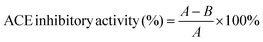Identification and the molecular mechanism of a novel myosin-derived ACE inhibitory peptide
Zhipeng
Yu
 ab,
Sijia
Wu
a,
Wenzhu
Zhao
*a,
Long
Ding
c,
David
Shiuan
a,
Feng
Chen
b,
Jianrong
Li
*a and
Jingbo
Liu
*c
ab,
Sijia
Wu
a,
Wenzhu
Zhao
*a,
Long
Ding
c,
David
Shiuan
a,
Feng
Chen
b,
Jianrong
Li
*a and
Jingbo
Liu
*c
aCollege of Food Science and Engineering, Bohai University, Jinzhou 121013, P.R. China. E-mail: zhaowenzhu777@163.com
bDepartment of Food Science and Human Nutrition, Clemson University, Clemson, SC 29634, USA
cLab of Nutrition and Functional Food, Jilin University, Changchun 130062, P.R. China
First published on 23rd November 2017
Abstract
The objective of this work was to identify a novel ACE inhibitory peptide from myosin using a number of in silico methods. Myosin was evaluated as a substrate for use in the generation of ACE inhibitory peptides using BIOPEP and ExPASy PeptideCutter. Then the ACE inhibitory activity prediction of peptides in silico was evaluated using the program peptide ranker, following the database search of known and unknown peptides using the program BIOPEP. In addition, the interaction mechanisms of the peptide and ACE were evaluated by DS. All of the tripeptides were predicted to be nontoxic. Results suggested that the tripeptide NCW exerted potent ACE inhibitory activity with an IC50 value of 35.5 μM. Furthermore, the results suggested that the peptide NCW comes into contact with Zn 701, Tyr 523, His 383, Glu 384, Glu 411, and His 387. The potential molecular mechanism of the NCW/ACE interaction was investigated. Results confirmed that the higher inhibitory potency of NCW might be attributed to the formation of more hydrogen bonds with the ACE's active site. Therefore, the in silico method is effective to predict and identify novel ACE inhibitory peptides from protein hydrolysates.
1. Introduction
Angiotensin I converting enzyme (ACE) as a crucial enzyme in the regulation of blood pressure elevates blood pressure by cleaving a dipeptide from the inactive decapeptide angiotensin I and converting it into the potent vasoconstrictor angiotensin II via the renin–angiotensin system (RAS).1 In addition, the ACE also converts the vasodilator bradykinin into an inactive peptide via the kallikrein–kinin systems (KKS). Thus, the inhibition of the ACE in the RAS and KKS as a potential strategy plays a critical role in the treatment of hypertension.2 ACE inhibitory peptides have been produced from a variety of food protein hydrolysates. Substantial evidence supports the contention that bioactive peptides are absorbed more rapidly than free amino acids. It has been documented that tripeptides should be absorbed from the intestine before entering into the blood circulation, and can exert ACE inhibitory effects. Additionally, the amino acid composition of the bioactive peptides also influences the rate of absorption. Thus, tripeptides with ACE inhibitory activity are attracting many investors. Some tripeptides have been prepared and isolated, such as VPP,3 IPP,4 IVY,5 IQW, LKP, VKP,6 IQA, VEP, TAY, FYN,7 and GFR.8 Currently, the classic approach for the discovery of ACE-inhibitory peptides from food proteins involved preparation (i.e., enzyme hydrolysis and microbial fermentation), isolation and characterization (LC-MS/MS).9 The preparation and isolation of ACE inhibitory peptides is mostly based on experiments, which needs a large amount of labor and cost.10,11 In order to circumvent some challenges of the classical approach, the virtual screening by the in silico experiment has been applied and may replace the traditional screening of antihypertensive peptides by laboratory efforts to some extent.The prediction and identification of bioactive peptides from natural sources in silico is becoming more efficient. In addition, BIOPEP (http://www.uwm.edu.pI/biochemia) is an open access database enabling us to hypothesize the potential biological activities of peptides based on the presence of specific amino acid sequences.12 A screening of the structures of these peptides with BIOPEP suggested that their structures were compatible with a potential ACE inhibitory activity. Mizuhopecten yessoensis is one of the most important aquaculture mollusca in China, which was first introduced into China from Japan.13,14 Nowadays, Mizuhopecten yessoensis has become a main aquaculture species in the Liaoning province. Myosin from Mizuhopecten yessoensis was selected for hydrolysis in this study, based on its abundance and sequence information for the parent protein.
The aim of this work was to generate and characterize a novel ACE inhibitory peptide from myosin. Firstly, myosin was hydrolysed with pepsin and trypsin in silico, and the in vitro potential ACE inhibitory activity of myosin-derived tripeptides was evaluated. Subsequently, the interaction of a bioactive peptide with the ACE was investigated using an in silico molecular model. Finally, the in silico potential toxicity of the identified tripeptides was assessed using ToxinPred.
2. Materials and methods
2.1 Materials and reagents
Angiotensin-converting enzyme (ACE) from rabbit lung, hippuryl-L-histidyl-L-leucine, captopril, and hippuric acid was purchased from Sigma Chemical Co. (St Louis, Mo. USA). HPLC grade acetonitrile, methanol and trifluoroacetic acid (TFA) were HPLC-grade and purchased from Fisher Scientific Company (Waltham, MA, USA).2.2 In silico hydrolysis of myosin in Mizuhopecten yessoensis
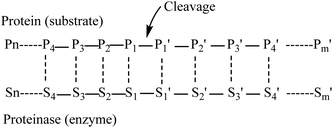
Myosin (GenBank: BAB40711.1) from Mizuhopecten yessoensis has been taken from the NCBI.Myosin was hydrolyzed independently using the in silico method with pepsin (pH 1.3) and low specificity chymotrypsin (C-term to [FYWML], not before P) as described previously.15The in silico hydrolysis of myosin was performed using the program ExPASy peptide cutter, available at http://web.expasy.org/peptide_cutter. Amino acid residues in a substrate undergoing cleavage are designated position 1(P1), position 2 (P2), position 3 (P3), position 4 (P4) etc. in the N-terminal direction from the cleaved bond. Likewise, the residues in the C-terminal direction are designated position 1′ (P1′), position 2′ (P2′), position 3′ (P3′), position 4′ (P4′). etc. (Fig. 1). Pepsin preferentially cleaves at Phe, Tyr, Trp and Leu in P1 or P1′.16 Negative effects on cleavage are exerted by Arg, Lys and His in P3 and Arg in P1. Pro has favorable effects when being located in P4 and P3, but unfavorable ones when found in P2 to P3′. Cleavage is more specific at pH 1.3. Chymotrypsin preferentially cleaves at Trp, Tyr and Phe in P1 (high specificity) and to a lesser extent (taken into account when dealing with low specificity chymotrypsin) at Leu, Met and His in P1 (http://web.expasy.org). Exceptions to these rules are the following: when Trp, Met and Pro are found in P1 at the same time, the cleavage is blocked. Furthermore, Pro in P1′ nearly fully blocks the cleavage independent of the amino acids found in P1. When Met is found in P1, the cleavage is blocked due to the presence of Tyr in P1′. Finally, when His is located in P1, the presence of Asp, Met or Trp also blocks the cleavage16 (http://web.expasy.org). These tripeptides of hydrolysates from myosin were collected for further study.
2.3 Prediction of ACE inhibitory activity for selected peptides
The database search of known and unknown peptides from myosin was performed using the program BIOPEP, available at http://www.uwm.edu.pl/biochemia/index.php/pl/biopep, peptideDB, available at http://www.peptides.be/. Subsequently, ACE inhibitory activity prediction in silico was evaluated using the program peptide ranker,17 available at http://bioware.ucd.ie/~compass/biowareweb/Server_pages/peptideranker.php. PeptideRanker is a server for the prediction of bioactive peptides based on a novel N-to-1 neural network. In the current work, PeptideRanker was trained at a threshold of 0.5 i.e. any peptide predicted over a 0.5 threshold is labelled as bioactive.2.4 Toxicity and physicochemical properties of bioactive peptides
One of the bottlenecks in peptide-based nutrients is their toxicity. Therefore, in the present study, the potential toxicity of these selected peptides was predicted in silico using ToxinPred available at http://www.imtech.res.in/raghava/toxinpred/. In addition, these peptides’ physicochemical properties (i.e., hydrophobicity, steric hindrance, amphipathicity, pI, and hydrophilicity) were determined.2.5 Solid phase synthesis of selected peptides
The peptide sequences derived from myosin in Mizuhopecten yessoensis were synthesized by the solid phase procedure using the FMOC protected amino acid synthesis methods with an AAPPTEC 396 Automated Peptide Synthesizer (Advanced Automated Peptide Protein Technologies, USA).18,19 These peptides were provided by Shanghai Science Peptide Biological Technology Corporation. The synthesized peptides were purified by HPLC on an ODS column. Their purity was verified by HPLC in our laboratory. The molecular masses of these synthesized peptides were determined by mass spectrometry (Dionex, MSQ). The synthetic peptides were used for further study.2.6 In vitro ACE inhibitory assay
The ACE-inhibitory activity was measured by performing high-performance liquid chromatography with some modifications as explained below. A reaction mixture in a volume of 60 μL contained 100 mM borate buffer (pH 8.3), 4 mM hippuryl-L-histidyl-L-leucine (HHL), 300 mM NaCl, and 10 milliunit ACE. All solutions were incubated at 37 °C for 30 min in a thermostatically controlled water bath prior to mixing, subsequently for an additional 30 min to react at the same temperature after mixing. The reaction was injected into the HPLC sample loop to quantify the hippuric acid produced by the enzymatic hydrolysis of the substrate HHL. The isocratic mobile phase consisted of 25% acetonitrile in deionizer water (v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) with 0.5% TFA. An aliquot of 10 μL of the reaction mixture was analyzed on a Shimadzu C18 column. HHL and hippuric acid (HA) were detected at 228 nm, as the mobile phase was controlled at 0.5 mL min−1 (ref. 20).
v) with 0.5% TFA. An aliquot of 10 μL of the reaction mixture was analyzed on a Shimadzu C18 column. HHL and hippuric acid (HA) were detected at 228 nm, as the mobile phase was controlled at 0.5 mL min−1 (ref. 20).
The inhibitory activity degree was calculated as follows:
2.7 The estimated free energy of binding between the peptide and ACE
The 3-D structure of the human ACE was imported from the Protein Data Bank (1O86.pdb, a crystal structure of the human ACE).21 The structure of the peptide was generated by using Discovery Studio (DS). The evaluation of the molecular docking was performed according to the scores and total potential energy in order to obtain the best pose of the peptide. The DS software was used to identify the hydrogen bonds as well as the hydrophobic, hydrophilic, electrostatic and coordination interactions between the residues present within the ACE's active site.22 “CDOCKER” is a scorning function using a CHARMm force field based molecular dynamics scheme to dock ligands into the receptor binding site.23 The energy was minimized with CHARMm force field. The DOCK ligands with the set parameters are as follows: Top Hit 10 and a pose cluster radius parameter value of 0.1. The evaluation of the molecular docking was performed according to the scores in order to obtain the best pose of the peptide.3. Results and discussion
3.1 In silico prediction of ACE inhibitory tripeptides from myosin
In in silico simulation, one hundred tripeptides were generated, and the score of those peptides was calculated (Table 1). The peptide ranker score is a prediction of how likely the peptide is to be bioactive. The scores of peptides NCW, ADF, EGF, GGH, VAF, ADM, and ANM were 0.97, 0.81, 0.69, 0.68, 0.56, 0.52, and 0.50, respectively, as the top 7 of 100 tripeptides. The search of known ACE inhibitory tripeptides was performed using the program BIOPEP, and peptides AQL and VKP have been found and are known as ACE inhibitory peptides with the corresponding IC50 values of 57.54 μM and 1.30 μM, respectively.6,24 In addition, the other tripeptides with the ACE inhibitory activity have been identified from natural food proteins (Table 2). Based on the previously published work, the results documented that peptides NCW, ADF, EGF, GGH, VAF, ADM, and ANM have not been identified in previous work. Moreover, some previous research also mentioned that the in silico approach is effective to predict DPP-IV inhibitory activities in vitro25 and antithrombotic activities in vitro23 of protein hydrolysates.| Peptide | Location | Score | Peptide | Location | Score |
|---|---|---|---|---|---|
| ADE | f 1503–1505, f 1748–1750, f 1763–1765, f 1851–1853 | 0.06 | EEL | f 326–328, f 1115–1117, f 1119–1121, f 1143–1145, f 1381–1383, f 1535–1537, f 1683–1685 | 0.04 |
| ADF | f 912–914, f 1254–1256 | 0.81 | EEM | f 772–774 | 0.07 |
| ADM | f 1335–1337 | 0.52 | EER | f 922–924, f 1904–1906 | 0.04 |
| ADR | f 1840–1842 | 0.26 | EGF | f 41–43 | 0.69 |
| AEL | f 685–687, f 1136–1138, f 1168–1170, f 1476–1478 | 0.10 | ENK | f 1498–1500 | 0.04 |
| AEM | f 1624–1626 | 0.21 | ENT | f 184–186, f 957–959 | 0.03 |
| AGV | f 1329–1331 | 0.18 | EPL | f 615–617 | 0.27 |
| AIN | f 1887–1889 | 0.16 | EQK | f 881–883 | 0.03 |
| ANM | f 89–91 | 0.50 | EQT | f 23–25, f 1027–1029 | 0.03 |
| AQH | f 1896–1898 | 0.15 | ESL | f 651–654 | 0.08 |
| AQL | f 916–918, f 1558–1560 | 0.23 | ESM | f 1592–1594 | 0.16 |
| CID | f 519–521 | 0.40 | ESR | f 1470–1472, f 1813–1815 | 0.08 |
| CNH | f 1016–1018 | 0.45 | ESY | f 1666–1668 | 0.08 |
| CSA | f 1408–1410 | 0.38 | EVA | f 1623–1625 | 0.03 |
| CTL | f 1675–1677 | 0.45 | GDK | f 898–900 | 0.20 |
| DEM | f 846–848 | 0.20 | GEL | f 1743–1745 | 0.21 |
| DER | f 776–778 | 0.08 | GEM | f 359–361, f 979–981 | 0.41 |
| DGK | f 735–737 | 0.21 | GER | f 1146–1148 | 0.19 |
| DTK | f 38–40, f 447–449 | 0.04 | GGH | f 936–938 | 0.68 |
| EAE | f 379–381, f 1122–1124, f 1167–1169, f 1543–4545, f 1599–1601, f 1796–1798, f 1881–1883, f 1902–1904 | 0.03 | GGK | f 632–634, f 1804–1806 | 0.48 |
| EAQ | f 915–917, f 1185–1187 | 0.05 | GQT | f 413–415 | 0.17 |
| EDM | f 86–88 | 0.17 | GSQ | f 1306–1308 | 0.21 |
| EDN | f 1034–1036 | 0.05 | IDK | f 1865–1867, f 1569–1571 | 0.09 |
| EEE | f 206–208, f 534–536, f 928–930, f 1118–1120, f 1345–1347, f 1577–1579, f 1655–1657 | 0.02 | IEE | f 1114–1117, f 1485–1487, f 1654–1656 | 0.03 |
| EEK | f 343–345 | 0.02 | IGV | f 456–458 | 0.17 |
| IQK | f 1809–1811 | 0.09 | SAR | f 104–106 | 0.28 |
| KPK | f 400–402 | 0.20 | SDR | f 865–867 | 0.24 |
| NAK | f 231–233, f 1405–1407, f 1869–1871 | 0.09 | SEK | f 1215–1217 | 0.04 |
| NAR | F 1126–1128, f 1257–1259 | SNL | f 1242–1244 | 0.24 | |
| NCW | f 33–35 | 0.97 | SQL | f 1286–1288, f 1303–1305, f 1307–1309 | 0.22 |
| NDL | f 884–886 | 0.21 | SSK | f 50–52, f 1090–1092 | 0.09 |
| NID | f 2–4 | 0.11 | SSQ | f 1285–1287, f 1774–1776 | 0.10 |
| NIR | f 815–817 | 0.22 | STH | f 663–665 | 0.09 |
| QAA | f 1538–1540 | 0.14 | TDQ | f 1510–1512 | 0.05 |
| QAH | f 787–789 | 0.17 | TEE | f 1380–1382 | 0.02 |
| QAK | f 1154–1156 | 0.09 | TVK | f 68–70 | 0.03 |
| QAR | f 1111–1113 | 0.26 | TVR | f 234–236, f 1564–1566 | 0.05 |
| QAS | f 1595–1597 | 0.09 | VAE | f 618–620 | 0.03 |
| QDK | f 551–553, f 965–967 | 0.07 | VAF | f 383–385 | 0.56 |
| QID | f 1159–1161 | 0.11 | VDL | f 860–862 | 0.11 |
| QIR | f 1646–1648, f 1791–1793 | 0.26 | VEK | f 904–906 | 0.02 |
| QNE | f 1327–1329 | 0.05 | VIM | f 1047–1049, f 1131–1133, f 1776–1778 | 0.29 |
| QNM | f 161–163, f 1399–1401 | 0.44 | VKP | f 835–837 | 0.13 |
| QQE | f 1329–1331 | 0.05 | VNE | f 480–482, f 1700–1702 | 0.03 |
| QSE | f 1461–1463 | 0.05 | VNK | f 442–444 | 0.04 |
| QSM | f 346–348 | 0.39 | VNS | f 1457–1459 | 0.05 |
| QTD | f 1735–1737 | 0.06 | VSK | f 1235–1237, f 1391–1393 | 0.04 |
| SAA | f 640–642, f 1935–1937 | 0.16 | VSV | f 1295–1297, f 1925–1927, f 1932–1934 | 0.04 |
| SAE | f 277–279, f 1475–1477 | 0.07 | VTK | f 410–412 | 0.03 |
| SAL | f 1419–1421, f 1687–1689 | 0.30 | VTY | f 272–274 | 0.06 |
| No. | Sequence | IC50 | Parent protein |
|---|---|---|---|
| 1 | IVY | 80.4 μM | Brewers’ spent grain protein |
| 2 | IVR | 0.81 μM | Soft-shelled turtle egg |
| 3 | VKP | 1.3 μM | Rhopilema esculentum hydrolysate |
| 4 | TAY | 16.7 μM | Oyster protein |
| 5 | FYN | 68.2 μM | Oyster protein |
| 6 | GFR | 94.25 μM | Sweet potato tryptic hydrolysates |
| 7 | VEV | 115.20 μM | Wheat germ |
| 8 | AMY | 5.86 μM | Wheat germ |
| 9 | LKY | 10.1 μM | Antarctic krill tail meat hydrolysate |
| 10 | VDF | 6.59 μM | Cooked eggs |
| 11 | LPF | 10.59 μM | Cooked eggs |
| 12 | MPF | 17.98 μM | Cooked eggs |
| 13 | IPF | 8.78 μM | Cooked eggs |
| 14 | TTI | 24.94 μM | Cooked eggs |
| 15 | VSV | 0.15 μM | Defatted canola meal |
| 16 | VRL | 208.6 μM | Sweet potato storage root |
| 17 | FHG | 52.9 μg ml−1 | Beef sarcoplasmic protein |
| 18 | PYY | 3.07 g ml−1 | Goat's milk casein |
| 19 | HPY | 1.68 μM | Bovine blood plasma protein |
| 20 | RIY | 28 μM | Rapeseed |
| 21 | LRP | 0.27 μM | ALPHA-ZEIN |
| 22 | LSP | 1.7 μM | ALPHA-ZEIN |
| 23 | LQP | 1.9 μM | ALPHA-ZEIN |
| 24 | LKP | 0.86 μM | Milk protein |
3.2 In silico prediction of the toxicity of ACE inhibitory tripeptides
Peptides have numerous advantages over small molecules that include high biological activity, high specificity, low production cost, and high penetration. However, toxicity remains the main concern in the development of peptides.26 The identified tripeptides (i.e., NCW, ADF, EGF, GGH, VAF, ADM, and ANM) were derived from Mizuhopecten yessoensis proteins, which are consumed as food. Thus, the in silico assessment of the toxicity of peptides should be carried out. In this study, the in silico assessment of toxicity and physiochemical properties of these tripeptides was carried out using ToxinPred and the results suggested that none of the peptides chosen was predicted to be toxic (Table 3). The prediction of the toxicity of peptides before their synthesis is very important for saving both time and money consumed in developing peptide-based functional foods. As mentioned previously, melittin, as a major peptide component of bee venom, is an attractive candidate for cancer therapy. However, its applicability to functional food and drugs has met with challenges due to several issues including its non-specific cytotoxicity.27| Peptide | Toxin | Hydrophobicity | Steric hindrance | pI | Hydrophilicity | Amphipathicity |
|---|---|---|---|---|---|---|
| NCW | Non | −0.08 | 0.63 | 5.85 | −1.40 | 0.00 |
| ADF | Non | 0.05 | 0.66 | 3.80 | 0.00 | 0.00 |
| EGF | Non | 0.05 | 0.69 | 4.00 | 0.17 | 0.42 |
| GGH | Non | −0.03 | 0.45 | 7.10 | −0.17 | 0.48 |
| VAF | Non | 0.47 | 0.64 | 5.88 | −1.50 | 0.00 |
| ADM | Non | −0.07 | 0.69 | 3.80 | 0.40 | 0.00 |
| ANM | Non | −0.04 | 0.69 | 5.88 | −0.53 | 0.00 |
3.3 Molecular docking
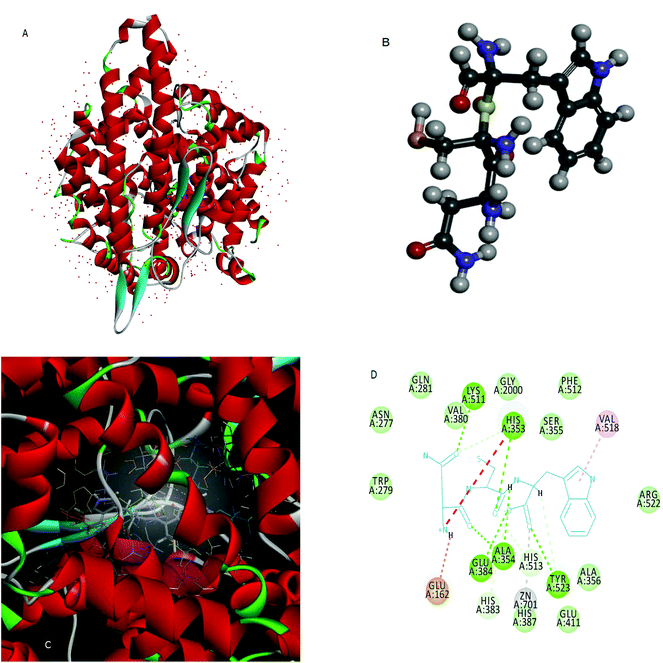
Molecular docking is used to predict the preferred orientation of tripeptides NCW, ADF, EGF, GGH, VAF, ADM, and ANM to ACE when they bound to each other to form a stable complex. The estimated free energy of binding between the peptide and ACE indicated the binding affinity. The higher the affinity of the peptide and ACE, the higher the value of “-the estimated free energy” will be. The values of “-the estimated free energy” predicted peptides showed different activities, in an increasing order as peptide ADM (f 922–924, f 1904–1906) (7.66 kcal mol−1), peptide EGF (f 41–43) (7.75 kcal mol−1), peptide ADF (f 772–774) (7.75 kcal mol−1), peptide GGH (f 936–938) (7.97 kcal mol−1), peptide ANM (f 89–91) (8.38 kcal mol−1), VAF (f 383–385) (9.44 kcal mol−1), and peptide NCW (f 33–35) (9.69 kcal mol−1). Moreover, the peptide NCW showed the highest affinity, which was 1.26-fold higher than that of peptide ADM. The previous work has also suggested that the free energy of the binding of ACE and peptide WTQRF represented the affinity between the two molecules described above.22 Subsequently, the 3D structures of ACE (Fig. 2A) and the peptide NCW (Fig. 2B) were shown based on the PDB database information. Furthermore, the analysis of ACE–NCW complexes revealed the binding site residue (Fig. 2C). The receptor–ligand interactions on a 2D diagram showed various interactions, such as carbon hydrogen bonds, salt bridges, conventional hydrogen bonds, metal–acceptor, van der Waals, unfavorable positive–positive, and pi-alkyl interactions between the surrounding residues and ligands (Fig. 2D). From the interaction mode of ACE–NCW complexes with the predicted active site, the molecular docking results showed that NCW comes into contact with residues His 353, His 383, Tyr 513, and His 523 through hydrogen bonds; Val 518 through pi-alkyl interactions; Tyr 523 through metal–acceptor interactions; Glu 162 through salt bridge interactions; Glu 384, Ala 354, Tyr 523, His 353, and Lys 511 through conventional hydrogen bond interactions; Trp 279, Gln 281, Asn 277, Val 380, Gly 2000, Ser 355, Phe 512, Arg 522, Ala 356,Glu 411, and His 387 by van der Waals interactions (Fig. 2D). Some previous studies also suggested that the peptide comes into contact with residues His 353, 383, Ser 355, Glu 384, and Val 518 through hydrophilic interactions.22 The active site of the ACE constituted of a zinc ion, His 383, Glu 384, His 387, and Glu 411. The present results also suggested that the peptide NCW comes into contact with Zn 701, Tyr 523, His 383, Glu 384, Glu 411, and His 387. These results indicated that the peptide NCW identified in silico method might have relatively potent inhibitory activity against ACE.3.4 ACE inhibitory activity of the peptide NCW
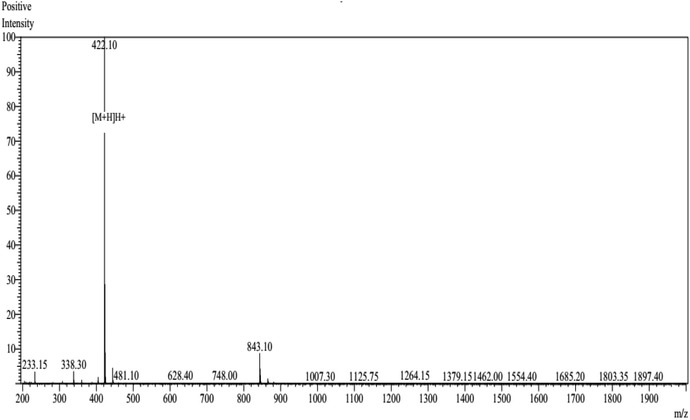
The peptide NCW predicted in silico was synthesized by conventional Fmoc solid-phase synthesis, and purified by HPLC on an ODS column. The purity of the peptide NCW was 93.25% and the molecular masses of the synthetic peptides were obtained according to the theoretical values (shown in Fig. 3). The inhibitory activity of the peptide was measured through HPLC, the value IC50 of the peptide NCW, which has potent ACE-inhibitory activity, was 35.5 μM. Compared to the previous identified tripeptides, which exerted potent ACE inhibitory activity, IVY with an IC50 value of 480.4 μM, was isolated from grain protein hydrolysates, and FYN with an IC50 of 68.2 μM was liberated from oyster protein hydrolysates. It should be noted that the novel tripeptide NCW showed a stronger ACE inhibition than that of the other ACE inhibitory peptides isolated from food proteins reported thus far.28,29 Recent studies have revealed that tripeptides with highly potent inhibitory activities have Trp (W), Phe (F), Tyr (Y), or Pro (P) at the C-terminus.30 However, the relationships between the structure and activity of ACE inhibitory peptides have not yet been completely established thus far. Additionally, due to decreased bioavailability with the increasing chain length of the peptides, further studies are undertaken to identify novel peptides containing more than three amino acids.314. Conclusions
The current work has demonstrated that the in silico method is reliable to predict the ACE inhibitory activities of food protein hydrolysates in vitro. A total of 100 tripeptides were identified from myosin in silico hydrolysis with pepsin and trypsin using the program ExPASy peptide cutter. Subsequently, the ACE inhibitory activity and potential toxicity of all those peptides were assessed, and the tripeptide NCW has exhibited potent activity against ACE, with an IC50 value of 35.5 μM. Therefore, the in silico method is helpful for researchers as a screening tool to screen ACE inhibitory peptides from food proteins. The present work also suggested that the peptide NCW comes into contact with Zn 701, Tyr 523, His 383, Glu 384, Glu 411, and His 387. Further studies are underway to confirm whether the peptide NCW has in vivo physiological relevance in blood pressure regulation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This paper was supported by The National Natural Science Funds of China (No. 31601479).References
- A. K. Rai, S. Sanjukta and K. Jeyaram, Production of angiotensin I converting enzyme inhibitory (ACE-I) peptides during milk fermentation and their role in reducing hypertension, Crit. Rev. Food Sci. Nutr., 2017, 57(13), 2789–2800 CrossRef CAS PubMed.
- R. Q. Yang, Y. Zou, N. J. Yu and Z. X. Gu, Accumulation and Identification of Angiotensin-Converting Enzyme Inhibitory Peptides from Wheat Germ, J. Agric. Food Chem., 2011, 59(8), 3598–3605 CrossRef CAS PubMed.
- Y. Sawada, et al., Milk-derived peptide Val-Pro-Pro (VPP) inhibits obesity-induced adipose inflammation via an angiotensin-converting enzyme (ACE) dependent cascade, Mol. Nutr. Food Res., 2015, 59(12), 2502–2510 CAS.
- A. M. Turpeinen, et al., A spread containing bioactive milk peptides Ile-Pro-Pro and Val-Pro-Pro, and plant sterols has antihypertensive and cholesterol-lowering effects, Food Funct., 2012, 3(6), 621–627 CAS.
- A. Connolly, M. B. O'Keeffe, C. O. Piggott, A. B. Nongonierma and R. J. FitzGerald, Generation and identification of angiotensin converting enzyme (ACE) inhibitory peptides from a brewers’ spent grain protein isolate, Food Chem., 2015, 176, 64–71 CrossRef CAS PubMed.
- J. Li, Q. Li, J. Li and B. Zhou, Peptides Derived from Rhopilema esculentum Hydrolysate Exhibit Angiotensin Converting Enzyme (ACE) Inhibitory and Antioxidant Abilities, Molecules, 2014, 19(9), 13587–13602 CrossRef PubMed.
- C. L. Xie, J. S. Kim, J. M. Ha, S. Y. Choung and Y. J. Choi, Angiotensin I-Converting Enzyme Inhibitor Derived from Cross-Linked Oyster Protein, BioMed. Res. Int., 2014, 379234 Search PubMed.
- G. J. Huang, et al., Sweet potato storage root defensin and its tryptic hydrolysates exhibited angiotensin converting enzyme inhibitory activity in vitro, Bot. Stud., 2011, 52(3), 257–264 CAS.
- B. Hernandez-Ledesma and C. C. Hsieh, Chemopreventive role of food-derived proteins and peptides: A review, Crit. Rev. Food Sci. Nutr., 2017, 57(11), 2358–2376 CrossRef CAS PubMed.
- S. Y. Lee and S. J. Hur, Antihypertensive peptides from animal products, marine organisms, and plants, Food Chem., 2017, 228, 506–517 CrossRef CAS PubMed.
- T. T. Chai, Y. C. Law, F. C. Wong and S. K. Kim, Enzyme-Assisted Discovery of Antioxidant Peptides from Edible Marine Invertebrates: A Review, Mar. Drugs, 2017, 15(2), 42 CrossRef PubMed.
- C. Lammi, C. Zanoni, A. Arnoldi and G. Vistoli, Peptides Derived from Soy and Lupin Protein as Dipeptidyl-Peptidase IV Inhibitors: In Vitro Biochemical Screening and in Silico Molecular Modeling Study, J. Agric. Food Chem., 2016, 64(51), 9601–9606 CrossRef CAS PubMed.
- Z. L. Hao, et al., Biochemical Components of Different Colored Strains of Cultured Japanese Scallop (Mizuhopecten yessoensis) Under Different Cultivation Systems, Isr. J. Aquacult-Bamid., 2015, 67, 8 Search PubMed.
- W. D. Liu, et al., Studies on Japanese scallop (Mizuhopecten yessoensis) germplasm resource in China, J. Biotechnol., 2008, 136, S552 CrossRef.
- T. Lafarga, P. O'Connor and M. Hayes, In silico methods to identify meat-derived prolyl endopeptidase inhibitors, Food Chem., 2015, 175, 337–343 CrossRef CAS PubMed.
- B. Keil, Proteolysis Data Bank: specificity of alpha-chymotrypsin from computation of protein cleavages, Protein Sequences Data Anal., 1987, 1(1), 13–20 CAS.
- C. Mooney, N. J. Haslam, G. Pollastri and D. C. Shields, Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity, PLoS One, 2012, 7(10), e45012 CAS.
- Z. Yu, et al., Short- and long-term antihypertensive effect of egg protein-derived peptide QIGLF, J. Agric. Food Chem., 2017, 97(2), 551–555 CrossRef CAS PubMed.
- Z. Yu, Y. Yin, W. Zhao, F. Chen and J. Liu, Antihypertensive Effect of Angiotensin-Converting Enzyme Inhibitory Peptide RVPSL on Spontaneously Hypertensive Rats by Regulating Gene Expression of the Renin-Angiotensin System, J. Agric. Food Chem., 2014, 62(4), 912–917 CrossRef CAS PubMed.
- Z. Yu, W. Zhao, L. Ding, Y. Yu and J. Liu, Anxiolytic effects of ACE inhibitory peptides on the behavior of rats in an elevated plus-maze, Food Funct., 2016, 7(1), 491–497 CAS.
- R. Natesh, S. L. U. Schwager, E. D. Sturrock and K. R. Acharya, Crystal structure of the human angiotensin-converting enzyme-lisinopril complex, Nature, 2003, 421(6922), 551–554 CrossRef CAS PubMed.
- T. Zhang, et al., Activity Prediction and Molecular Mechanism of Bovine Blood Derived Angiotensin I-Converting Enzyme Inhibitory Peptides, PLoS One, 2015, 10(3), e0119598 Search PubMed.
- M. L. Tu, et al., Sequence analysis and molecular docking of antithrombotic peptides from casein hydrolysate by trypsin digestion, J. Funct. Foods, 2017, 32, 313–323 CrossRef CAS.
- J. P. Wu and R. E. Aluko, Quantitative structure-activity relationship study of bitter di- and tri-peptides including relationship with angiotensin I-converting enzyme inhibitory activity, J. Pept. Sci., 2007, 13(1), 63–69 CrossRef CAS PubMed.
- T. Y. Wang, et al., A study to evaluate the potential of an in silico approach for predicting dipeptidyl peptidase-IV inhibitory activity in vitro of protein hydrolysates, Food Chem., 2017, 234, 431–438 CrossRef CAS PubMed.
- S. Gupta, et al., In Silico Approach for Predicting Toxicity of Peptides and Proteins, PLoS One, 2013, 8(9), e73957 CAS.
- I. Rady, I. A. Siddiqui, M. Rady and H. Mukhtar, Melittin, a major peptide component of bee venom, and its conjugates in cancer therapy, Cancer Lett., 2017, 402, 16–31 CrossRef CAS PubMed.
- X. D. Lan, et al., Rapid purification and characterization of angiotensin converting enzyme inhibitory peptides from lizard fish protein hydrolysates with magnetic affinity separation, Food Chem., 2015, 182, 136–142 CrossRef CAS PubMed.
- S. Pan, S. Wang, L. Jing and D. Yao, Purification and characterisation of a novel angiotensin-I converting enzyme (ACE)-inhibitory peptide derived from the enzymatic hydrolysate of Enteromorpha clathrata protein, Food Chem., 2016, 211, 423–430 CrossRef CAS PubMed.
- T. Toopcham, J. J. Mes, H. J. Wichers, S. Roytrakul and J. Yongsawatdigul, Bioavailability of angiotensin I-converting enzyme (ACE) inhibitory peptides derived from Virgibacillus halodenitrificans SK1-3-7 proteinases hydrolyzed tilapia muscle proteins, Food Chem., 2017, 220, 190–197 CrossRef CAS PubMed.
- S. Rudolph, D. Lunow, S. Kaiser and T. Henle, Identification and quantification of ACE-inhibiting peptides in enzymatic hydrolysates of plant proteins, Food Chem., 2017, 224, 19–25 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |