One-step analysis of glucose and acetylcholine in water based on the intrinsic peroxidase-like activity of Ni/Co LDHs microspheres†
Li
Su
*ab,
Xinai
Yu
ab,
Wenjie
Qin
c,
Wenpei
Dong
ab,
Chengke
Wu
ab,
Yan
Zhang
ab,
Guojiang
Mao
ab and
Suling
Feng
*ab
aCollaborative Innovation Center of Henan Province for Green Manufacturing of Fine Chemicals, Key Laboratory of Green Chemical Media and Reactions, Ministry of Education, Henan Normal University, Xinxiang, Henan 453007, P. R. China. E-mail: suli@htu.edu.cn
bHenan Key Laboratory of Green Chemical Media and Reactions, School of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang, Henan 453007, P. R. China
cQinghai Institute for Products Quality Supervision & Test, Xining, Qinghai 810000, P. R. China
First published on 12th November 2016
Abstract
In the present study, a simple strategy was developed for Ni/Co layered double hydroxides (LDHs) as a substitute for natural peroxidase. The obtained Ni/Co LDHs exhibited ease of preparation, low-cost, and water-solubility; importantly, this material showed high catalytic activity in neutral pH solutions (phosphate buffer, Tris–HCl buffer, and even water). Benefitting from Ni/Co LDHs having a similar pH and temperature with specificity oxidase, such as glucose oxidase, choline oxidase, acetylcholinesterase, etc., a novel one-step method for a biosensor was developed in water. Glucose detection was selected as an application model system to evaluate the performance of this method, which showed a linear detection range from 0.5 μM to 100 μM with a detection limit (DL) of 0.1 μM. We also extended the one-step method to detect acetylcholine (ACh) by taking advantage of the specific catalytic reaction of acetylcholinesterase (AChE) and choline oxidase (ChOx). The linear detection range was from 10 μM to 150 μM with the DL of 1.62 μM. The proposed method had ease of operation, simple steps, and was rapid for glucose and ACh detection in real samples. On the basis of these advantages and virtues, Ni/Co LDHs could become attractive nanozymes in biotechnology and bioassays, and create a great influence on the next generation of enzyme mimetic systems.
1. Introduction
Natural enzymes possess high catalytic capacity and substrate specificity under mild conditions.1 However, the practical application of natural enzymes is hampered by their disadvantages such as high cost, low operational stability (denaturation and deactivation), and inability to tolerate harsh reaction conditions.2 Therefore, artificial enzymes that mimic the complexity and function of natural enzymes have received attention over the past two decades.3 Nanozymes, which are nanomaterials with enzyme-like activity, have attracted enormous interest from researchers due to their unique properties such as large surface area for further modification, size- (shape-, composition-) dependent catalytic activities, smart response to external stimuli, and the capability for self-assembly.4–7 To date, several nanomaterials, such as metal nanomaterials,8–13 metal oxide nanomaterials,14–20 carbon nanomaterials,21–27 disulfide-based nanomaterials,28,29 and BiOI hierarchical nanoflowers,30 have been discovered to possess peroxidase-like activity. As compared to horse radish peroxidase (HRP), the nanozymes may serve as promising candidates with advantages of controlled synthesis with low cost, tunability in catalytic activities, and high stability against stringent conditions.However, despite these favorable properties, one of the main shortcomings that these reported peroxidase mimetics is that its optimum reaction occurs in acidic solutions, which results in the peroxidase mimetics and natural enzyme working separately to catalyze single isolated reactions rather than coupled tandem reactions. For example, Fe3O4 magnetic nanoparticles,15 Au nanoparticles,8 carbon nanodots,24 and WS2 nanosheet31 were used as peroxidase mimics to detect glucose. Their strategies for the detection of glucose were all carried out through a separate two-step method. In the first step, glucose oxidase was used to catalyze glucose with molecular oxygen to produce H2O2 under mild conditions (pH = 7, 37 °C). In the second step, the peroxidase mimic and a colorimetric substrate were added to the above solution and incubated under acidic conditions. Similar to the glucose detection, the analysis of xanthine,32 acetylcholine (ACh),33 sarcosine,34 and D-alanine35 based on peroxidase mimics was also carried out through the separate two-step method. Though this analytical approach exhibited good sensitivity, selectivity, and accuracy, such a separation step was tedious, complex, time-consuming, and even resulting in limited overall efficiency of the enzymatic reactions.
Therefore, great efforts have been devoted to overcome the abovementioned limitations. Only a few studies about nanozymes exhibiting peroxidase-like activity at neutral pH have been reported. For example, Qu et al. reported a GO–AuNCs hybrid exhibiting high peroxidase-like catalytic activity over a broad pH range,36 and Liu and co-workers reported that Au@Ag heterogeneous nanorods had a high catalytic activity in nearly neutral pH and a one-pot method for the detection of glucose was developed.37 However, these noble metal-based nanozymes are typically expensive and complex. Thus, it is still necessary to develop simple and cheap peroxidase mimics that are able to exhibit catalytic activity in neutral solutions.
To tackle these challenges, herein, we report a facile chemical co-precipitation method to prepare Ni/Co layered double hydroxides (LDHs), which exhibit peroxidase-like activity at neutral pH (phosphate buffer, Tris–HCl buffer, and even water). Although other LDHs, including Co–Al LDHs,38 DNA–LDH,39 and Fe–Ni LDHs-hemin40 possessing peroxidase-like activity have been reported, the preparation process of these LDHs was complex. As peroxidase mimics, Ni/Co LDHs showed several advantages, such as ease of preparation, low-cost, and water-solubility. To demonstrate the promising application of the synthesized Ni/Co LDHs, a novel one-step method for glucose detection was developed in water. Furthermore, we also extended this one-step method to the detection of ACh by taking advantage of the specific catalytic reaction of acetylcholinesterase (AChE) and choline oxidase (ChOx). The promising one-step method had ease of operation, simple steps, and a rapid processing time.
2. Experimental
2.1. Materials
30% H2O2, Co(NO3)2·6H2O, Ni(NO3)2·6H2O, and NH3·H2O were purchased from Tianjin Guangfu Chemical Reagent Factory (Tianjin, China). 3,3′,5,5′-Tetramethylbenzidine (TMB), 2,2′-azinobis(3-ethylbenzothiazo-line-6-sulfonic acid)diammonium salt (ABTS), o-phenylenediamine (OPD), glucose oxidase (GOx, 50 KU), acetylcholinesterase (AChE, from Electrophorus electricus) and choline oxidase (ChOx, from Alcaligenes sp.) were purchased from Sigma-Aldrich. Glucose, fructose, lactose and maltose were purchased from Sangon Biotech (Shanghai). Acetylcholine chloride (ACh) was purchased from Acros. Glycine, cystine, tryptophan, threonine, and proline were purchased from Adamas.All other chemicals were of analytical reagent grade and used without further purification. Deionized water was used throughout the experiments.
2.2. Apparatus and characterization
A TU-1900 double beam UV-vis spectrophotometer (Beijing Purkine General Instrument Co. Ltd, China) was used to implement kinetic experiments and measure absorbance. Transmission electron microscopy (TEM) images of Ni/Co LDHs were obtained using a Tecnai G2 F30 instrument. The surface morphology of Ni/Co LDHs was observed using field emission scanning electron microscopy (FE-SEM Hitachi S-4800, Japan). The X-ray diffraction (XRD) pattern of Ni/Co LDHs was obtained with a X'Pertpro Philips X-ray diffractometer with Cu Kα radiation (λ = 0.154056). Electron Spin Resonance (ESR) measurements were acquired using a JES-FA200 electron spin resonance spectrometer.2.3. Preparation of Ni/Co LDHs
The Ni/Co LDHs microspheres were fabricated by a facile one-step chemical co-precipitation method. 10 mL 0.1 mol L−1 Co(NO3)2·6H2O and 20 mL 0.1 mol L−1 Ni(NO3)2·6H2O were uniformly mixed, and 1 mL 35% NH3·H2O was added into the abovementioned mixed solution drop by drop. The precursor was sealed in a 50 mL glass bottle for 3 h at room temperature. The sample was collected and washed by centrifugation for several cycles and the as-prepared Ni/Co LDHs sample was obtained by washing with deionized water and absolute ethanol several times; then, the obtained precipitate was dried in a vacuum oven at 60 °C overnight.2.4. Kinetic analysis
The peroxidase activity of Ni/Co LDHs was observed in 3 mL of reaction solution with 0.03 mg mL−1 Ni/Co LDHs at 37 °C. The catalytic experiments were performed when the concentration of substrate ABTS was fixed and the concentration of H2O2 was varied or vice versa. Kinetic measurements were carried out in time course mode by monitoring the absorbance change at 417 nm for 5 min on a TU-1900 UV-vis spectrophotometer. The Michaelis–Menten constant was calculated using a Lineweaver–Burk plot:| 1/ν = Km/Vmax(1/[S] + 1/Km) | (1) |
2.5. Assay of glucose
Glucose detection was performed by a facile one-step method. The mixed solution, which included 5 μL 10 mg mL−1 GOx, 100 μL of different concentrations of glucose, 50 μL 10 mg mL−1 ABTS, 100 μL 1 mg mL−1 Ni/Co LDHs and 2.75 mL deionized water, was incubated at 37 °C for 30 min and absorbance at 417 nm was measured.The serum samples were treated by ultrafiltration on a 10 kDa ultrafiltration membrane and centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 40 min, and then the supernatant was diluted 40 times before measurement.
000 rpm for 40 min, and then the supernatant was diluted 40 times before measurement.
2.6. Assay of ACh
The detection of ACh was also performed by the one-step method. The coupled reaction was carried out by adding 40 μL 2 U mL−1 ChOx, 60 μL 2 U mL−1 AChE, 100 μL of different concentrations of ACh, 50 μL 10 mg mL−1 ABTS, 100 μL 1 mg mL−1 Ni/Co LDHs and 2.65 mL deionized water at 37 °C for 1 h; then, the absorbance at 417 nm was measured.The milk sample was prepared as follows: 20 g milk was added into 10 mL of 3.0 M HCl and mixed uniformly. The mixture was incubated at 70 °C for 3 h and shaken every half an hour. Then, the solution was adjusted to pH 3.5–4 with NaOH. After filtration with an ultrafiltration membrane having 0.45 μm pore size, the solution was quantitatively diluted with water to 50 mL; then the solution containing milk was diluted 40 times before measurement.
3. Results and discussion
3.1. Characterization of Ni/Co LDHs
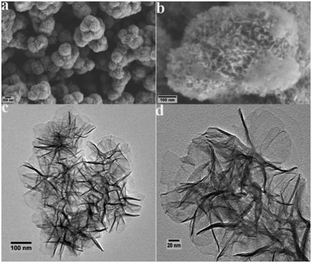
The crystalline structure of the as-obtained Ni/Co LDHs was examined by XRD characterization. From the XRD curves (Fig. S1, ESI†), the four characteristic peaks at 11.1°, 22.5°, 34.3°, and 60.9° were successfully attributed to the (003), (006), (012), and (110) lattice planes of the layered hydroxides phase, which are in accordance with the standard XRD peaks for Ni/Co LDHs.41,42 The SEM morphologies of Ni/Co LDHs are shown in Fig. 1a, which indicates that the products consist of uniform microspheres with the average size of about 470 nm. The detailed feature of the products is shown in Fig. 1b, indicating the 3D flower-like hierarchical architectures of the microspheres. To obtain more structural insight for the Ni/Co LDHs, TEM results were recorded for the as-synthesized products. In Fig. 1c, it can be seen that Ni/Co LDHs samples are spherical and consist of thin and crinkly nanosheets, which grow out from the core of the microspheres and form flower-like features. Details of the flower-like features are given in Fig. 1d, which indicate that the hybrid ultrathin nanosheets show flat, compact, and transparent features. The SAED pattern (Fig. S2, ESI†) shows well defined rings, which are an indication of the polycrystalline nature of Ni/Co LDHs. The EDX spectrum in Fig. S3 (ESI†) reveals that the single LDHs nanosheet mainly contains Ni, Co, and O elements. Furthermore, a Brunauer–Emmett–Teller (BET) analysis of Ni/Co LDHs gives a specific surface area of 77.8 m2 g−1 (Fig. S4, ESI†). This confirms that the LDHs possess a relative large specific surface area caused by the unique flower-like hierarchical configuration, which is beneficial to the catalytic property.3.2. The peroxidase-like catalytic activity of Ni/Co LDHs
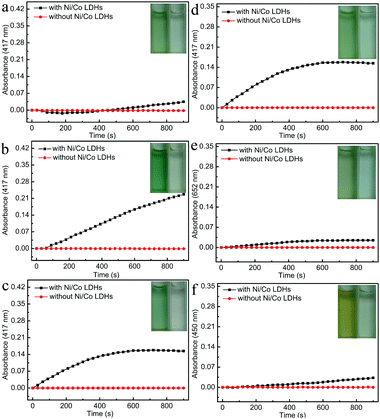
To demonstrate proof of principle, the peroxidase-like catalytic activity of Ni/Co LDHs was investigated under three different neutral pH solutions and compared with that of the absence of Ni/Co LDHs. As shown in Fig. 2a–c, the absorbance at 417 nm increased in the presence of Ni/Co LDHs, whereas there was no absorbance at 417 nm in the absence of Ni/Co LDHs, indicating Ni/Co LDHs possesses peroxidase-like activity at neutral pH. In order to determine the influence of neutral pH solution on the peroxidase-like activity of Ni/Co LDHs, phosphate buffer (PBS), Tris–HCl buffer and deionized water were selected for the contrast experiment. Firstly, as shown in Fig. 2a, Ni/Co LDHs could catalyze the reaction of the peroxidase substrate 2,2-azino-bis(3-ethylbenzothiazoline-6-sulfonicacid) (ABTS) in the presence of H2O2 at pH 7.0 PBS. However, the rate of catalysis was very slow and changed near 200 s. This was probably because the Co and Ni ions in the surface of Ni/Co LDHs conjugated with PO43− from PBS and exhibited strong coordination and low solubility (Co3(PO4)2Ksp = 10−35, Ni3(PO4)2Ksp = 10−32), which may spoil the native structure of Ni/Co LDHs and decrease the catalytic performance. Secondly, the influence of Tris–HCl buffer (pH = 7.0) on the peroxidase-like activity of Ni/Co LDHs is shown in Fig. 2b. The absorbance at 417 nm always increased linearly in 900 s in the presence of Ni/Co LDHs; however, the rate of catalysis was fast in Tris–HCl buffer. A terminating agent was added in order to terminate the on-going reaction, which led to a complex operational protocol. Lastly, the influence of deionized water on the peroxidase-like activity of Ni/Co LDHs is shown in Fig. 2c. The absorbance at 417 nm always increased in 600 s and then remained unchanged for the next 300 s, which indicated that the catalytic reaction was terminated without adding the terminating agent. This also reduced the complexity of the experiment. Furthermore, at the beginning of 360 s, the rate of catalysis was faster in deionized water than that in Tris–HCl buffer. Given all this, deionized water was selected as the optimal reaction medium for the catalysis reaction by Ni/Co LDHs.To further characterize the peroxidase-like activity of the Ni/Co LDHs, other typical peroxidase substrates were used in place of ABTS including 3,3′,5,5′-tetramethylbenzidine (TMB) and o-phenylenediamine (OPD). As shown in Fig. 2d–f, the oxidation of ABTS, TMB and OPD catalyzed by the Ni/Co LDHs formed green, blue and orange products and the maximum absorbance of the oxidation of ABTS, TMB and OPD was 417, 652 and 450 nm, respectively. The absorbance at 417, 652 and 450 nm, respectively, increased with time in the presence of Ni/Co LDHs, whereas there was no absorbance in the absence of Ni/Co LDHs. All results confirmed that the as-prepared Ni/Co LDHs exhibited intrinsic peroxidase-like activity. Furthermore, Fig. 2d–f also compares the catalytic activity of the reaction of ABTS, TMB and OPD with H2O2 using Ni/Co LDHs. Certainly, the reaction rate of ABTS with H2O2 was faster than that of TMB and OPD. This is because ABTS contains negatively charged sulfonic groups, giving a strong electrostatic affinity toward the positively charged Ni/Co LDHs.43 Therefore, ABTS, as the optimal peroxidase substrate and colorimetric indicator, was used to investigate the peroxidase-like activity of the Ni/Co LDHs and detect glucose and ACh.
Similarly to natural HRP, the catalytic activity of Ni/Co LDHs was also dependent on pH and temperature. Because deionized water was selected as the reaction medium, we only examined the peroxidase-like activity of Ni/Co LDHs varying the temperature from 10 °C to 65 °C. As shown in Fig. S5 (ESI†), the catalytic performance of Ni/Co LDHs was retained (above ∼80%) from 10 °C to 40 °C and it attained its highest catalytic activity at 20 °C. Therefore, the Ni/Co LDHs exhibited high activity over the broad temperature range. However, in order to make the detection of glucose and ACh easy to carry out, 37 °C was set as the optimal temperature.
3.3. Kinetic study and mechanism of Ni/Co LDHs
To investigate the mechanism of the peroxidase-like activity of Ni/Co LDHs, the apparent steady-state kinetic parameters of the reaction of ABTS and H2O2 were determined by changing the concentration of H2O2 and ABTS, respectively (Fig. S6a and b, ESI†). The enzyme kinetic parameters, including Michaelis–Menten constant (Km) and maximum initial velocity (Vmax), were obtained according to the Lineweaver–Burk equation. The results are shown in Table S1 (ESI†). The apparent Km values for the Ni/Co LDHs with H2O2 and ABTS were all larger than that of HRP, suggesting that the Ni/Co LDHs had a lower affinity for H2O2 and ABTS than HRP. To further investigate the mechanism of Ni/Co LDHs catalysis, the peroxidase-like activity of Ni/Co LDHs was measured under standard reaction conditions by varying the concentration of H2O2 at a fixed concentration of ABTS or vice versa. The double reciprocal plots of initial velocity against the concentration of one substrate were obtained over a range of concentrations of the second substrate (Fig. S6c and d, ESI†). The slopes of the lines were parallel, which was characteristic of a ping–pong mechanism, as observed for HRP.44 This indicated that Ni/Co LDHs bind and react with the first substrate and then release the first product before reacting with the second substrate. The possible product in the present system might originate from Ni/Co LDHs' catalytic ability toward the decomposition of H2O2 to generate reactive oxygen species (ROS). Then, the electron spin resonance (ESR) spectroscopy (as the direct analytical method) was used for the detection of ROS. The signal of 5,5-dimethyl-1-pyrroline N-oxide (DMPO)/˙OH was observed in the presence of Ni/Co LDHs (Fig. 3a), which indicating that Ni/Co LDHs could decompose H2O2 to generate ˙OH radicals, which could oxidize the peroxidase substrate ABTS. In addition, the signal of 2,2,6,6-tetramethyl-4-piperidine (TEMP)/1O2 was also observed in the presence Ni/Co LDHs (Fig. 3b), which verified that 1O2 was formulated via the addition of Ni/Co LDHs. Then, even in water, Ni/Co LDHs could catalyze the decomposition of H2O2 to generate ˙OH and 1O2, which resulted in the oxidation of ABTS.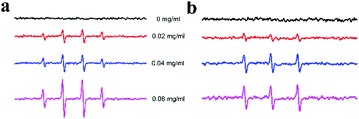 | ||
| Fig. 3 ESR spectra of the formation of ˙OH (a) and 1O2 (b) from the decomposition of H2O2 catalyzed using different concentrations of Ni/Co LDHs. | ||
3.4. One-step detection glucose using Ni/Co LDHs
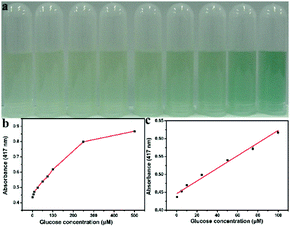
Since the peroxidase-like activity of Ni/Co LDHs is H2O2 concentration dependent and H2O2 is the main product of the glucose oxidase (GOx)-catalyzed reaction, a colorimetric detection of glucose could be developed using Ni/Co LDHs instead of HRP. Most studies have reported glucose detection based on peroxidase mimics, and most peroxidase mimics possess catalytic activity in acidic solutions; however, GOx would be denatured in an acid solution. Then, the two-step procedure was carried out in the following ways: GOx and glucose were incubated under neutral conditions for about 30 min, and then peroxidase mimic and colorimetric substrate were added to the above solution and incubated under acid conditions for about 20 min again. Despite the fact that the two-step method could avoid denaturing GOx, the procedure was tedious, complex and time-consuming. Therefore, the method developed using Ni/Co LDHs benefits from operation under physiological conditions and the presented colorimetric assay for glucose can be completed in only one-step. The detailed procedure of this one-step method was described in Experimental section 2.5. The results of glucose detection are shown in Fig. 4. Fig. 4b shows a typical glucose concentration–response curve. The linear range for glucose was from 0.5 μM to 100 μM (Fig. 4c) with a detection limit (DL) of 0.1 μM. This detection of glucose method based on the Ni/Co LDHs system not only gave a wider linear range and lower DL, but also showed easy reaction conditions, rapid process and simpler steps compared with many other nanozymes-based glucose sensors (Table 1). To test the specificity of this method, control experiments were conducted using fructose, lactose and maltose instead of glucose (Fig. S7, ESI†). As shown in Fig. S7 (ESI†), the absorbance of glucose is evidently higher than the glucose analogues, although the concentration of these analogs are about 5-fold higher than that of glucose due to the specificity of GOx. Comparing to fructose and lactose, maltose has less interference in the detection of glucose.39,45| Enzyme mimic | Optimum condition | Method | Time | Linear range (μM) | LD (μM) | Ref. |
|---|---|---|---|---|---|---|
| C60 | pH 3.5 | Two-step | 2 h 30 min | 1.0–40 | 0.5 | 25 |
| 45 °C | ||||||
| WS2 nanosheets | pH 6.9 | Two-step | 1 h | 5–300 | 2.9 | 31 |
| 60 °C | ||||||
| Carbon nitrides | pH 3.0 | Three-step | 1 h 55 min | 5–100 | 1.0 | 27 |
| 60 °C | ||||||
| CoFe LDHs | pH 4.0 | Two-step | 1 h 30 min | 1–10 | 0.6 | 46 |
| 30 °C | ||||||
| Fe-MIL-88NH2 MOFs | pH 4.0 | Two-step | 50 min | 2–300 | 0.48 | 47 |
| 45 °C | ||||||
| MoS2/graphene oxide hybrid | pH 4.0 | Two-step | 1 h 30 min | 1–50 | 0.83 | 48 |
| 37 °C | ||||||
| Heterogeneous Au@Ag | pH 6.5 | One-step | 30 min | 50–20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
39 | 37 |
| 25 °C | ||||||
| Ni/Co LDHs | Water | One-step | 30 min | 0.5–100 | 0.1 | This work |
| 37 °C |
To confirm the practicability, the method was used in different human serum samples. According to the calibration curve, the results by our method were in good agreement with the readings measured by the GOD-PAP biochemical analyzer in the hospital (Table S2, ESI†).
3.5. One-step detection ACh using Ni/Co LDHs
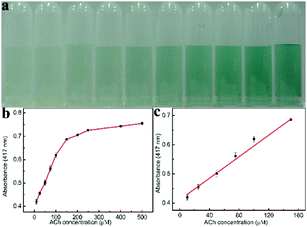
Furthermore, we also extended the one-step method to detect ACh. Since H2O2 could be generated in the presence of both AChE and ChOx in the solution containing ACh, ACh was also detected via the peroxidase mimic Ni/Co LDHs. Based on the peroxidase-like activity of Ni/Co LDHs under physiological conditions, we employed a one-step process to detect ACh. This is because the optimum conditions for the peroxidase-like activity of Ni/Co LDHs agree with that of the activity of AChE and ChOx. In this one-step process, ACh was firstly converted to choline by AChE, and the ChOx could further catalytically oxidize choline to produce H2O2, which could oxidize ABTS in the presence of Ni/Co LDHs to produce the green-colored ox-ABTS. Therefore, the color change and the absorbance of ox-ABTS were employed to measure ACh content. Fig. 5b shows a typical ACh concentration–response curve. The linear range for ACh was from 10 μM to 150 μM with the DL of 1.62 μM (Fig. 5c). This one-step method based on Ni/Co LDHs gave a lower DL than the two-step method using platinum nanoparticles (2.84 μM).33 The selectivity of the colorimetric sensing of ACh was investigated by detecting in the presence of some interfering substances including glycine, cystine, tryptophan, threonine, and proline (Fig. S8, ESI†). As shown in Fig. S8 (ESI†), ACh induced an increased absorbance. While, except that cysteine and proline induced less interference, all of the other substance showed lesser interference. This method was employed to detect ACh in milk in order to evaluate its applicability. The results of the determination and recovery are shown in Table S3 (ESI†). From Table S3 (ESI†), it can be seen that the average recoveries for ACh ranged from 90.2 to 104.8%, indicating that the present colorimetric assay has great potential for ACh detection in food inspection applications.4. Conclusions
In summary, by a facile one-step chemical co-precipitation method, we have obtained Ni/Co LDHs that exhibit peroxidase-like activity. Ni/Co LDHs, as a peroxidase mimic, exhibits several advantages. Firstly, compared to natural enzymes, Ni/Co LDHs has advantages of ease of preparation, low-cost, and water-solubility. Secondly, Ni/Co LDHs showed high catalytic activity over a broad temperature range, from 10 °C to 40 °C. Lastly, but most importantly, Ni/Co LDHs had high catalytic activity at neutral pH. Because of the similar optimal conditions (neutral pH, 37 °C) for the catalytic activity of Ni/Co LDHs with GOx, AChE and ChOx, we developed a one-step method for the detection of glucose and ACh in water, which showed shorter reaction times, simpler steps and ease of operation. Our study will facilitate the utilization of the intrinsic peroxidase activity of Ni/Co LDHs in biotechnology and bioassays, and make a great influence on the next generation of enzyme mimetic systems.Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (21405034, 21505032). Program for Innovative Research Team in University of Henan Province (17IRTSTHN001). Key scientific research project of Education Department of Henan Province (14B150020). Key scientific research project of Higher Education of Henan Province (15A150016). Dr start-up project funding of Henan Normal University (qd13003).Notes and references
- R. Wolfenden and M. J. Snider, Acc. Chem. Res., 2001, 34, 938–945 CrossRef CAS PubMed.
- E. Shoji and M. S. Freund, J. Am. Chem. Soc., 2001, 123, 3383–3384 CrossRef CAS PubMed.
- Y. Murakami, J.-i. Kikuchi, Y. Hisaeda and O. Hayashida, Chem. Rev., 1996, 96, 721–758 CrossRef CAS PubMed.
- Y. Lin, J. Ren and X. Qu, Adv. Mater., 2014, 26, 4200–4217 CrossRef CAS PubMed.
- Y. Song, W. Wei and X. Qu, Adv. Mater., 2011, 23, 4215–4236 CrossRef CAS PubMed.
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- J. Xie, X. Zhang, H. Wang, H. Zheng and Y. Huang, TrAC, Trends Anal. Chem., 2012, 39, 114–129 CrossRef CAS.
- Y. Jv, B. X. Li and R. Cao, Chem. Commun., 2010, 46, 8017–8019 RSC.
- W. W. He, Y. Liu, J. S. Yuan, J. J. Yin, X. C. Wu, X. N. Hu, K. Zhang, J. B. Liu, C. Y. Chen, Y. L. Ji and Y. T. Guo, Biomaterials, 2011, 32, 1139–1147 CrossRef CAS PubMed.
- H. Jiang, Z. H. Chen, H. Y. Cao and Y. M. Huang, Analyst, 2012, 137, 5560–5564 RSC.
- J. B. Liu, X. N. Hu, S. Hou, T. Wen, W. Q. Liu, X. Zhu, J. J. Yin and X. C. Wu, Sens. Actuators, B, 2012, 166, 708–714 CrossRef.
- Y. Nangia, B. Kumar, J. Kaushal and C. R. Suri, Anal. Chim. Acta, 2012, 751, 140–145 CrossRef CAS PubMed.
- S. Cai, C. Qi, Y. Li, Q. Han, R. Yang and C. Wang, J. Mater. Chem. B, 2016, 4, 1869–1877 RSC.
- L. Hong, A.-L. Liu, G.-W. Li, W. Chen and X.-H. Lin, Biosens. Bioelectron., 2013, 43, 1–5 CrossRef CAS PubMed.
- H. Wei and E. Wang, Anal. Chem., 2008, 80, 2250–2254 CrossRef CAS PubMed.
- W. Chen, J. Chen, A. L. Liu, L. M. Wang, G. W. Li and X. H. Lin, ChemCatChem, 2011, 3, 1151–1154 CrossRef CAS.
- J. S. Mu, Y. Wang, M. Zhao and L. Zhang, Chem. Commun., 2012, 48, 2540–2542 RSC.
- Q. Y. Liu, Y. T. Yang, H. Li, R. R. Zhu, Q. Shao, S. G. Yang and J. J. Xu, Biosens. Bioelectron., 2015, 64, 147–153 CrossRef CAS PubMed.
- W. Chen, J. Chen, Y. B. Feng, L. Hong, Q. Y. Chen, L. F. Wu, X. H. Lin and X. H. Xia, Analyst, 2012, 137, 1706–1712 RSC.
- C. Ray, S. Dutta, S. Sarkar, R. Sahoo, A. Roy and T. Pal, J. Mater. Chem. B, 2014, 2, 6097–6105 RSC.
- R. Zhang, S. He, C. Zhang and W. Chen, J. Mater. Chem. B, 2015, 3, 4146–4154 RSC.
- L. Lin, X. Song, Y. Chen, M. Rong, T. Zhao, Y. Wang, Y. Jiang and X. Chen, Anal. Chim. Acta, 2015, 869, 89–95 CrossRef CAS PubMed.
- Y. J. Song, K. G. Qu, C. Zhao, J. S. Ren and X. G. Qu, Adv. Mater., 2010, 22, 2206–2210 CrossRef CAS PubMed.
- W. B. Shi, Q. L. Wang, Y. J. Long, Z. L. Cheng, S. H. Chen, H. Z. Zheng and Y. M. Huang, Chem. Commun., 2011, 47, 6695–6697 RSC.
- R. M. Li, M. M. Zhen, M. R. Guan, D. Q. Chen, G. Q. Zhang, J. C. Ge, P. Gong, C. R. Wang and C. Y. Shu, Biosens. Bioelectron., 2013, 47, 502–507 CrossRef CAS PubMed.
- S. Y. Zhu, X. E. Zhao, J. M. You, G. B. Xu and H. Wang, Analyst, 2015, 140, 6398–6403 RSC.
- T. Lin, L. Zhong, J. Wang, L. Guo, H. Wu, Q. Guo, F. Fu and G. Chen, Biosens. Bioelectron., 2014, 59, 89–93 CrossRef CAS PubMed.
- T. Lin, L. Zhong, L. Guo, F. Fu and G. Chen, Nanoscale, 2014, 6, 11856–11862 RSC.
- S. Cai, Q. Han, C. Qi, Z. Lian, X. Jia, R. Yang and C. Wang, Nanoscale, 2016, 8, 3685–3693 RSC.
- P. Ju, Y. Xiang, Z. Xiang, M. Wang, Y. Zhao, D. Zhang, J. Yu and X. Han, RSC Adv., 2016, 6, 17483–17493 RSC.
- T. R. Lin, L. S. Zhong, Z. P. Song, L. Q. Guo, H. Y. Wu, Q. Q. Guo, Y. Chen, F. F. Fu and G. N. Chen, Biosens. Bioelectron., 2014, 62, 302–307 CrossRef CAS PubMed.
- X. X. Wang, Q. Wu, Z. Shan and Q. M. Huang, Biosens. Bioelectron., 2011, 26, 3614–3619 CrossRef CAS PubMed.
- S. B. He, G. W. Wu, H. H. Deng, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Biosens. Bioelectron., 2014, 62, 331–336 CrossRef CAS PubMed.
- J. Lan, W. Xu, Q. Wan, X. Zhang, J. Lin, J. Chen and J. Chen, Anal. Chim. Acta, 2014, 825, 63–68 CrossRef CAS PubMed.
- W. Haider, A. Hayat, Y. Raza, A. A. Chaudhry, R. Ihtesham Ur and J. L. Marty, RSC Adv., 2015, 5, 24853–24858 RSC.
- Y. Tao, Y. Lin, Z. Huang, J. Ren and X. Qu, Adv. Mater., 2013, 25, 2594–2599 CrossRef CAS PubMed.
- L. Han, C. Li, T. Zhang, Q. Lang and A. Liu, ACS Appl. Mater. Interfaces, 2015, 7, 14463–14470 CAS.
- L. Chen, B. Sun, X. Wang, F. Qiao and S. Ai, J. Mater. Chem. B, 2013, 1, 2268–2274 RSC.
- L. Chen, K. Sun, P. Li, X. Fan, J. Sun and S. Ai, Nanoscale, 2013, 5, 10982–10988 RSC.
- F.-T. Zhang, X. Long, D.-W. Zhang, Y.-L. Sun, Y.-L. Zhou, Y.-R. Ma, L.-M. Qi and X.-X. Zhang, Sens. Actuators, B, 2014, 192, 150–156 CrossRef CAS.
- T. Li, G. Li, L. Li, L. Liu, Y. Xu, H. Ding and T. Zhang, ACS Appl. Mater. Interfaces, 2016, 8, 2562–2572 CAS.
- H. Chen, F. Cai, Y. Kang, S. Zeng, M. Chen and Q. Li, ACS Appl. Mater. Interfaces, 2014, 6, 19630–19637 CAS.
- L. Qian, Z. Lu, T. Xu, X. Wu, Y. Tian, Y. Li, Z. Huo, X. Sun and X. Duan, Adv. Energy Mater., 2015, 5, 1500245 CrossRef.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- W. Zhang, D. Ma and J. Du, Talanta, 2014, 120, 362–367 CrossRef CAS PubMed.
- Y. W. Zhang, J. Q. Tian, S. Liu, L. Wang, X. Y. Qin, W. B. Lu, G. H. Chang, Y. L. Luo, A. M. Asiri, A. O. Al-Youbi and X. P. Sun, Analyst, 2012, 137, 1325–1328 RSC.
- Y. L. Liu, X. J. Zhao, X. X. Yang and Y. F. Li, Analyst, 2013, 138, 4526–4531 RSC.
- J. Peng and J. Weng, Biosens. Bioelectron., 2017, 89, 652–658 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S8 and Tables S1–S3. See DOI: 10.1039/c6tb02273a |
| This journal is © The Royal Society of Chemistry 2017 |