study describes a systematic functional investigation of the MonAzPs cluster from Monascus ruber M7 using a combination of gene knockouts, heterologous gene expression, and in vitro chemical and enzymatic reactions. Our results revise and correct some of the long-standing assumptions on MonAzPs biosynthesis. Notably, we show that yellow MonAzPs are the primary products of the shunt pathway, followed by their enzymatic conversion to the orange pigments. Finally, we show that the various red MonAzPs derived from their orange counterparts by non-enzymatic reactions with amines.
Results and discussion
MonAzPs production in M. ruber M7
Wild type M. ruber M7 produces at least ten MonAzPs congeners. These include two orange pigments (rubropunctatin 3 and monascorubrin 4), six yellow pigments (monascin 1, ankaflavin 2, monasfluol A 7, monasfluol B 8, monasfluore A 9, and monasfluore B 10), and two frequent red pigments (rubropunctamine 5 and monascorubramine 6) (see ESI† Section 1). It should be noted that the variety of red MonAzPs produced is highly dependent on the ingredients of the culture media.Comparative bioinformatic analysis of MonAzPs biosynthetic gene clusters
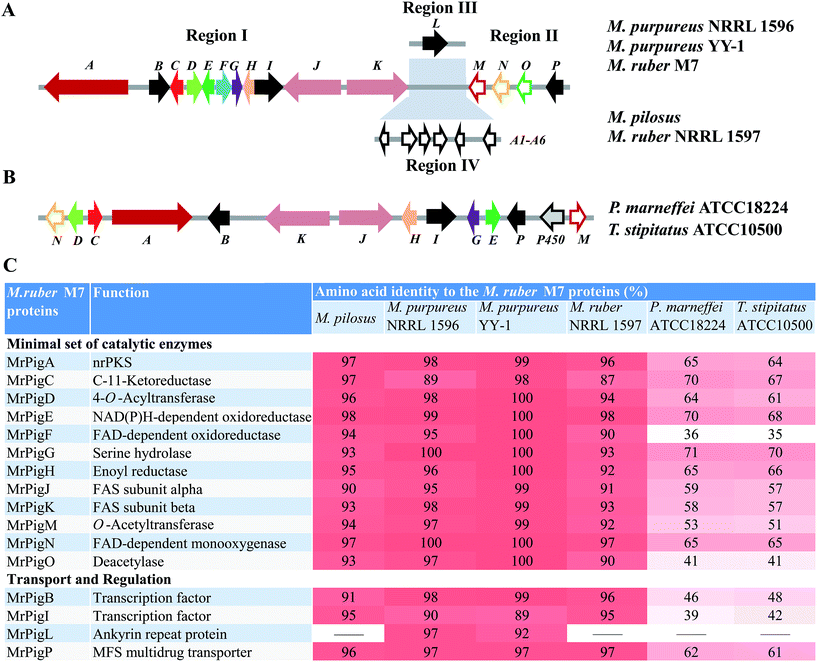
We began by analyzing the five known gene clusters for MonAzPs biosynthesis from various Monascus species, including the clusters from M. pilosus,27M. purpureus YY-1 (ref. 12) and NRRL 1596 (http://genome.jgi.doe.gov/Monpu1/Monpu1.home.html), and M. ruber M7 (ref. 1) and NRRL 1597 (http://genome.jgi.doe.gov/Monru1/Monru1.home.html). These clusters could be divided into four regions (I–IV) based on their gene contents and synteny (Fig. 1; see ESI† Section 2 for detailed gene annotations). Regions I and II are highly conserved, in fact, nearly identical amongst the MonAzPs gene clusters from all the different Monascus strains, indicating that these regions encode the core enzymes for MonAzPs biosynthesis. Region III, present in M. ruber M7 and the two M. purpureus strains, features only an ankyrin repeat protein-encoding gene. Region IV is present only in M. ruber NRRL 1597 and M. pilosus and contains six genes related to transport and signal transduction. We also compared the Monascus MonAzPs gene clusters with those from Penicillium (Talaromyces) marneffei and its close relative Talaromyces stipitatus (see ESI† Section 3). The comparison indicated that most biosynthetic genes in regions I and II are conserved across all these fungi. However, the physical order of the genes in the clusters appears different in Monascus versus Talaromyces.Next, we investigated the functional roles of the minimal set of conserved MonAzPs biosynthetic genes that encode putative catalytic enzymes (Fig. 1). We used a combination of gene knockouts in M. ruber M7 (ESI† Sections 4–6), heterologous expression of selected genes in Saccharomyces cerevisiae and Aspergillus oryzae (ESI† Section 7), and in vitro reconstitution of selected reactions with isolated enzymes and intermediates (ESI† Sections 8–9). Isolation and structure elucidation of the MonAzPs intermediates and shunt products afforded by these strains and reactions allowed us to compile a comprehensive biosynthetic pathway for MonAzPs (Scheme 2).
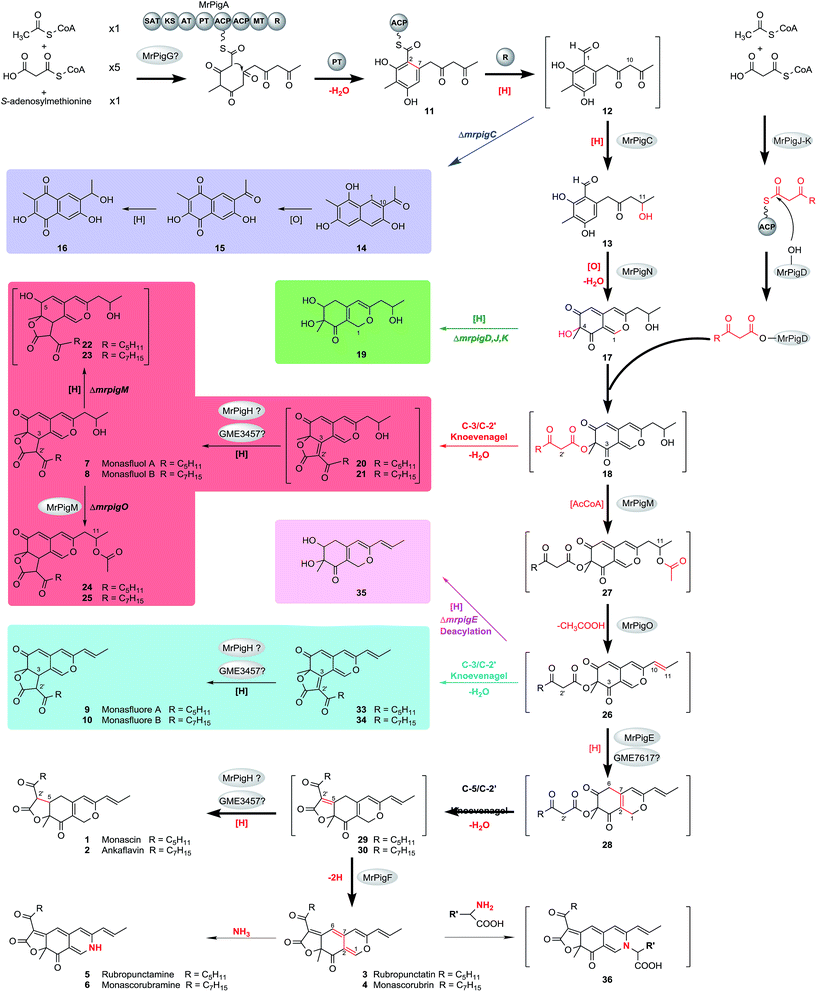 | ||
| Scheme 2 Proposed biosynthetic pathway for MonAzPs in M. ruber M7. Yellow pigments: 1, 2, 7–10, 13–17, 19–25, 29, 30, and 33–35. Orange pigments: 3, and 4. Red pigments: 5, 6, and 36. 36 is a model for amino acid derivatives of red MonAzPs. The spectral data is kept in ESI† Sections 6 and 9. 12, 18, and 26–28 are too reactive to isolate, whereas other compounds in square brackets were detected by MS but were not present in high enough concentration for NMR analysis. The main MonAzPs pathway is emphasized with bold arrows. Shunt pathways that contribute to the diversity of yellow pigments are boxed in color. See text for abbreviations. Red atoms and bonds indicate the chemical changes. | ||
Biosynthesis of the first stable enzyme-free intermediate
The biosynthesis of MonAzPs begins with the assembly of a hexaketide intermediate by the nonreducing PKS (nrPKS) MrPigA (see ESI† Section 10 for domain arrangement). As expected, knockout of mrpigA in M. ruber M7 completely abolishes the production of all MonAzPs-related substances (Fig. 2A).30 The biosynthetic programming of MrPigA is similar to its orthologue AzaA (identity 57%), the previously characterized azanigerone synthase nrPKS from Aspergillus niger.31 The starter acyl transferase (SAT) domain of MrPigA selects an acetyl-CoA starter unit, and the ketoacyl synthase (KS)–acyl transferase (AT)–acyl carrier protein (ACP) domains extend this starter unit five times with malonyl-CoA in five successive decarboxylative Claisen condensation cycles. The methyltransferase (MT) domain of MrPigA conducts a single C-methylation at C-4, most likely at the pentaketide stage. The reactive hexaketide chain then undergoes a product template (PT) domain-mediated C-2 to C-7 aldol cyclization to afford the first aromatic ring 11, followed by reductive release of the putative intermediate 12 by the NADPH-dependent reductive release (R) domain of MrPigA. This very reactive benzaldehyde is intercepted by the MrPigC ketoreductase that reduces the ω-1 carbonyl to the alcohol to provide compound 13, the first stable enzyme-free MonAzPs intermediate (Scheme 2). In the absence of MrPigC, 12 undergoes a spontaneous aldol cyclization in the C-1 to C-10 register to afford the trihydroxynaphthalene shunt metabolite 14 (blue shunt pathway, Scheme 2) and its spontaneously oxidized quinone derivative 15, as seen during the efficient heterologous expression of the MrPigA nrPKS in S. cerevisiae BJ5464-NpgA32 or in A. oryzae M-2-3 (ref. 33) hosts (ESI† Section 7). In contrast, co-expression of MrPigA and MrPigC in the S. cerevisiae host yields the expected on-pathway benzaldehyde 13 (ESI† Section 7). Similarly, the ΔmrpigC mutant of M. ruber M7 biosynthesizes 14, 15 and 16 (Fig. 2B). While 16 features the ω-1 alcohol, we propose that its formation is the result of an adventitious reduction of the off-pathway product 15 by other enzymes of M. ruber M7.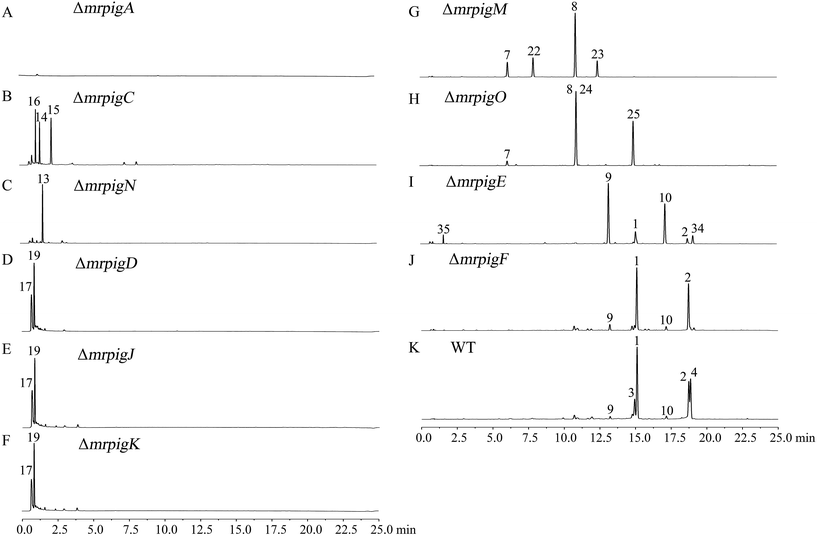 | ||
| Fig. 2 MonAzPs produced by the wild type M. ruber M7 and its gene knockout mutants. Representative UPLC profiles recorded at 370 nm are shown for fermentation extracts from the indicated knockouts and WT (the wild type M. ruber M7). See ESI† Sections 5 and 6 for the isolation, quantification, and structure elucidation for MonAzPs and their intermediates. | ||
The role of the MrPigG-type putative serine hydrolases in azaphilone polyketide biosynthesis is controversial. MppD, the M. purpureus orthologue of MrPigG, has recently been suggested to play an obligate but uncharacterized accessory role for the nrPKS MpPKS5. A ΔmppD mutant was severely reduced in MonAzPs formation, while co-expression of mppD with the nrPKS was necessary for product formation in a yeast heterologous expression system.34 Another orthologue of MrPigG, CitA from M. ruber M7 (53% identity to MrPigG) was also necessary for high level citrinin biosynthesis in both the native producer fungus and in an A. oryzae heterologous production system.35 On the other hand, a MrPigG orthologue is missing from the biosynthetic gene cluster of the azaphilone chaetoviridin, and it is not present elsewhere in the genome of the producer Chaetomium globosum strain.34 In spite of our best attempts, a ΔmrpigG mutant of M. ruber M7 could not be created. However, co-expression of MrPigG with MrPigA in S. cerevisiae BJ5464-NpgA32 led to no improvement in the yields of 14 and 15, nor did it afford any additional metabolites (see ESI† Section 7). This is similar to the case of AfoC, another putative esterase that is present in the biosynthetic gene cluster of asperfuranone, a related azaphilone: AfoC also fails to contribute to the production of the nrPKS product pre-asperfuranone upon heterologous expression in yeast.36 An orthologue of MrPigG is present in the genome of A. oryzae (XP_001823616, 53% identity to MrPigG), and hence this protein may contribute to product formation in that host. However, no orthologous or even similar proteins to MrPigG are encoded in the S. cerevisiae genome, thus metabolic cross-complementation with a host protein is less likely. Taken together, we propose that MrPigG plays at most a supplementary role in the formation of the putative benzaldehyde 12 by the MrPigA nrPKS.
Formation of the acylated pyran ring system
MrPigN, a FAD-dependent monooxygenase, is an orthologue of TropB (47% identity) and AzaH (56% identity), which have confirmed roles in benzaldehyde hydroxylation and subsequent pyran ring formation during tropolone and azanigerone biosynthesis, respectively.31,37 In particular, AzaH has been demonstrated to be the only enzyme required to convert 13 to 17in vitro.31 Thus, we propose that MrPigN hydroxylates the benzaldehyde intermediate 13 at C-4, which triggers the formation of the pyran ring to afford 17 (Scheme 2). Correspondingly, the mrpigN knockout mutant was found incapable of producing benzopyran MonAzPs but accumulated benzaldehyde 13 (Fig. 2C).MrPigJ and MrPigK form the two subunits of a dedicated fungal FAS that produces the side chain fatty acyl moiety of MonAzPs. Although orthologous to the dedicated FAS that is encoded in aflatoxin/sterigmatocystin biosynthetic gene clusters,38–40 the MrPigJ-MrPigK FAS produces a β-keto fatty acid. In addition, the chain length control of the MrPigJ-MrPigK pair is somewhat flexible as MonAzPs features either a β-ketooctanoic or a β-ketodecanoic acid moiety. The β-ketoacyl-ACP probably serves as the substrate for the AT MrPigD that directly transfers the fatty acyl chain to the C-4 alcohol of 17 to produce the putative intermediate 18, via a mechanism analogous to that of the AzaD AT in azanigerone biosynthesis, an orthologue of MrPigD (48% identity).31 As expected, the mrpigD, mrpigJ, and mrpigK knockout mutants all accumulate the same pyran intermediate 17 and its reduced product monascusone A 19 (green shunt pathway, Scheme 2), both devoid of the medium-chain fatty acyl moiety (Fig. 2D–F).
To confirm the catalytic function of this AT, we expressed the mrpigD gene in E. coli and reconstituted the reaction in vitro using the purified enzyme. We used the purified pyran 17 as the acyl acceptor and a synthetic N-acetylcysteamine thioester derivative of 3-ketooctanoic acid as a stand-in for the proposed 3-ketooctanoyl-ACP acyl donor (see ESI† Section 8 for the detailed experimental procedures and results). The expected intermediate 20 (red shunt pathway, Scheme 2), derived by a spontaneous intramolecular C3–C2′ Knoevenagel cyclization of the proposed intermediate 18, was then detected by HPLC-MS, although it was present in low titer, denying the possibility of NMR analysis.
Elimination of the ω-1 alcohol
MrPigM and MrPigO are homologous hypothetical proteins (mutual identity: 30%) whose orthologous protein relatives are widespread in ascomycete fungi. MrPigM and MrPigO, as well as their database orthologues, feature regions with similarity to the PapA5 (PRK09294) or the Tri3 (pfam07428) AT domains, respectively. Surprisingly, the pigment profiles of the mrpigM and the mrpigO knockout mutants are related: the two types of mutants share the angular tricyclic shunt intermediates monasfluol A 7 and monasfluol B 8 that retain the ω-1 alcohol (red shunt pathway, Scheme 2). Nevertheless, these two mutants differ in their production of additional shunt metabolites. Thus, mrpigM knockout mutants also produce low concentrations of compounds corresponding to the C-5 reduced monasfluol derivatives 22 and 23, still containing the ω-1 alcohol, whereas ΔmrpigO mutants yield the new C-11 acetylated shunt products 24 and 25 (Fig. 2G and H) in higher titers, which allowed full NMR analysis. MrPigM was previously suggested to mediate the dehydration of the ω-1 alcohol of 18 to yield the putative intermediate 26.25 Herein, we extend this hypothesis to propose that dehydration of 18 to 26 on the main pathway requires a two-step acetylation-elimination sequence catalyzed by MrPigM and MrPigO. In this model, MrPigM acts as an O-acetyltransferase that synthesizes the putative O-11 acetyl intermediate 27. Knockout of mrpigM arrests the pathway at compound 18 and leads to the production of compounds 7, 8, 22 and 23, angular tricyclic shunt products that retain the ω-1 hydroxyl group and result from a spontaneous C3–C2′ Knoevenagel aldol condensation (red shunt pathway, Scheme 2).Moreover, on the main pathway, MrPigO eliminates acetic acid from 27 to yield the putative intermediate 26 with the C10(11) double bond. Knockout of mrpigO or cultivation of the wild type M. ruber M7 with the sirtuin deacetylase inhibitor 3,4-dihydrocoumarin41 (data to be published elsewhere) then shunts the pathway towards 24 and 25. Similar enzymatic acetylation-elimination sequences that yield alkene functionalities have been described for the biosynthesis of tetronate and spirotetronate antibiotics in bacteria42,43 and for the production of the indole diterpene penitrem in Penicillium spp.44
Knoevenagel cyclizations yield yellow MonAzPs
Intramolecular Knoevenagel aldol condensations of pathway intermediates 18, 26, and 28 establish the tricyclic ring systems of various yellow on-pathway and shunt product MonAzPs (Scheme 2; see ESI† Section 11 for mechanism of Knoevenagel cyclization). On the main pathway, the dehydration of the C-11 alcohol of 18 is followed by the reduction of the C6(7) double bond of intermediate 26 by the NAD(P)H-dependent oxidoreductase MrPigE.22,24 This reduction increases the electrophilicity of the C-5 ketone of the resulting acyl benzopyran 28 by eliminating the π-conjugated system present in intermediate 26. This in turn sets up the C-5 ketone (instead of the C-3 ketone) for an intramolecular Knoevenagel aldol condensation with the C-2′ enol of the side chain. This condensation affords the characteristic linear tricyclic carbon skeletons of the on-pathway yellow pigments 29 and 30 that serve as the common precursors for the classical yellow, orange and red MonAzPs 1–6. An analogous bias for regiospecific cyclization has also been observed during the biosynthesis of chaetoviridins.45In contrast, shunt pathways branching out from intermediates 18 and 26 yield yellow pigments with angular tricyclic carbon skeletons. Thus, intermediate 18 produced by the ΔmrpigM or ΔmrpigO mutants undergoes a spontaneous Knoevenagel cyclization in the C-3 to C-2′ register, derailing the pathway towards the angular pigments 7/8via intermediates 20/21 (red shunt pathway, Scheme 2), which were detected in trace concentrations. Because a C-5 to C-2′ condensation is not favored for intermediate 18, the corresponding linear tricyclic hypothetical pigments 31 and 32 or their derivatives have never been found amongst Monascus fermentation products (Scheme 3A). Similarly, a C-5 to C-2′ condensation is not favored for intermediate 26 either, thus 26 cannot serve as the direct precursor for the classical orange pigments 3 and 4 (Scheme 3A). Accordingly, ΔmrpigE mutants do not produce orange or red pigments. Instead, intermediate 26 produced by the ΔmrpigE mutant is shunted towards the angular pigments monasfluores 9/10via intermediates 33/34 as a result of a spontaneous cyclization in the C-3 to C-2′ register (cyan shunt pathway, Scheme 2). Small amounts of intermediate 26 may also undergo deacylation and reduction to yield shunt product 35 in this strain (magenta shunt pathway, Scheme 2; Fig. 2I).
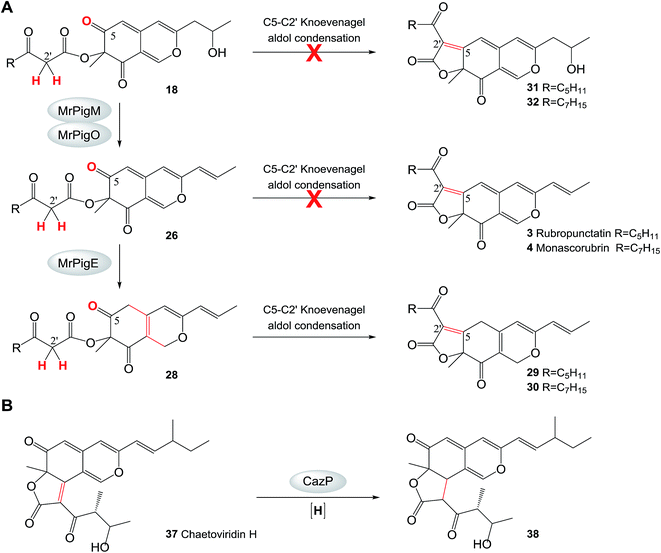 | ||
| Scheme 3 Reductive transformations channel azaphilone pigment biosynthesis. A. Intramolecular Knoevenagel aldol condensation at the C-5 to C-2′ register is not favored in intermediates 18 and 26 in M. ruber M7. B. The reaction catalyzed by CazP.45 Red crosses indicate reactions that are not observed. | ||
Compounds 20/21 and 33/34 are the necessary intermediates for the angular tricyclic shunt compounds. While small amounts of 33/34 have been identified from the ΔmrpigE mutant here and from the equivalent ΔmppC strains of M. purpureus,2420/21 have not been observed to date in Monascus fermentations. However, thorough scrutiny of our MS data allowed us to detect compounds corresponding to 20 and 21 in trace amounts in extracts of M. ruber M7 fermentations (see ESI† Section 6). Just as with the analogous intermediate chaetoviridin H, the low abundances of 20/21 and 33/34 could be explained by the efficient reduction of the C-3(2′) double bond in these intermediates (Scheme 2). In the chaetoviridin producer, this reduction is catalyzed by the dehydrogenase CazP, an enzyme with a relaxed substrate specificity (Scheme 3B).45 The M. ruber M7 genome encodes an orthologue of CazP (GME3457 [GenBank accession: KY270501], 46% amino acid identity). However, unlike cazP, which is part of the chaetoviridin gene cluster, the corresponding M. ruber M7 gene is encoded outside of the MonAzPs cluster. Saturation of the C-3(2′) double bond may also be facilitated by MrPigH (see below).
While majority of MonAzPs are ushered through the main pathway towards the linear tricyclic pigments 1–6 in M. ruber, control over the Knoevenagel condensation regioselectivity is by no means absolute. Thus, the angular pigments 9/10 are also produced at low levels in the wild type M7 strain, presumably by small amounts of intermediate 26 escaping C6(7) reduction by MrPigE (cyan shunt pathway, Scheme 2). Conversely, small amounts of the linear pigments 1 and 2 are also detectable in the mrpigE deletion mutant that produces the angular pigments 9/10 as its main product. We hypothesize that the necessary reduction of the C6(7) double bond of intermediate 26 may be carried out in this strain by GME7617 (GenBank accession: KY491645), a NAD(P)H-dependent oxidoreductase that is orthologous to MrPigE (39% identity), but is encoded in a different genomic locus of M. ruber M7.
Orange pigments originate from yellow MonAzPs
Pigments 29 and 30 are the expected linear tricyclic products of the directed Knoevenagel condensation on the main pathway. While these pigments could be detected in the mass spectra (see ESI† Section 6), they were present in amounts that are below the detection limit of UPLC, probably because of their instability and high turnover into various other MonAzPs, and they could not be characterized by NMR. We propose that these yellow pigments are in fact the key branch point intermediates for the synthesis of the classical MonAzPs.Starting with the deduced intermediates 29 and 30, the main pathway branches and leads to the classical yellow pigments monascin 1 and ankaflavin 2 by reduction of the C-5/C-2′ double bond. This reduction may be carried out by the CazP orthologue encoded by GME3457, as discussed above. However, reduced production of 1 and 2 in an M. purpureus strain with a deletion in the mppE gene (an orthologue of the mrpigH gene of M. ruber M7) and increased production of the same pigments in the mppE overexpressing strain suggest that MrPigH may also contribute to the saturation of the C5(2′) double bond of 29 and 30.46 In spite of our best efforts, a ΔmrpigH strain of M. ruber M7 could not be isolated.
The second branch of the main pathway yields the classical orange pigments rubropunctatin 3 and monascorubrin 4 by restoring the C6(7) double bond. We found that ΔmrpigF strains of M. ruber M7 are deficient in this branch of the pathway: these mutants produce no orange or red pigments but still accumulate large amounts of the classical yellow pigments monascin 1 and ankaflavin 2 (Fig. 2J). MrPigF, a FAD-dependent oxidoreductase, was previously suggested to play a role in pyran ring closure.27 However, considering the MonAzPs profile of the ΔmrpigF mutants, we propose that MrPigF desaturates C6(7) of putative intermediates 29 and 30 to afford the orange pigments rubropunctatin 3 and monascorubrin 4 featuring an extended π-conjugated system.
Red MonAzPs derive from the orange pigments 3 and 4
The classical red MonAzPs rubropunctamine 5 and monascorubramine 6 and their derivatives originate by the direct reaction of rubropunctatin 3 and monascorubrin 4 with endogenous amines, especially amino acids, present in Monascus cultures. We performed in vitro chemical reactions with 18 amines as the nucleophiles and the orange MonAzPs monascorubrin 4 rubropunctatin 3 or the yellow pigment monascin 1 as the substrates (see ESI† Section 9). UPLC-MS confirmed that the amines directly react with the orange pigments 3 and 4 to afford the red-colored γ-vinylogous pyridines 5 and 6 and their derivatives. However, monascin 1 is unreactive in these conditions, indicating that the classical yellow pigments are not the direct precursors of the red pigments. Monascorubrin 4 and rubropunctatin 3 preferentially react with ammonia and with basic amino acids such as arginine. Alkaline environments also promote this reaction because the nucleophilic character of the unprotonated amine group is more conducive to the O-to-N substitution. More than 42 red MonAzPs have been identified from Monascus spp.,18 most of which are various amino acid adducts of monascorubrin 4 and rubropunctatin 3. The facile reaction between orange azaphilones and amines is the main cause of the wide diversity of red MonAzPs.Conclusions
MonAzPs are a large family of structurally related secondary metabolites isolated from Monascus fungi. Tens of thousands of tons of MonAzPs are used every year for various dyeing and coloring applications all over the world, and demand for these pigments is rapidly growing. This study describes a systematic analysis of the MonAzPs biosynthetic pathway and defines each major step leading to the classical yellow, orange and red pigment constituents. In addition, this study provides an insight into how MonAzPs chemical diversity is generated by a complex biosynthetic pathway. Sequence analysis of the known MonAzPs biosynthetic gene clusters shows that the genes are highly conserved in various host organisms, but the organization of the clusters differs. Regions I and II (Fig. 1) are represented in all species. While regions I and II are contiguous in P. marneffei and T. stipitatus, they are split by insertion of unrelated genes (regions III and IV) in M. purpureus, M. pilosus and M. ruber. This insertion may reflect recent genomic rearrangements in the Monascus species and is unlikely to affect the biosynthesis.During MonAzPs biosynthesis, highly reactive intermediates are ushered down the main pathway by key redox enzymes. Failure of these enzymes to intercept the intermediates opens various shunt pathways (highlighted in color in Scheme 2) that yield varied products. The value of this work is illustrated by the knockout of mrPigF, which selectively produces 1 and 2 in high titers and indicates that rational redirection of the MonAzPs pathway is now possible. As MonAzPs finds wide uses in food production and preservation, our study illustrating how these compounds can be engineered via renewable fermentation technology is likely to be of direct benefit to sustainable food production. The findings in this study will underpin future advances in the characterization of the biosynthetic genes and enzymes involved in the production of varied MonAzPs and provide a roadmap towards the selective, directed biosynthesis of desired MonAzPs constituents. In addition, the MonAzPs pathway also serves as a useful model system for many other fungal polyketide biosynthetic pathways.
Acknowledgements
The authors express special thanks to members of the Chen, Cox and Molnár laboratories for their contributions and useful discussions. This study was supported by the Major Program of National Natural Science Foundation of China (No. 31330059 to F. C.), the National Natural Science Foundation of China (No. 31171649 to F. C., 31271834 to Y. S., 31371824 to Y. Z., 31401631 to M. L., and 31601446 to W. C.), the National Institutes of Health (NIGMS R01GM114418-01A1 to I. M.), and Deutsche Forschungsgemeinschaft (INST 187/621-1, LCMS equipment, to R. J. C). I. M. has a disclosed financial interest in TEVA Pharmaceuticals Hungary, which is unrelated to the subject of the research presented here. All the other authors declare no competing financial interests.Notes and references
- W. Chen, Y. He, Y. Zhou, Y. Shao, Y. Feng, M. Li and F. Chen, Compr. Rev. Food Sci. Food Saf., 2015, 14, 555–567 CrossRef CAS.
- F. Vendruscolo, R. M. M. Bühler, J. C. de Carvalho, D. de Oliveira, D. E. Moritz, W. Schmidell and J. L. Ninow, Appl. Biochem. Biotechnol., 2016, 178, 211–223 CrossRef CAS PubMed.
- M. E. Mostafa and M. S. Abbady, Global J. Biotechnol. Biochem., 2014, 9, 1–13 CAS.
- L. Dufossé, P. Galaup, A. Yaron, S. M. Arad, P. Blanc, K. N. Chidambara Murthy and G. A. Ravishankar, Trends Food Sci. Technol., 2005, 16, 389–406 CrossRef.
- C. E. Fabre, A. L. Santerre, M. O. Loret, R. Baberian, A. Pareilleux, G. Goma and P. J. Blanc, J. Food Sci., 1993, 58, 1099–1102 CrossRef CAS.
- J. Fink-Gremmels, J. Dresel and L. Leistner, Fleischwirtschaft, 1991, 71, 1184–1186 CAS.
- A. Bakosova, D. Mate, A. Laciakova and M. Pipova, Bull. Vet. Inst. Pulawy, 2001, 45, 111–116 Search PubMed.
- S. Marcinčák, P. Popelka, J. Nagy, D. Máté, M. Setláková and D. Šebestová, Slov. Vet. Čas., 2002, 27, 84–87 Search PubMed.
- N. Tsuyoshi, R. Fudou, S. Yamanaka, T. Furunaga, K. Sato and Y. Kondo, US Pat. Appl., US 20040150702, 2004.
- P. Velmurugan, S. Kamala-Kannan, V. Balachandar, P. Lakshmanaperumalsamy, J. C. Chae and B. T. Oh, Carbohydr. Polym., 2010, 79, 262–268 CrossRef CAS.
- A. Takayuki and S. Hiroshi, Japan Pat., JP 6329576, 1994.
- Y. Yang, B. Liu, X. J. Du, P. Li, B. Liang, X. Z. Cheng, L. C. Du, D. Huang, L. Wang and S. Wang, Sci. Rep., 2015, 5, 8331 CrossRef CAS PubMed.
- Y. Feng, Y. Shao and F. Chen, Appl. Microbiol. Biotechnol., 2012, 96, 1421–1440 CrossRef CAS PubMed.
- H. Salomon and P. Karrer, Helv. Chim. Acta, 1932, 15, 18–22 CrossRef CAS.
- P. S. Manchand, W. Whalley and F. C. Chen, Phytochemistry, 1973, 12, 2531–2532 CrossRef CAS.
- F. Chen, P. Manchard and W. Whalley, J. Chem. Soc. D, 1969, 130–131 RSC.
- S. Kumasaki, K. Nakanishi, E. Nishikawa and M. Ohashi, Tetrahedron, 1962, 18, 1171–1184 CrossRef CAS.
- Y. Feng, Y. Shao, Y. Zhou, W. Chen and F. Chen, in Industrial Biotechnology of Vitamins, Biopigments, and Antioxidants, Wiley-VCH Verlag GmbH & Co. KGaA, 2016, pp. 497–536 Search PubMed.
- A. Birch, A. Cassera, P. Fitton, J. Holker, H. Smith, G. Thompson and W. Whalley, J. Chem. Soc., 1962, 3583–3586 RSC.
- M. Kurono, K. Nakanishi, K. Shindo and M. Tada, Chem. Pharm. Bull., 1963, 11, 359–362 CrossRef CAS.
- J. Hadfield, J. Holker and D. Stanway, J. Chem. Soc. C, 1967, 751–755 RSC.
- Q. Liu, N. Xie, Y. He, L. Wang, Y. Shao, H. Zhao and F. Chen, Appl. Microbiol. Biotechnol., 2014, 98, 285–296 CrossRef CAS PubMed.
- J. Liu, Y. Zhou, T. Yi, M. Zhao, N. Xie, M. Lei, Q. Liu, Y. Shao and F. Chen, Appl. Microbiol. Biotechnol., 2016, 100, 7037–7049 CrossRef CAS PubMed.
- B. Bijinu, J. W. Suh, S. H. Park and H. J. Kwon, RSC Adv., 2014, 4, 59405–59408 RSC.
- B. Balakrishnan, C. C. Chen, T. M. Pan and H. J. Kwon, Tetrahedron Lett., 2014, 55, 1640–1643 CrossRef CAS.
- H. Hajjaj, A. Klaébé, G. Goma, P. J. Blanc, E. Barbier and J. François, Appl. Environ. Microbiol., 2000, 66, 1120–1125 CrossRef CAS PubMed.
- B. Balakrishnan, S. Karki, S. H. Chiu, H. J. Kim, J. W. Suh, B. Nam, Y. M. Yoon, C. C. Chen and H. J. Kwon, Appl. Microbiol. Biotechnol., 2013, 97, 6337–6345 CrossRef CAS PubMed.
- N. Xie, Q. Liu and F. Chen, Biotechnol. Lett., 2013, 35, 1425–1432 CrossRef CAS PubMed.
- S. A. S. Mapari, U. Thrane and A. S. Meyer, Trends Biotechnol., 2010, 28, 300–307 CrossRef CAS PubMed.
- N. Xie, Y. Zhang and F. Chen, Acta Microbiol. Sin., 2015, 55, 863–872 Search PubMed.
- A. O. Zabala, W. Xu, Y. H. Chooi and Y. Tang, Chem. Biol., 2012, 19, 1049–1059 CrossRef CAS PubMed.
- S. M. Ma, J. W. H. Li, J. W. Choi, H. Zhou, K. K. M. Lee, V. A. Moorthie, X. K. Xie, J. T. Kealey, N. A. Da Silva, J. C. Vederas and Y. Tang, Science, 2009, 326, 589–592 CrossRef CAS PubMed.
- K. Pahirulzaman, K. Williams and C. M. Lazarus, Methods Enzymol., 2012, 517, 241–260 CAS.
- B. Balakrishnan, R. Chandran, S. H. Park and H. J. Kwon, J. Microbiol. Biotechnol., 2015, 25, 1648–1652 CrossRef CAS PubMed.
- Y. He and R. J. Cox, Chem. Sci., 2016, 7, 2119–2127 RSC.
- J. Bai, Y. Y. Lu, Y. M. Xu, W. Zhang, M. Chen, M. Lin, A. A. L. Gunatilaka, Y. Q. Xu and I. Molnar, Org. Lett., 2016, 18, 1262–1265 CrossRef CAS PubMed.
- J. Davison, A. al Fahad, M. H. Cai, Z. S. Song, S. Y. Yehia, C. M. Lazarus, A. M. Bailey, T. J. Simpson and R. J. Cox, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 7642–7647 CrossRef CAS PubMed.
- T. S. Hitchman, E. W. Schmidt, F. Trail, M. D. Rarick, J. E. Linz and C. A. Townsend, Bioorg. Chem., 2001, 29, 293–307 CrossRef CAS PubMed.
- J. Yu, P.-K. Chang, K. C. Ehrlich, J. W. Cary, D. Bhatnagar, T. E. Cleveland, G. A. Payne, J. E. Linz, C. P. Woloshuk and J. W. Bennett, Appl. Environ. Microbiol., 2004, 70, 1253–1262 CrossRef CAS PubMed.
- J. Yu, Toxins, 2012, 4, 1024–1057 CrossRef CAS PubMed.
- A. J. Olaharski, J. Rine, B. L. Marshall, J. Babiarz, L. Zhang, E. Verdin and M. T. Smith, PLoS Genet., 2005, 1, e77 Search PubMed.
- C. Kanchanabanca, W. Tao, H. Hong, Y. Liu, F. Hahn, M. Samborskyy, Z. Deng, Y. Sun and P. F. Leadlay, Angew. Chem., 2013, 125, 5897–5900 CrossRef.
- L.-F. Wu, H.-Y. He, H.-X. Pan, L. Han, R. Wang and G.-L. Tang, Org. Lett., 2014, 16, 1578–1581 CrossRef CAS PubMed.
- C. Liu, K. Tagami, A. Minami, T. Matsumoto, J. C. Frisvad, H. Suzuki, J. Ishikawa, K. Gomi and H. Oikawa, Angew. Chem., 2015, 127, 5840–5844 CrossRef.
- M. Sato, J. M. Winter, S. Kishimoto, H. Noguchi, Y. Tang and K. Watanabe, Org. Lett., 2016, 18, 1446–1449 CrossRef CAS PubMed.
- B. Balakrishnan, S. H. Park and H. J. Kwon, Biotechnol. Lett., 2017, 39, 163–169 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Complete description of methods and additional results; and tables and figures, including structure elucidation for compounds 1–10, 13–16, 19–25, 29–30 and 33–35. See DOI: 10.1039/c7sc00475c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |