DOI:
10.1039/C7FO01381G
(Paper)
Food Funct., 2018,
9, 355-363
In vitro inhibition of pancreatic α-amylase by spherical and polygonal starch nanoparticles†
Received
6th September 2017
, Accepted 22nd November 2017
First published on 23rd November 2017
Abstract
Nanoparticles are novel and fascinating materials for tuning the activities of enzymes. In this study, we investigated the influence of spherical and polygonal starch nanoparticles (SNPs) on α-amylase activity and revealed the reaction mechanisms by ultraviolet-visible spectrophotometry, fluorescence spectroscopy, and circular dichroism (CD) spectroscopy. We discovered that both spherical and polygonal SNPs could inhibit the α-amylase activity, with half-inhibitory concentration values of 0.304 and 0.019 mg mL−1, respectively. Furthermore, spherical and polygonal SNPs followed competitive and mixed competitive inhibition mechanisms, respectively. The fluorescence data indicated that static quenching was dominant in the interaction between SNPs and α-amylase. The CD results demonstrated that the inhibition of α-amylase by SNPs was accompanied by the decreased intensity of the CD spectra of α-amylase. Our findings provide a novel strategy to inhibit α-amylase to reduce the digestion of starch, thus managing blood glucose levels.
1. Introduction
Type 2 diabetes is a complex metabolic and endocrine disorder characterized by a high blood sugar level, which results from the absolute or the relative lack of insulin secretion from pancreatic β cells.1–3 Type 2 diabetes rates have been reported to be increasing all over the world, thus causing grave consequences to human health.4,5 Moreover, type 2 diabetes initiates complications, such as cardiovascular disease, heart attack, and stroke, thereby seriously reducing the patients’ quality of life and even leading to death.6,7 One of the main sources of postprandial glucose in the blood is the absorption of glucose digested from starch by α-amylase and α-glucosidase.8 The postprandial starch digestive enzymes with high activity digest the carbohydrates in our food and convert them to glucose, which is absorbed in the bloodstream. Therefore, the inhibition of starch digestive enzymes is an attractive strategy for the control of postprandial hyperglycemia.
Drugs such as acarbose and metformin are available for inhibiting α-amylase and therefore decreasing blood glucose values. Natural amylase inhibitors, such as polyphenols, flavonoids, and polysaccharides, have drawn considerable interest in both academia and industry because of the potential side effects of the synthesized drugs.9,10 Recently, green tea extract has been reported to inhibit the α-amylase activity with an inhibitory percentage of 63.5% and a half-inhibitory concentration (IC50) value of 2.07 mg ml−1.11 Fei et al. (2014) reported that the IC50 values of Oolong tea polyphenols, epigallocatechin gallate, and epigallocatechin 3-O-(3-O-methyl) gallate isolated from Oolong tea against α-amylase were 0.375, 0.350, and 0.572 mg mL−1, respectively.12 Tannic acid, chlorogenic acid, and caffeic acid in young apple polyphenols showed inhibitory effects against α-amylase with IC50 values of 0.30, 1.96, and 3.69 mg mL−1, respectively.13 Although polyphenols exhibited excellent properties to inhibit α-amylase activity, natural polyphenols are expensive and tend to decompose easily in the gastrointestinal tract, thus limiting their application.14,15 Therefore, seeking natural, cost-effective, and efficient α-amylase inhibitors is a challenging task.
In recent years, a novel method of enzyme inhibition through the addition of nanoparticles has attracted the interest of researchers. Many inorganic nanomaterials, such as silica, fullerene, and zinc oxide, have been thoroughly examined. For example, Vertegel et al. (2004)16 reported that the adsorption of lysozymes onto the surface of silica nanoparticles with a diameter of 4–100 nm resulted in the loss of their activity. Song et al. (2010)17 found that fullerene nanoparticles effectively inhibited the polymerase activity. Cha et al. (2015)18 reported that zinc oxide nanoparticles exhibited a strong inhibiting activity of β-galactosidase. Dhobale et al. (2008)19 found that the inhibitory efficacy of zinc oxide nanoparticles against α-amylase was up to 49% at a concentration of 20 μg mL−1.
Nanoparticles prepared from natural biopolymers have many significant advantages, such as renewable nature, low cost, biodegradability, and biocompatibility. Thus, biopolymer nanoparticles are widely studied in many fields, including food, agriculture, medicine, and cosmetics.20 Specifically, the interaction between natural polymer nanoparticles and enzymes has attracted considerable technological interest. Previously, we reported that chitin nanowhiskers as adsorbents caused substantial improvements in the enzymatic activity of lysozymes alone.21 By contrast, starch nanoparticles (SNPs) are potential inhibitors against tyrosinase, with the IC50 values of hollow nanoparticles, amylopectin nanoparticles, corn starch nanoparticles, and tapioca starch nanoparticles of 0.308, 0.669, 1.490, and 4.774 μM, respectively.22 However, as far as we know, the effect of SNPs on α-amylase activity has never been evaluated. Thus, this research aimed to determine the effect of SNPs on α-amylase activity and to explore the reaction mechanisms by fluorescence emission spectroscopy, transmission electron microscopy (TEM), confocal laser scanning microscopy (CLSM), Fourier transform infrared spectroscopy (FTIR), and circular dichroism (CD) spectroscopy.
2. Results and discussion
2.1 Morphology of SNPs and α-amylase/SNPs
TEM images (Fig. 1) revealed that spherical SNPs exhibited spherical shapes with a particle size of 30–50 nm, and polygonal SNPs showed irregularly angular shapes with a size range of 40–100 nm. Note that the morphology of spherical SNPs or polygonal SNPs showed some differences by incubating with α-amylase for 2 h. These halos appeared around nanoparticles’ edges, which was probably due to the coating of SNPs by α-amylase (Fig. 1B and E). The other amylase/SNPs appeared to have a porous structure (Fig. 1C and F). The result may be attributed to the adsorption of the amount of enzymes on the SNP surface, thus leading to the hydrolysis of SNPs. A similar result was reported by Liu et al.23 in which SNPs digested with α-amylase and amyloglucosidase for 120 min exhibited random holes. To further demonstrate the combination of α-amylase and SNPs, the CLSM technique was used in this work. As shown in Fig. 2B and C, SNPs displayed obvious green fluorescence from the FITC dye, and α-amylase showed red fluorescence from the Rhodamine B dye. α-Amylase/SNPs exhibited yellow fluorescence (Fig. 2D), which indicated that α-amylase was adsorbed on the surface of SNPs.
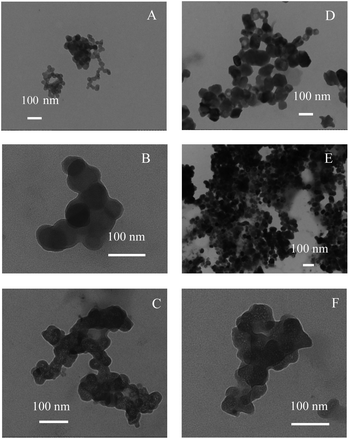 |
| | Fig. 1 TEM images of spherical SNPs (A) and polygonal SNPs (D), and images of α-amylase/spherical SNPs (B and C) and α-amylase/polygonal SNPs (E and F). | |
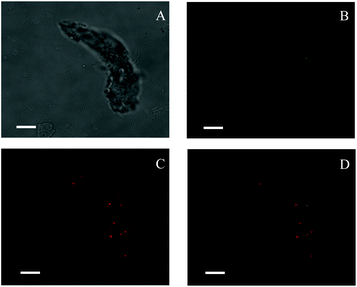 |
| | Fig. 2 Confocal laser scanning micrographs of enzyme-bound SNPs: (A) optical microscopy image under a bright field; (B) green fluorescence showing FITC-labeled SNPs; (C) red fluorescence showing Rhodamine B labeling α-amylase; (D) a merged micrograph showing both. The enzyme-bound SNPs were prepared with 0.25 mg mL−1 α-amylase and 1.0 mg mL−1 SNPs in the mixture system. All scale bars are 7.5 μm. | |
2.2 Effect of starch nanoparticles on the activity of α-amylase
The inhibition effects of different concentrations of spherical and polygonal SNPs on α-amylase were investigated. Both SNPs exhibited potential inhibition against α-amylase (Fig. 3). Enhanced inhibition against α-amylase could be achieved by increasing the concentrations of SNPs. Soluble starch was substituted with spherical or polygonal SNPs and the enzyme activity of α-amylase was 6.6 U mg−1 and 5.4 U mg−1, respectively. The results suggested that the hydrolysis activity of α-amylase on SNPs was low, thus the enzyme activity of α-amylase on SNPs was not taken into account in the following experiments. The IC50 values for the α-amylase activity inhibited by spherical and polygonal SNPs were determined to be 304 μg mL−1 and 19 μg mL−1 (Table 1), respectively. The relative inhibitory activity against α-amylase of polygonal SNPs is more efficient than that of spherical SNPs. The enhanced inhibitory activity for polygonal SNPs may be ascribed to a relatively loose structure. The polygonal SNPs could not only compete with the substrate but also entrap amylase in the polygonal SNPs effectively. Thus, the effective amount of α-amylase reacting with soluble starch decreased and resulted in the low activity of α-amylase. In addition, the enhanced inhibitory activity compared with spherical SNPs may also be originated from the specific apex geometry. Similarly, Cha et al. also reported that pyramidal zinc oxide nanoparticles exhibited greater inhibition against β-galactosidase than spherical nanoparticles because of their better geometrical match with the enzyme surface and sharper apexes and edges.18
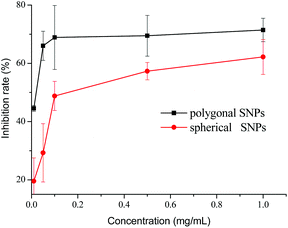 |
| | Fig. 3 Inhibitory effects of spherical and polygonal starch nanoparticles (SNPs) on α-amylase. | |
Table 1 Comparison of various samples for the half-inhibitory concentration (IC50) of α-amylase
| Inhibitor |
IC50 |
Ref. |
| Spherical SNPs |
304 ± 40 μg mL−1 |
Our paper |
| Polygonal SNPs |
19 ± 0.34 μg mL−1 |
| Acarbose |
6.9 ± 0.8 μg mL−1 |
Yilmazer-Musa et al. (2012)26 |
| ZnO nanoparticles |
20 μg mL−1 |
Dhobale et al. (2008)19 |
| Oolong tea polyphenols |
375 μg mL−1 |
Fei Qunqin et al. (2014)25 |
| EGCG |
350 μg mL−1 |
| EGCG3′′Me |
572 μg mL−1 |
| Proanthocyanidin-rich sumac sorghum bran extract |
1.4 μg mL−1 |
Hargrove et al. (2010)27 |
| Proanthocyanidin-free black sorghum bran extract |
11.4 μg mL−1 |
| Black sorghum bran extract |
18.8 μg mL−1 |
| Condensed tannins I |
105 ± 4 μM |
Gonçalves et al. (2010)28 |
| Condensed tannins II |
38 ± 4 μM |
| Condensed tannins III |
25 ± 4 μM |
| Green tea extract |
2070 μg mL−1 |
Miao Ming et al. (2015)11 |
| Tannic acid |
300 μg mL−1 |
Sun Lijun et al. (2015)13 |
| Chlorogenic acid |
1960 μg mL−1 |
| Caffeic acid |
3690 μg mL−1 |
| Grape seed extract |
8.7 ± 0.8 μg mL−1 |
Yilmazer-Musa et al. (2012)26 |
| Green tea extract |
34.9 ± 0.9 μg mL−1 |
| White tea extract |
378 ± 134 μg mL−1 |
The anti-amylase activity of the two kinds of SNPs was not as efficient as acarbose, which has ever been reported,24 but the SNPs exhibited exceptional inhibitory ability compared with other inhibitors.11,13,19,25 Recently, Miao et al. reported that the green tea extract inhibited α-amylase activity with an IC50 value of 2.07 mg ml−1.11 Fei et al. (2014) also reported that the IC50 values of Oolong tea polyphenols, epigallocatechin gallate, and epigallocatechin 3-O-(3-O-methyl) gallate against α-amylase were 0.375, 0.350, and 0.572 mg mL−1, respectively.25 Therefore, our results demonstrated that SNPs showed more potential inhibition ability against α-amylase than most of the natural polyphenols mentioned above. We also evaluated the effect of chitin nanowhiskers and found that they also played a role despite being weaker than SNPs (Fig. S1†). This might be attributed to the weak binding interaction between α-amylase and chitin nanowhiskers. In addition, α-amylase was unable to hydrolyze the β-1,4 glucan linkage of chitin, so, chitin nanowhiskers could act as an inhibitor of α-amylase.
2.3 Inhibition types of α-amylase activity by spherical starch nanoparticles and polygonal starch nanoparticles
The inhibition type of the spherical SNPs and polygonal SNPs in the α-amylase action was obtained through the analysis of the modified Dixon plots.29 The Dixon plots for spherical SNPs yielded a family of straight lines and the lines were parallel, thus indicating that spherical SNPs were a competitive-type inhibitor (Fig. 4A). By contrast, polygonal SNPs inhibited α-amylase in a mixed-competitive manner because the modified Dixon plots yielded a group of lines that intercepted in the second quadrant (Fig. 4B). The reason was probably that spherical SNPs with a dense double helix could immediately combine with the active domain of α-amylase to form α-amylase-SNPs, thus the combination of α-amylase and soluble starch was hampered. However, polygonal SNPs with a relatively loose structure could not only compete with the substrate but also entrap α-amylase in the polygonal SNPs effectively. Therefore, the two kinds of SNPs displayed different types of enzyme inhibition. A similar behaviour reported that EGCG3 and Oolong tea polyphenols inhibited α-amylase in a competitive mode.25 Dhital et al. showed that cellulose is a mixed competitive inhibitor of α-amylase.30
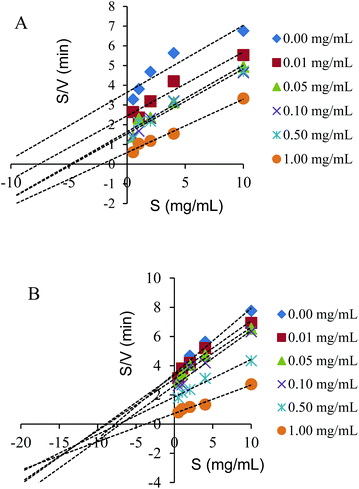 |
| | Fig. 4 Modified Dixon plots for the inhibition of spherical SNPs (A) and polygonal SNPs (B) on α-amylase. | |
2.4 UV spectral analysis
UV spectroscopy is an efficient tool to monitor the perturbations around the tryptophan and tyrosine residues and to probe the complex formation between proteins and ligands. For spherical SNPs, the spectral intensity of α-amylase/SNPs increased with the increase in the spherical SNP concentration (Fig. 5A), thus indicating the complex formation between amylase and spherical SNPs. The maximum absorption was slightly red-shifted (from 282 to 285 nm), and this outcome implied that the environment of the tryptophan and tyrosine residues of α-amylase changed and that the hydrophobicity of α-amylase increased. Khan et al. discovered that the absorbance maxima of protein at 280 nm increased with the increasing concentration of SnO2 nanoparticles.31 Conversely, for polygonal SNPs, the intensity of α-amylase/SNPs decreased compared with that of pure α-amylase (Fig. 5B). This result could be due to the penetration of enzymes into the polygonal SNPs. In agreement with this hypothesis, our data showed that the relative crystallinity of spherical SNPs was 55%, which formed a relatively dense double helix. By contrast, the crystallinity intensity of single-helical polygonal SNPs was weak with a relative crystallinity of 25% (Fig. S2†).
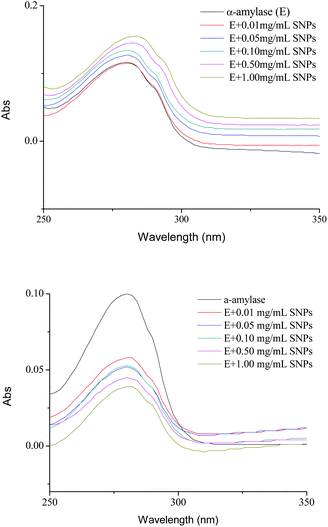 |
| | Fig. 5 UV spectra of α-amylase in the absence and presence of different concentrations of spherical SNPs (A) and polygonal SNPs (B). The final spectra were baseline-corrected by subtracting the corresponding SNPs spectra obtained under the same conditions. | |
2.5 Fluorescence quenching analysis of α-amylase with SNPs
Fluorescence quenching is widely applied to investigate the interactions between ligands and proteins.32 The fluorescence emission spectra of α-amylase in the absence and presence of SNPs with an excitation wavelength of 280 nm are shown in Fig. 4. SNPs at the determined concentrations (0.01–1.00 mg mL−1) in the experiment did not exhibit any fluorescence intensity (data not shown). The fluorescence intensity of α-amylase at around 335 nm decreased with the increasing concentration of SNPs (Fig. 6A and B). The extent of quenching of polygonal SNPs was even more significant. The plots of F0/F versus the concentrations of spherical and polygonal SNPs were determined to be linear (Fig. 6C and D). The values of KSV were equal to 1.01 × 108 L mol−1 s−1 and 2.10 × 107 L mol−1 s−1, respectively, according to eqn (1). The calculated kq for spherical SNPs and polygonal SNPs were equal to 1.77 × 1016 L mol−1 s−1 and 3.68 × 1015 L mol−1 s−1, respectively, according to eqn (2). kq was observed to be greater than the maximum scatter collision quenching content of various quenchers with biopolymers at 2.0 × 1010 L mol−1 s−1.33 Thus, static quenching was dominant in the interaction between spherical SNPs and polygonal SNPs and α-amylase. Similarly, Gonçalves et al. showed that condensed tannins associated with α-amylase were of a static type involving a stable interaction between them.28
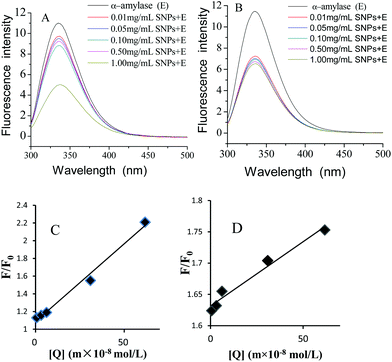 |
| | Fig. 6 Changes in the intrinsic α-amylase fluorescence at different concentrations of spherical SNPs (A) and polygonal SNPs (B), the Stern–Volmer curves of α-amylase quenched by spherical SNPs, (C) and polygonal SNPs (D). | |
2.6 FTIR spectroscopy analysis
FTIR was applied to investigate the functional groups of SNPs, α-amylase, and α-amylase/SNPs (Fig. 7). The enzyme showed a band at 1625 cm−1, which was ascribed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching (VC
O stretching (VC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, amide I), and a band at 1520 cm−1, which corresponded to the N–H bending (VN–H, amide II).34 The characteristic bands of amide I and amide II were observed for α-amylase and all α-amylase/SNPs. Hydroxyl groups were confirmed by the broad peaks appearing between 3000 and 3800 cm−1. The addition of SNPs resulted in a red-shift of the peak at 3000–3800 cm−1, indicating the formation of hydrogen bonding between α-amylase and SNPs. The band at 2927 cm−1 was due to the C–H stretching associated with the anhydroglucose of α-amylase/SNPs. The peak at 1364 cm−1 belonged to the C–O–H stretching vibration and shifted toward a low frequency with the increase in the spherical SNP concentration, thereby indicating an interaction of α-amylase with SNPs. Previously, researchers reported that the shifts in the intensity of 3300 cm−1 (N–H stretching), 1600–1690 cm−1 (C
O, amide I), and a band at 1520 cm−1, which corresponded to the N–H bending (VN–H, amide II).34 The characteristic bands of amide I and amide II were observed for α-amylase and all α-amylase/SNPs. Hydroxyl groups were confirmed by the broad peaks appearing between 3000 and 3800 cm−1. The addition of SNPs resulted in a red-shift of the peak at 3000–3800 cm−1, indicating the formation of hydrogen bonding between α-amylase and SNPs. The band at 2927 cm−1 was due to the C–H stretching associated with the anhydroglucose of α-amylase/SNPs. The peak at 1364 cm−1 belonged to the C–O–H stretching vibration and shifted toward a low frequency with the increase in the spherical SNP concentration, thereby indicating an interaction of α-amylase with SNPs. Previously, researchers reported that the shifts in the intensity of 3300 cm−1 (N–H stretching), 1600–1690 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), and 1480–1575 cm−1 (C–N stretching and N–H bending) indicated the secondary structural conformation of enzyme changes.35 Compared with those in the pure α-amylase, the changes in these regions demonstrated the alterations in the molecular microenvironment of chromophores of α-amylase when it interacted with the SNPs.
O stretching), and 1480–1575 cm−1 (C–N stretching and N–H bending) indicated the secondary structural conformation of enzyme changes.35 Compared with those in the pure α-amylase, the changes in these regions demonstrated the alterations in the molecular microenvironment of chromophores of α-amylase when it interacted with the SNPs.
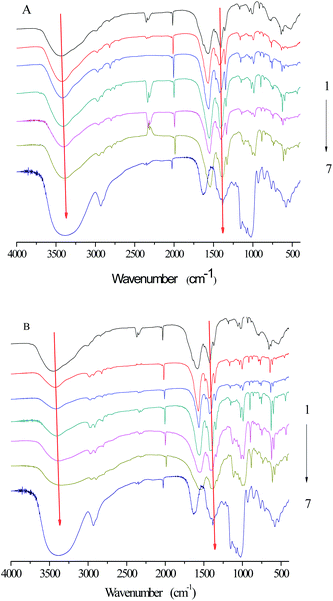 |
| | Fig. 7 FTIR spectra of SNPs, α-amylase, and α-amylase/SNPs prepared by spherical SNPs (A) and polygonal SNPs (B). 1–7: α-amylase (0.25 mg mL−1); α-amylase with SNP concentrations of 0.01, 0.05, 0.10, 0.50, and 1.00 mg mL−1; SNPs. | |
2.7 CD spectroscopy
CD spectroscopy is a powerful analytical tool for studying the interaction of proteins with other molecules in solution or adsorbed onto other molecules.36 To further explore the role of SNPs in the effects of α-amylase, the spectra of native α-amylase and α-amylase/SNPs were recorded between 190 nm and 250 nm (Fig. 8). SNPs at the determined concentrations (0.01–1.00 mg mL−1) in the experiment did not exhibit any CD spectroscopy intensity (data not shown). The typical signal of α-helix was observed among α-amylase and α-amylase/SNPs. However, the intensity of the negative ellipticities of α-amylase decreased with the increased concentration of SNPs. This observation suggested the formation of a complex between SNPs and α-amylase.
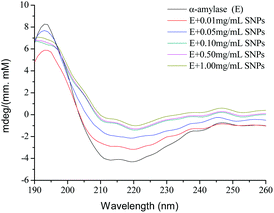 |
| | Fig. 8 CD spectra of α-amylase with different concentrations of polygonal starch nanoparticles. | |
2.8 Schematic illustration of the inhibition mechanism of SNPs
As shown in Fig. 9, small-sized SNPs, when mixed with α-amylase, could immediately combine with the active domain of α-amylase to form α-amylase-SNPs, and the combination of α-amylase and soluble starch was hampered by SNPs. Thus, the effective amount of α-amylase reacting with soluble starch decreased and resulted in the low activity of α-amylase. This result was similar to that of Patel, who reported that retrograded starch acts as a competitive inhibitor of amylase and decreased the catalytic efficiency of α-amylase.37 Polygonal SNPs with a relatively loose structure not only could compete with the substrate but also could entrap amylase in the polygonal SNPs effectively, thus leading to the reduction of hydrolysis of soluble starch.
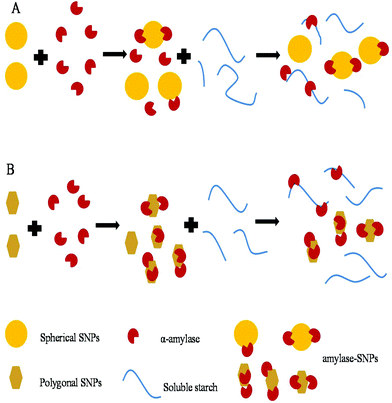 |
| | Fig. 9 Suggested inhibition mechanism of spherical SNPs (A) and polygonal SNPs (B) against α-amylase. | |
3. Experimental
3.1 Reagents and materials
Corn starch was supplied by Ingredient China Ltd (Guangdong, China). Soluble starch prepared from native potato starch was supplied by Tianjin BASF Chemical Co. Ltd (Tianjin, China). α-Amylase from porcine pancreas with an activity of 45 U mg−1 (E.C.3.2.1.1) was purchased from Sigma-Aldrich (China). One unit of α-amylase activity was defined as the amount of enzyme that released 1 μmol of reducing sugars per minute at 37 °C. Rhodamine B and fluorescein isothiocyanate (FITC) were obtained from Beijing Solarbio Science & Technology Co., Ltd (Beijing, China). Pullulanase (E.C.3.2.1.41, 27 U mg−1) was obtained from Novozymes Investment Co. Ltd (Beijing, China). Pullulanase activity was assayed following previously described procedures.38 3,5-Dinitrosalicylic acid (DNS) and other analytical grade chemicals were purchased from Sinopharm Group Co., Ltd (Shanghai, China).
3.2 Preparation of starch nanoparticles
In this work, spherical and polygonal SNPs were prepared by using retrogradation39 and anti-solvent40 methods, respectively. In brief, waxy corn starch was cooked in boiling water and stirred vigorously for 30 min, and then the cooked waxy corn starch was debranched by pullulanase at 58 °C for 8 h. After centrifugation, the supernatant was heated to stop the reaction; then, the solution was kept at 4 °C for 8 h. The obtained nanoparticles were washed several times with distilled water and then freeze-dried to obtain the SNPs. The as-prepared SNPs exhibited spherical shapes with a particle size of approximately 30–50 nm, and the relative crystallinity was 33.41%. To prepare the polygonal SNPs, corn starch suspensions (1%, w/v) were fully gelatinized, and then four times the volume of ethanol was added dropwise to the gelatinized starch solution and incubated at 25 °C for another 4 h. The suspensions were centrifuged (3000g, 5 min) and rinsed with absolute ethanol. The SNPs were collected by vacuum freeze-drying. The obtained SNPs showed irregularly angular particles with a size range of 40–100 nm, and the relative crystallinity was 23.34%.
3.3 Sample preparation
Different concentrations of chitin nanowhiskers or SNP dispersions (0, 0.02, 0.10, 0.20, 1.00, and 2.00 mg mL−1) and α-amylase (0.50 mg mL−1) solutions were prepared in 20 mM phosphate buffer solution (pH 5.8). Then, the mixture of SNPs and α-amylase with an equal volume was constantly stirred at ambient temperature for 2 h.
3.4 Ultraviolet (UV) absorption measurements
The UV spectra of the samples were obtained using a UV-visible spectrophotometer (TU-1810, Beijing, China) in the continuous range of 240–320 nm. The final spectra were baseline corrected by subtracting the corresponding SNP spectra obtained under the same conditions.
3.5 Fluorescence spectroscopy measurements
Fluorescence measurements of α-amylase and α-amylase/SNP mixtures were carried out using a fluorescence spectrophotometer (F-7000, Hitachi, Japan). The fluorescence emission spectra of the samples were recorded at wavelengths of 300–500 nm upon excitation at 280 nm.
The Stern–Volmer equation (eqn (1))41 was used to analyze the fluorescence quenching of the samples:
| | 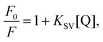 | (1) |
where
F0 and
F are the fluorescence intensities of α-amylase in the absence and presence of quenchers (SNPs), [Q] is the concentration of SNPs, and
KSV is the Stern–Volmer quenching constant.
kq is calculated by using
eqn (2):
where
kq is the biomolecular rate constant of the quenching reaction, and
τ0 is the average integral fluorescence lifetime of tryptophan (Trp) of approximately 5.7 × 10
−9 s.
42
3.6 TEM
TEM images of SNPs and α-amylase/SNP mixtures were recorded using a 7700 transmission electron microscope (Hitachi, Tokyo, Japan) operating at an acceleration voltage of 80 kV. A tiny drop of SNP or the α-amylase/SNP mixture (0.04%, w/v) dispersion was spread on a carbon-coated copper grid, and extra water was blotted with filter paper. Freeze-dried samples were observed.
3.7 CLSM
The microstructure of the α-amylase/SNP mixture was visualized using a confocal laser scanning microscope (Leica TCS SP5, Leica Microsystems Inc., Heidelberg, Germany). Before preparing the α-amylase/SNP mixture, two drops of 0.1 mg mL−1 FITC in ethanol were used to stain the SNPs (1%, w/v) by stirring for 10 h at 25 °C. Extra FITC was rinsed with absolute ethanol, and the SNPs (1 mg mL−1) were diluted with water. Then, 0.25 mg mL−1 α-amylase was added, stirring for 1 h to prepare the α-amylase/SNP mixture. One drop of Rhodamine B was added before the observation. The FITC and Rhodamine B fluorescent dyes were excited at 488 nm and 568 nm, respectively.
3.8 FTIR spectroscopy
The FTIR spectra of the SNPs, α-amylase, and the α-amylase/SNP mixture were recorded using an FTIR spectrophotometer (NEXUS-870; Thermo Nicolet Corporation, Madison, WI, USA). The spectra were collected under ambient conditions in the transmittance mode from an accumulation of 64 scans at a 4 cm−1 resolution over the 4000–400 cm−1 regions.
3.9 CD spectroscopy
CD spectroscopy was applied to analyze the changes that occurred in the secondary structure of α-amylase. α-Amylase (0.50 mg mL−1) was individually incubated with various concentrations of SNPs (0.00–2.00 mg mL−1) for 2 h. The CD spectra were recorded using a Jasco Spectropolarimeter (Tokyo, Japan) in the far-UV (190–260 nm) region under constant nitrogen flush. The path length was 0.1 mm for the far-UV region. Ellipticity was recorded at a 100 nm min−1 speed, 0.5 nm resolution, 20 accumulations, and 2.0 nm bandwidth.
3.10
In vitro α-amylase inhibition assay
The in vitro α-amylase inhibition was assayed by measuring the reducing sugar released.12 The mixture of SNPs and α-amylase with an equal volume (1 mL) was constantly stirred at ambient temperature for 2 h. Subsequently, 1% soluble starch solution as the substrate was added. After incubation at 37 °C for 3 min, the reaction was stopped by adding 0.5 mL of DNS reagent (1% DNS, 0.2% phenol, 0.05% Na2SO3, and 1% NaOH in aqueous solution) to the reaction mixture and boiling at 100 °C for 8 min. After cooling to room temperature, the absorbance (Abs) at 540 nm was recorded by using a spectrophotometer. The inhibition rate of α-amylase was calculated as the percentage of the control (without SNPs):| | | % inhibition = [(OD of control − OD of test)/OD of control]/100. | (3) |
3.11 X-ray diffraction (XRD)
The X-ray patterns of SNPs were analyzed using an X-ray diffractometer (AXS D8 ADVANCE; Bruker, Karlsruhe, Germany) with Cu Kα radiation at a voltage of 40 kV and 30 mA. The samples were scanned with a 2θ angle range of 3–40°. The crystallinity of the samples was determined by plotting the peak baseline on the diffractogram and calculating the area using the software spectrum viewer (Version 2.6). The area above and under the curve corresponded to crystalline domains and amorphous regions, respectively. The ratio of the upper area to total area was taken as relative crystallinity:| | 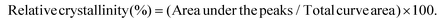 | (4) |
3.11 Statistical analysis
Triplicate samples of all quantitative results were obtained. Statistical analysis was performed through Duncan's multiple range tests using the SPSS V.17 statistical software package (SPSS Inc., Chicago, IL, USA). Differences were considered at a significance test level of 95% (p < 0.05).
4. Conclusions
In this study, the interactions between α-amylase and SNPs were explored in detail. The results suggested that the enzymatic activity of α-amylase decreased as the concentration of SNPs increased. The relative inhibitory activity against α-amylase of polygonal SNPs was more efficient than that of spherical SNPs. SNPs were hypothesized to bind to α-amylase, thereby inhibiting starch hydrolysis. Our results further showed that spherical SNPs were a competitive type inhibitor and that polygonal SNPs were a competitive–uncompetitive mixed-type inhibitor of α-amylase. CD analysis indicated that the secondary structure of α-amylase with SNPs changed. These results indicated that SNPs showed promise as a potent α-amylase inhibitor and were important in delaying the digestion of starch. SNPs could help type 2 diabetes patients with enzymatic activity control and have potential in food and medical applications.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
This work was supported by Special Funds from Shandong Province Taishan Scholars Project-China and Shandong Provincial Natural Science Foundation-China (ZR2017MC044).
References
- D. Yabe and Y. Seino, Type 2 diabetes via β-cell dysfunction in east Asian people, Lancet Diabetes Endocrinol., 2015, 4, 2–3 CrossRef PubMed.
- A. A. Tahrani, C. J. Bailey, P. S. Del and A. H. Barnett, Management of type 2 diabetes: new and future developments in treatment, Lancet, 2012, 378, 182–197 CrossRef.
- S. Puri and M. Hebrok, Diabetic beta Cells: To Be or Not To Be?, Cell, 2012, 150, 1103–1104 CrossRef CAS PubMed.
- H. Nyambe-Silavwe, J. A. Villa-Rodriguez, I. Ifie, M. Holmes, E. Aydin, J. M. Jensen and G. Williamson, Inhibition of human α-amylase by dietary polyphenols, J. Funct. Foods, 2015, 19, 723–732 CrossRef CAS.
- C. Funnell, M. M. Doyle-Waters, S. Yip and T. Field, What is the relationship between type 2 diabetes mellitus status and the neuroradiological correlates of cerebral small vessel disease in adults? Protocol for a systematic review, Syst. Rev., 2017, 6, 7 CrossRef PubMed.
- G. J. Biessels, S. Staekenborg, E. Brunner, C. Brayne and P. Scheltens, Risk of dementia in diabetes mellitus: a systematic review, Lancet Neurol., 2006, 5, 64 CrossRef PubMed.
- D. G. Bruce, Type 2 diabetes and cognitive function: many questions, few answers, Lancet Neurol., 2015, 14, 241–242 CrossRef PubMed.
- G. Williamson, Possible effects of dietary polyphenols on sugar absorption and digestion, Mol. Nutr. Food Res., 2013, 57, 48–57 CAS.
- J. Xiao, X. Ni, G. Kai and X. Chen, A review on structure-activity relationship of dietary polyphenols inhibiting α-amylase, Crit. Rev. Food Sci. Nutr., 2013, 53, 497–506 CrossRef CAS PubMed.
- J. Wu, S. Shi, H. Wang and S. Wang, Mechanisms underlying the effect of polysaccharides in the treatment of type 2 diabetes: A review, Carbohydr. Polym., 2016, 144, 474–494 CrossRef CAS PubMed.
- M. Miao, B. Jiang, H. Jiang, T. Zhang and X. Li, Interaction mechanism between green tea extract and human α-amylase for reducing starch digestion, Food Chem., 2015, 186, 20–25 CrossRef CAS PubMed.
- Q. Fei, Y. Gao, X. Zhang, Y. Sun, B. Hu, L. Zhou, S. Jabbar and X. Zeng, Effects of oolong tea polyphenols, EGCG, and EGCG3 ′′Me on pancreatic α-amylase activity in vitro, J. Agric. Food Chem., 2014, 62, 9507–9514 CrossRef CAS PubMed.
- L. Sun, W. Chen, Y. Meng, X. Yang, Y. Li and Y. Guo, Interactions between polyphenols in thinned young apples and porcine pancreatic α-amylase: Inhibition, detailed kinetics and fluorescence quenching, Food Chem., 2016, 208, 51–60 CrossRef CAS PubMed.
- Z. Li, H. Jiang, C. Xu and L. Gu, A review: Using nanoparticles to enhance absorption and bioavailability of phenolic phytochemicals, Food Hydrocolloids, 2015, 43, 153–164 CrossRef CAS.
- E. Frankel, A. Bakhouche, J. Lozanosánchez, A. Seguracarretero and A. Fernándezgutiérrez, Literature review on production process to obtain extra virgin olive oil enriched in bioactive compounds. Potential use of byproducts as alternative sources of polyphenols, J. Agric. Food Chem., 2013, 61, 5179–5188 CrossRef CAS PubMed.
- A. A. Vertegel, R. W. Siegel and J. S. Dordick, Silica Nanoparticle Size Influences the Structure and Enzymatic Activity of Adsorbed Lysozyme, Langmuir, 2004, 20, 6800–6807 CrossRef CAS PubMed.
- M. Song, G. Jiang, J. Yin and H. Wang, Inhibition of polymerase activity by pristine fullerene nanoparticles can be mitigated by abundant proteins, Chem. Commun., 2010, 46, 1404–1406 RSC.
- S. H. Cha, J. Hong, M. Mcguffie, B. Yeom, J. S. Vanepps and N. A. Kotov, Shape-Dependent Biomimetic Inhibition of Enzyme by Nanoparticles and Their Antibacterial Activity, ACS Nano, 2015, 9, 9097–9105 CrossRef CAS PubMed.
- S. Dhobale, T. Thite, S. L. Laware, C. V. Rode, S. J. Koppikar, R. K. Ghanekar and S. N. Kale, Zinc oxide nanoparticles as novel alpha-amylase inhibitors, J. Appl. Phys., 2008, 104, 3027 CrossRef.
- X. Cheng and R. J. Lee, The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery, Adv. Drug Delivery Rev., 2016, 99, 129–137 CrossRef CAS PubMed.
- S. Jiang, Y. Qin, J. Yang, M. Li, L. Xiong and Q. Sun, Enhanced antibacterial activity of lysozyme immobilized on chitin nanowhiskers, Food Chem., 2016, 1507–1513 Search PubMed.
- J. Yang, R. Chang, S. Ge, M. Zhao, C. Liang, L. Xiong and Q. Sun, The inhibition effect of starch nanoparticles on tyrosinase activity and its mechanism, Food Funct., 2016, 7, 4804–4815 CAS.
- C. Liu, S. Jiang, Z. Han, L. Xiong and Q. Sun, Invitro digestion of nanoscale starch particles and evolution of thermal, morphological, and structural characteristics, Food Hydrocolloids, 2016, 61, 344–350 CrossRef CAS.
- M. Yilmazermusa, A. M. Griffith, A. J. Michels, E. Schneider and B. Frei, Grape seed and tea extracts and catechin 3-gallates are potent inhibitors of α-amylase and α-glucosidase activity, J. Agric. Food Chem., 2012, 60, 8924–8929 CrossRef CAS PubMed.
- Q. Fei, Y. Gao, X. Zhang, Y. Sun, B. Hu, L. Zhou, S. Jabbar and X. Zeng, Effects of Oolong tea polyphenols, EGCG, and EGCG3′′Me on pancreatic α-amylase activity in vitro, J. Agric. Food Chem., 2014, 62, 9507 CrossRef CAS PubMed.
- M. Yilmazer-Musa, A. M. Griffith, A. J. Michels, E. Schneider and B. Frei, Grape seed and tea extracts and catechin 3-gallates are potent inhibitors of α-amylase and α-glucosidase activity, J. Agric. Food Chem., 2012, 60, 8924–8929 CrossRef CAS PubMed.
- J. L. Hargrove, P. Greenspan, D. K. Hartle and C. Dowd, Inhibition of aromatase and α-amylase by flavonoids and proanthocyanidins from Sorghum bicolor bran extracts, J. Med. Food, 2011, 14, 799–807 CrossRef CAS PubMed.
- R. Gonçalves, N. Mateus and V. D. Freitas, Inhibition of α-amylase activity by condensed tannins, Food Chem., 2011, 125, 665–672 CrossRef.
- A. Cornish-Bowden, A simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors, Biochem. J., 1974, 137, 143–144 CrossRef CAS PubMed.
- S. Dhital, M. J. Gidley and F. J. Warren, Inhibition of α-amylase activity by cellulose: Kinetic analysis and nutritional implications, Carbohydr. Polym., 2015, 123, 305–312 CrossRef CAS PubMed.
- M. J. Khan, S. Qayyum, F. Alam and Q. Husain, Effect of tin oxide nanoparticle binding on the structure and activity of α-amylase from Bacillus amyloliquefaciens, Nanotechnology, 2011, 22, 455708–455714 CrossRef PubMed.
- U. Anand, C. Jash, R. K. Boddepalli, A. Shrivastava and S. Mukherjee, Exploring the mechanism of fluorescence quenching in proteins induced by tetracycline, J. Phys. Chem. B, 2011, 115, 6312–6320 CrossRef CAS PubMed.
- W. Sun, Y. Du, J. Chen, J. Kou and B. Yu, Interaction between titanium dioxide nanoparticles and human serum albumin revealed by fluorescence spectroscopy in the absence of photoactivation, J. Lumin., 2009, 129, 778–783 CrossRef CAS.
- R. Ahmad, M. Mohsin, T. Ahmad and M. Sardar, Alpha amylase assisted synthesis of TiO2 nanoparticles: Structural characterization and application as antibacterial agents, J. Hazard. Mater., 2015, 283, 171–177 CrossRef CAS PubMed.
- E. Vinita, G. Sekar, M. Amitava and C. Natarajan, Enhanced activity of lysozyme-AgNP conjugate with synergic antibacterial effect without damaging the catalytic site of lysozyme, Artif. Cells, Nanomed., Biotechnol., 2014, 42, 336–343 CrossRef PubMed.
- A. J. Miles and B. A. Wallace, Circular dichroism spectroscopy of membrane proteins, Chem. Soc. Rev., 2016, 45, 4859–4872 RSC.
- H. Patel, P. G. Royall, S. Gaisford, G. R. Williams, C. H. Edwards, F. J. Warren, B. M. Flanagan, P. R. Ellis and P. J. Butterworth, Structural and enzyme kinetic studies of retrograded starch: Inhibition of α-amylase and consequences for intestinal digestion of starch, Carbohydr. Polym., 2017, 164, 154–161 CrossRef CAS PubMed.
- X. Wang, Y. Nie, X. Mu, Y. Xu and R. Xiao, Disorder prediction-based construct optimization improves activity and catalytic efficiency of Bacillus naganoensis pullulanase, Sci. Rep., 2016, 6, 24574 CrossRef CAS PubMed.
- Q. Sun, G. Li, L. Dai, N. Ji and X. Liu, Green preparation and characterisation of waxy maize starch nanoparticles through enzymolysis and recrystallisation, Food Chem., 2014, 162, 223–228 CrossRef CAS PubMed.
- Y. Qin, C. Liu, S. Jiang, L. Xiong and Q. Sun, Characterization of starch nanoparticles prepared by nanoprecipitation: Influence of amylose content and starch type, Ind. Crops Prod., 2016, 87, 182–190 CrossRef CAS.
- K. Shanmugaraj, S. Anandakumar and M. Ilanchelian, Probing the binding interaction of thionine with lysozyme: A spectroscopic and molecular docking investigation, Dyes Pigm., 2015, 112, 210–219 CrossRef CAS.
- S. T. Yang, H. Wang, L. Guo, Y. Gao, Y. Liu and A. Cao, Interaction of fullerenol with lysozyme investigated by experimental and computational approaches, Nanotechnology, 2008, 19, 395101 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7fo01381g |
|
| This journal is © The Royal Society of Chemistry 2018 |
Click here to see how this site uses Cookies. View our privacy policy here.  *
*


![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching (VC
O stretching (VC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, amide I), and a band at 1520 cm−1, which corresponded to the N–H bending (VN–H, amide II).34 The characteristic bands of amide I and amide II were observed for α-amylase and all α-amylase/SNPs. Hydroxyl groups were confirmed by the broad peaks appearing between 3000 and 3800 cm−1. The addition of SNPs resulted in a red-shift of the peak at 3000–3800 cm−1, indicating the formation of hydrogen bonding between α-amylase and SNPs. The band at 2927 cm−1 was due to the C–H stretching associated with the anhydroglucose of α-amylase/SNPs. The peak at 1364 cm−1 belonged to the C–O–H stretching vibration and shifted toward a low frequency with the increase in the spherical SNP concentration, thereby indicating an interaction of α-amylase with SNPs. Previously, researchers reported that the shifts in the intensity of 3300 cm−1 (N–H stretching), 1600–1690 cm−1 (C
O, amide I), and a band at 1520 cm−1, which corresponded to the N–H bending (VN–H, amide II).34 The characteristic bands of amide I and amide II were observed for α-amylase and all α-amylase/SNPs. Hydroxyl groups were confirmed by the broad peaks appearing between 3000 and 3800 cm−1. The addition of SNPs resulted in a red-shift of the peak at 3000–3800 cm−1, indicating the formation of hydrogen bonding between α-amylase and SNPs. The band at 2927 cm−1 was due to the C–H stretching associated with the anhydroglucose of α-amylase/SNPs. The peak at 1364 cm−1 belonged to the C–O–H stretching vibration and shifted toward a low frequency with the increase in the spherical SNP concentration, thereby indicating an interaction of α-amylase with SNPs. Previously, researchers reported that the shifts in the intensity of 3300 cm−1 (N–H stretching), 1600–1690 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), and 1480–1575 cm−1 (C–N stretching and N–H bending) indicated the secondary structural conformation of enzyme changes.35 Compared with those in the pure α-amylase, the changes in these regions demonstrated the alterations in the molecular microenvironment of chromophores of α-amylase when it interacted with the SNPs.
O stretching), and 1480–1575 cm−1 (C–N stretching and N–H bending) indicated the secondary structural conformation of enzyme changes.35 Compared with those in the pure α-amylase, the changes in these regions demonstrated the alterations in the molecular microenvironment of chromophores of α-amylase when it interacted with the SNPs.