Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Structural diversity of six metal–organic frameworks from a rigid bisimidazole ligand and their adsorption of organic dyes†
Siyu Liua,
Mingming Guoa,
Huadong Guo *a,
Yingying Suna,
Xianmin Guo*a,
Shaowen Suna and
Eugeny V. Alexandrov
*a,
Yingying Suna,
Xianmin Guo*a,
Shaowen Suna and
Eugeny V. Alexandrov *b
*b
aDepartment of Chemistry, Changchun Normal University, Changchun, 130032, P. R. China. E-mail: hdxmguo@163.com; xian_min@hotmail.com; Fax: +86-431-86168210; Tel: +86-431-86168210
bSamara Center for Theoretical Materials Science (SCTMS), Samara National Research University, Ac. Pavlov St 1, Samara, Russia 443011. E-mail: aleksandrov_ev1@mail.ru; Fax: +7-846-3356798; Tel: +7-846-3356798
First published on 23rd January 2018
Abstract
Six metal–organic frameworks, namely, [Cd2(odc)2(2,6-bin)2]·(CH3)2NH (1), [Cd2(tdc)2(2,6-bin)2(H2O)2] (2), [Cd2(bzdc)2(2,6-bin)2]·4DMF (3), [Cd2(hfdc)2(2,6-bin)2]·H2O (4), [Cd3(tpo)2(2,6-bin)3(H2O)4]·2DMF·2H2O (5) and [Zn3(btb)2(2,6-bin)]·8DMF (6) {2,6-bin = 2,6-bisimidazoylnaphthalene, H2odc = 4,4′-oxybisbenzoic acid, H2tdc = thiophene-2,5-dicarboxyl acid, H2bzdc = benzophenone-4,4′-dicarboxylic acid, H2hfdc = 2,2′-bis(4-carboxyphenyl)hexafluoropropane, H3tpo = tris-(4-carboxylphenyl)phosphine oxide and H3btb = 4,4′,4′′-benzene-1,3,5-triyl-tribenzoic acid} have been synthesized and structurally characterized. Compound 1 exhibits a three-fold interpenetration of 4T25 network. Compound 2 presents a five-fold interpenetration of dia network. Compound 3 displays a 2D → 2D three-fold interpenetration of sql layers. Compound 4 features a 2D → 3D parallel polycatenation of sql layers. Compound 5 exhibits an unusual 3,4,6-connected self-catenated network of 3,4,6T206 topology. Compound 6 shows a rare 3,8-connected self-catenated network of 3,8T72 topology. Their adsorption behaviors to organic dyes have been studied.
Introduction
Organic dyes have been widely used in medicines and the textile, paper, and printing industries, and the polluted water from dyes poses a significant threat to the environment and human health.1 Among various techniques for the removal of water pollutants, adsorption is the most attractive method owing to its simplicity and efficiency.2 Consequently, numerous adsorption materials have been extensively investigated, including carbon nanotubes, clay, zeolites, sol–gel adsorbents and polymer resins.3 However, these adsorbents often face problems like low adsorption capacity, slow adsorption kinetics, complex preparation process, and chemical or thermal instability. The development of an effective adsorbent that possesses good adsorption ability and selectivity toward dye removal is still a challenge.Recently, metal–organic frameworks (MOFs) constructed using multifunctional organic ligands as linkers and metal/metal clusters as nodes, have received considerable attention. Owing to their versatile structures, controllable pore size, high surface area and thermal stability, MOFs have been extensively investigated as a new class of solid adsorbents.4 In view of the big size of dye molecules, only MOFs with large pore size can adsorb dye molecules into the pores;5 while for others, dyes are mostly adsorbed on the external surface of materials. Thus, the surface physicochemical interactions between the dye molecules and MOF external surfaces, including hydrogen bonding, electrostatic interactions, π–π interaction, play a vital role in the adsorption of dyes.6 In this work, we chosed 2,6-bis(imidazole-1-yl)naphthalene (2,6-bin) as the building unit to construct new MOFs, because its rigid aromatic cycle can definitely promote the π–π interaction with dye molecules. Meanwhile, by the mixed-ligand strategy, a series of polycarboxylic acids with different conformations were introduced (Scheme 1), in order to evaluate the variable coordination environment and electroaffinity of metal ions to the adsorption of dyes. Successfully, we have synthesized six new MOFs, namely [Cd2(odc)2(2,6-bin)2]·(CH3)2NH (1), [Cd2(tdc)2(2,6-bin)2(H2O)2] (2), [Cd2(bzdc)2(2,6-bin)2]·4DMF (3), [Cd2(hfdc)2(2,6-bin)2]·H2O (4), [Cd3(tpo)2(2,6-bin)3(H2O)4]·2DMF·2H2O (5) and [Zn3(btb)2(2,6-bin)]·8DMF (6). These compounds have been characterized by elemental analysis, thermogravimetric analysis (TGA) and X-ray crystallography. Meanwhile, their adsorption behaviors to organic dyes have been studied.
Experimental section
All the starting materials were of analytic grade and used as received without further purification. Elemental analyses (C, H, N) were performed with a Perkin-Elmer 240c elemental analyzer. TGA was performed on a Perkin-Elmer TG-7 analyzer heated from 30 to 800 °C under nitrogen. Powder X-ray diffraction (PXRD) data were recorded on a Bruker D2 Phaser. UV-vis adsorption spectra were collected on a Shimadzu UV-3101PC spectrophotometer to monitor the adsorption progress.Synthesis of 2,6-bin
A mixture of Cu2O (0.07 g, 0.5 mmol), 2,6-dibromonaphthalene (2.86 g, 10 mmol), imidazole (2.72 g, 40 mmol), and K2CO3 (5.52 g, 40 mmol) in anhydrous DMF (30 mL) in a 100 mL two-necked round-bottom flask under N2 was stirred at 150 °C for 48 h. The reaction mixture was filtered, and then the filtrate was added into 300 mL of H2O. The deposit was filtered and washed with water and dried in vacuum to afford the product in 54%. Anal. calcd for C16H12N4 (%): C, 73.83; H, 4.65; N, 21.52. Found: C, 73.75; H, 4.60; N, 21.65.Synthesis of [Cd2(odc)2(2,6-bin)2]·(CH3)2NH (1)
A mixture of Cd(NO3)2·4H2O (0.0308 g, 0.1 mmol), H2odc (0.0258 g, 0.1 mmol), 2,6-bin (0.0260 g, 0.1 mmol), DMF (8 ml) and H2O (2 ml) was placed in a Teflon reactor (20 ml) and heated at 80 °C for 2 days. After gradually cooled to room temperature at a rate of 10 °C h−1, colorless crystals of 1 were obtained with 60% yield based on 2,6-bin. Anal. calcd for C62H47N9O10Cd2: C, 57.16; H, 3.64; N, 9.68. Found: C, 57.30; H, 3.69; N, 9.50.Synthesis of [Cd2(tdc)2(2,6-bin)2(H2O)2] (2)
Complex 2 was synthesized following the same synthetic procedure as that for complex 1 except that H2tdc was used instead of H2odc. Colorless crystals of 2 were obtained with 54% yield based on 2,6-bin. Anal. calcd for C44H32N8O10S2Cd2: C, 47.11; H, 2.88; N, 9.99. Found: C, 47.24; H, 2.74; N, 9.83.Synthesis of [Cd2(bzdc)2(2,6-bin)2]·4DMF (3)
Complex 3 was synthesized following the same synthetic procedure as that for complex 1 except that H2bzdc was used instead of H2odc. Colorless crystals of 3 were obtained with 56% yield based on 2,6-bin. Anal. calcd for C74H68Cd2N12O14: C, 56.46; H, 4.35; N, 10.68. Found: C, 56.60; H, 4.39; N, 10.54.Synthesis of [Cd2(hfdc)2(2,6-bin)2]·H2O (4)
Complex 4 was synthesized following the same synthetic procedure as that for complex 1 except that H2hfdc was used instead of H2odc. Colorless crystals of 4 were obtained with 52% yield based on 2,6-bin. Anal. calcd for C66H42F12N8O9Cd2: C, 51.35; H, 2.74; N, 7.26. Found: C, 51.46; H, 2.65; N, 7.40.Synthesis of [Cd3(tpo)2(2,6-bin)3(H2O)4]·2DMF·2H2O (5)
Complex 5 was synthesized following the same synthetic procedure as that for complex 1 except that H3tpo was used instead of H2odc. Colorless crystals of 5 were obtained with 48% yield based on 2,6-bin. Anal. calcd for C96H86Cd3N14O22P2: C, 52.72; H, 3.96; N, 8.97. Found: C, 52.86; H, 3.79; N, 9.11.Synthesis of [Zn3(btb)2(2,6-bin)]·8DMF (6)
Complex 6 was synthesized following the same synthetic procedure as that for complex 1 except that H3btb was used instead of H2odc and Zn(NO3)2·6H2O was used instead of Cd(NO3)2·4H2O. Colorless crystals of 6 were obtained with 49% yield based on 2,6-bin. Anal. calcd for C94H98N12O20Zn3: C, 59.05; H, 5.17; N, 8.79. Found: C, 59.15; H, 5.25; N, 8.68.X-ray crystallography
Single-crystal XRD data for compounds 1–6 were recorded on a Bruker Apex II diffractometer with graphite monochromatized Mo Kα radiation (λ = 0.71073 Å) at 285(2) K. Absorption corrections were applied using the multiscan technique. All the structures were solved by Direct Method of SHELXS-97 (ref. 7) and refined by the full-matrix least-squares techniques by using the SHELXL-97 (ref. 8) program within WINGX. No-hydrogen atoms were refined with anisotropic temperature parameters. The hydrogen atoms of the organic ligands were refined as rigid groups. For the high vibration, the disordered Ow1 in compound 4 was refined with a total occupancy of 1 and handled by isotropic refinement. The hydrogen atoms of water molecules in compounds 2, 4, 5 are not added. It should be noted that the guest molecules in 6 are highly disordered and could not be modeled properly so the diffused electron densities resulting from them were removed by the SQUEEZE routine in PLATON.9 The detailed crystallographic data and structure refinement parameters for 1–6 are summarized in Table 1.| Parameters | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Formula | C62H47Cd2N9O10 | C44H32Cd2N8O10S2 | C74H68Cd2N12O14 | C66H42Cd2F12N8O9 | C96H86Cd3N14O22P2 | C94H98N12O20Zn3 |
| fw | 1302.89 | 1121.70 | 1574.20 | 1543.88 | 2186.93 | 1911.95 |
| Space group | C2/c | P21/c | P21/c | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
C2/c |
| a | 22.0150(10) | 11.376(2) | 11.6567(5) | 9.9983(5) | 10.6823(7) | 30.643(3) |
| b | 12.2707(6) | 17.593(3) | 27.1818(11) | 10.2280(5) | 15.8149(9) | 10.5532(10) |
| c | 21.9979(15) | 11.651(2) | 11.3231(5) | 14.6829(8) | 15.9776(10) | 31.782(4) |
| α (deg) | 90 | 90 | 90 | 92.2540(10) | 63.8890(10) | 90 |
| β (deg) | 115.6130(10) | 114.192(4) | 99.4190(10) | 97.6100(10) | 79.1640(10) | 116.594(2) |
| γ (deg) | 90 | 90 | 90 | 92.0170(10) | 72.1500(10) | 90 |
| V | 5358.6(5) | 2127.0(7) | 3539.4(3) | 1485.90(13) | 2302.6(2) | 9190.4(17) |
| Z | 4 | 2 | 2 | 1 | 1 | 4 |
| Dcalcd (g cm−3) | 1.615 | 1.751 | 1.477 | 1.725 | 1.577 | 1.382 |
| F(000) | 2632 | 1120 | 1608 | 770 | 1110 | 3984 |
| GOF on F2 | 1.070 | 1.020 | 1.084 | 1.025 | 1.061 | 0.925 |
| R1/wR2[I ≥ 2sigma(I)] | 0.0266/0.0712 | 0.0390/0.0698 | 0.0367/0.0841 | 0.0352/0.0744 | 0.0472/0.1230 | 0.0446/0.1055 |
| R1/wR2 (all data) | 0.0329/0.0746 | 0.0724/0.0820 | 0.0488/0.0889 | 0.0484/0.0814 | 0.0578/0.1295 | 0.0704/0.1114 |
Results and discussion
Structure description of 1
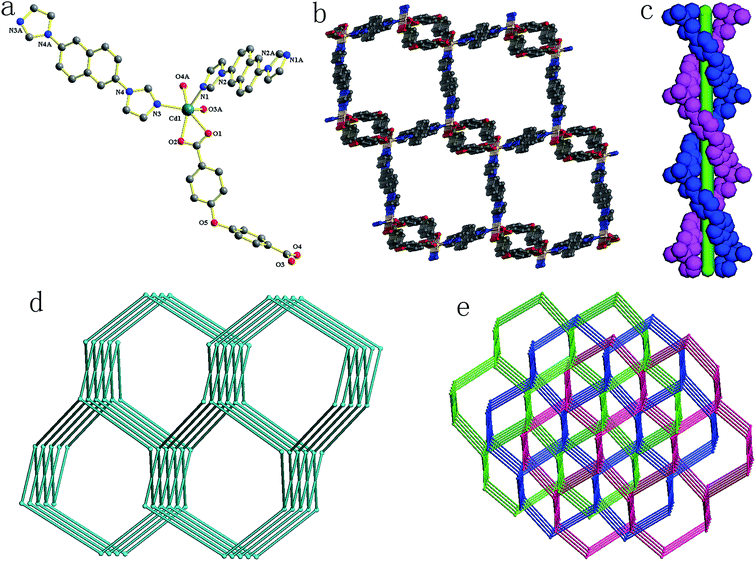
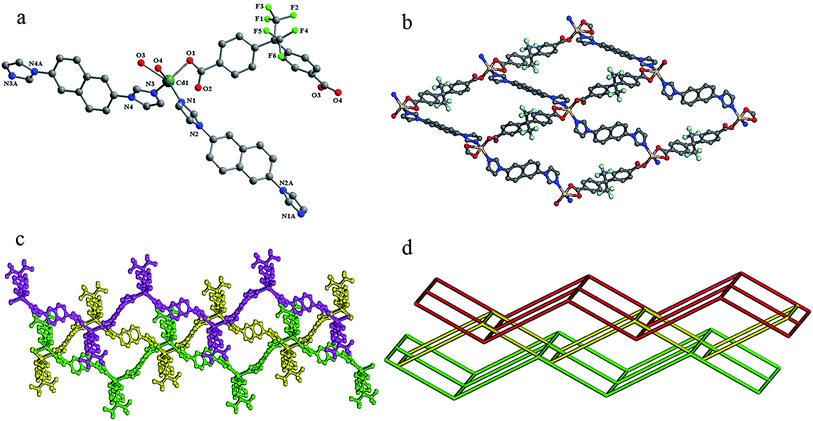
Compound 1 crystallizes in a monoclinic space group P21/n. In the asymmetric unit, there exists one crystallographically unique Cd(II) atom, one odc2− anion, two half 2,6-bin ligands and one free (CH3)2NH. As shown in Fig. 1a, the Cd(II) atom is ligated by four oxygen atoms from two odc2− anions (Cd–O, 2.2854(16)–2.5380(16) Å) and two nitrogen atoms from two 2,6-bin ligands (Cd–N, 2.2370(17)–2.2594(18) Å). The Cd(II) to O/N distances and bond angles are within the normal range (Table S1 in the ESI†). The odc2− anion connects two Cd(II) atoms through two chelate carboxlylates. The 2,6-bin ligand acts as a linker to bridge two Cd(II) atoms. In compound 1, every two odc2− anions are connected by Cd(II) atoms to give a 1D helical chain with a pitch of 24.54 Å, corresponding to the double length of b axis. Two such chains interweave together to assemble a double helix (Fig. 1c). These helixes are bridged by the 2,6-bin to build a 3D framework (Fig. 1b). Based on the concept of topology,10 the framework can be simplified into a 4-connected 4T25 network, with the point symbol of (65·8) (Fig. 1d). The potential voids are filled via mutual interpenetration of two independent equivalent networks, giving a rise to a 3-fold interpenetrating network (Fig. 1e). The 4T25 features self-catenation,11 which is detected as catenation of strong 10-rings and 14-rings of the same net, when odc2− ligand is presented by 2-coordinated node to avoid edge-crossings (Fig. S7†). The catenation in the net originates from interweaving of the helical chains [Cd(odc)]. The same topology was found before only in one compound, [Zn(tpdc)(bpmp)] (H2tpdc = 1,1′:3′,1′′-terphenyl-4,4′′-dicarboxylic acid; bpmp = 1,4-bis(3-pyridylmethyl)piperazine).12Structure description of 2
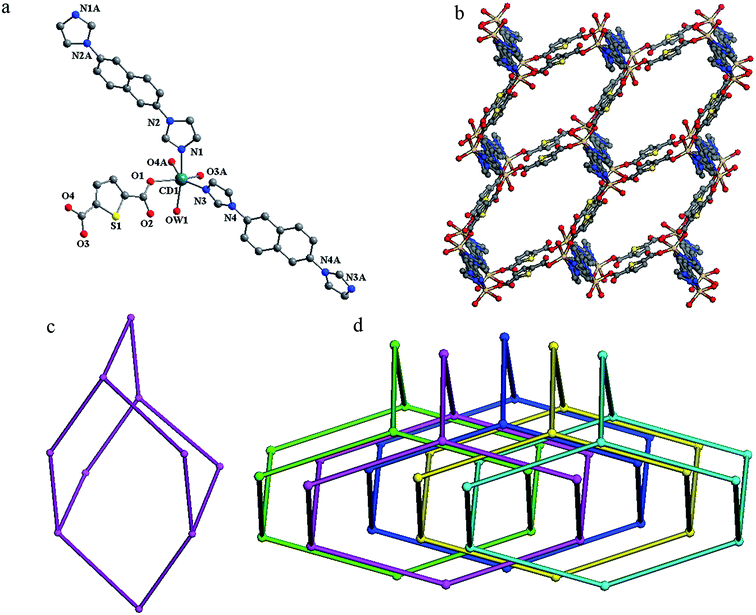
Compound 2 crystallizes in a monoclinic space group P21/c. In the asymmetric unit, there exists one crystallographically unique Cd(II) atom, one tdc2− anion, two half 2,6-bin ligands and one coordinated water molecule. As shown in Fig. 2a, the Cd(II) atom is surrounded by four oxygen atoms from two tdc2− anions and one water molecule (Cd–O, 2.256(3)–2.333(2) Å), and two nitrogen atoms from two 2,6-bin ligands (Cd–N, 2.274(3)–2.276(3) Å). The Cd(II) to O/N distances and bond angles are within the normal range (Table S2 in the ESI†). The tdc2− anion connects two Cd(II) atoms through one monodentate and one chelate carboxlylate. The 2,6-bin ligand acts as a linker to bridge two Cd(II) atoms. Thus, the Cd(II) atoms are joined by the organic ligands into framework of dia topology (Fig. 2b). Within the framework, the neighboring Cd(II) atoms are separated by distances of 15.67 Å, 16.23 Å (through 2,6-bin) and 10.91 Å (via tdc2−). A single adamantanoid cage is illustrated in Fig. 2c, which exhibits maximum dimensions of 40.33 Å × 34.04 Å × 32.01 Å × 29.30 Å (corresponding to the longest intracage Cd⋯Cd distances). Because of the spacious nature of the single network, the potential voids are filled via mutual interpenetration of the other four independent equivalent frameworks, generating a five-fold interpenetrating architecture (Fig. 2d).Structure description of 3
Compound 3 crystallizes in a monoclinic space group P21/c. In the asymmetric unit, there exists one crystallographically unique Cd(II) atom, one bzdc2− anion, two half 2,6-bin ligands and two uncoordinated DMF molecules. As shown in Fig. 3a, the Cd(II) atom is coordinated by four oxygen atoms from two bzdc2− anions (Cd–O, 2.2787(18)–2.4132(18) Å) and two nitrogen atoms from two 2,6-bin ligands (Cd–N, 2.238(2)–2.256(2) Å). The Cd(II) to O/N distances and bond angles are within the normal range (Table S3 in the ESI†). The bzdc2− anion connects two Cd(II) atoms through two chelate carboxylates. The 2,6-bin ligands act as linkers to join two Cd(II) atoms. In compound 3, the Cd(II) atoms are bridged by organic ligands to assemble a 2D undulated framework of sql topology (Fig. 3b). The neighboring Cd(II) atoms are separated by distances of 15.59 Å, 16.28 Å (through 2,6-bin) and 14.86 Å (via bzdc2−). The void space in the single framework is so large that three identical frameworks having one middle plane catenate each other to form a 2D → 2D polycatenane of frequently observed pattern, which can be described by 8L23 extended ring net (Fig. 3d).13 For the mean planes of these three layers are parallel and coincident, we can see that along the a axis there still exist 1D open channels, which are occupied by the uncoordinated DMF molecules (Fig. 3e). The void space accounts for approximately 32.7% of the whole crystal volume as obtained by PLATON analysis.Structure description of 4
Compound 4 crystallizes in a triclinic space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) . In the asymmetric unit, there exists one crystallographically unique Cd(II) atom, one hfdc2− anion and two half 2,6-bin ligands. As shown in Fig. 4a, the Cd(II) atom is coordinated by three oxygen atoms from two hfdc2− anions (Cd–O, 2.2175(17)–2.403(3) Å) and two nitrogen atoms from two 2,6-bin ligands (Cd–N, 2.235(2)–2.245(2) Å). The Cd(II) to O/N distances and bond angles are within the normal range (Table S4 in the ESI†). The hfdc2− anion connects two Cd(II) atoms by one monodentate and one chelate carboxylates. The 2,6-bin ligands act as linkers to bridge two Cd(II) atoms. In compound 4, the Cd(II) atoms are joined by organic ligands to assemble a framework of sql topology (Fig. 4b). Within the framework, the neighboring Cd(II) atoms are separated by distances of 15.53 Å, 15.57 Å (through 2,6-bin) and 14.68 Å (by hfdc2−). In order to minimize the space vacuum and stabilize the framework, each undulated layer catenates with two others from above and below. Although the catenated nets in 4 also have parallel mean planes, different from 3, these mean planes are not coincident and the neighbors are separated by distances of 4.53 Å, thus giving a 2D → 3D polycatenane. This polycatenated system can be described by extended ring net with topology hex frequent for 2D coordination polymers (Fig. 4c and d).13
. In the asymmetric unit, there exists one crystallographically unique Cd(II) atom, one hfdc2− anion and two half 2,6-bin ligands. As shown in Fig. 4a, the Cd(II) atom is coordinated by three oxygen atoms from two hfdc2− anions (Cd–O, 2.2175(17)–2.403(3) Å) and two nitrogen atoms from two 2,6-bin ligands (Cd–N, 2.235(2)–2.245(2) Å). The Cd(II) to O/N distances and bond angles are within the normal range (Table S4 in the ESI†). The hfdc2− anion connects two Cd(II) atoms by one monodentate and one chelate carboxylates. The 2,6-bin ligands act as linkers to bridge two Cd(II) atoms. In compound 4, the Cd(II) atoms are joined by organic ligands to assemble a framework of sql topology (Fig. 4b). Within the framework, the neighboring Cd(II) atoms are separated by distances of 15.53 Å, 15.57 Å (through 2,6-bin) and 14.68 Å (by hfdc2−). In order to minimize the space vacuum and stabilize the framework, each undulated layer catenates with two others from above and below. Although the catenated nets in 4 also have parallel mean planes, different from 3, these mean planes are not coincident and the neighbors are separated by distances of 4.53 Å, thus giving a 2D → 3D polycatenane. This polycatenated system can be described by extended ring net with topology hex frequent for 2D coordination polymers (Fig. 4c and d).13
Structure description of 5
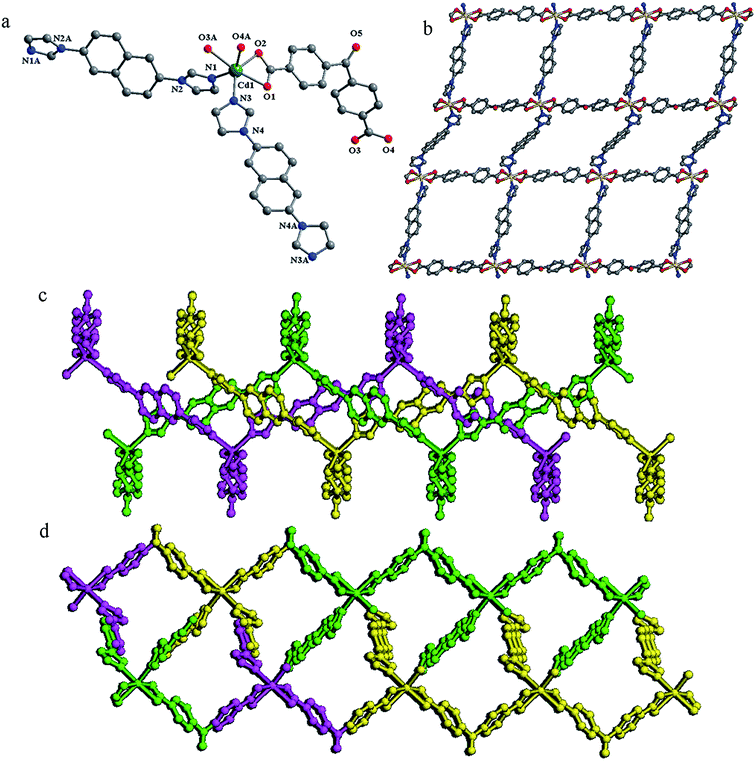
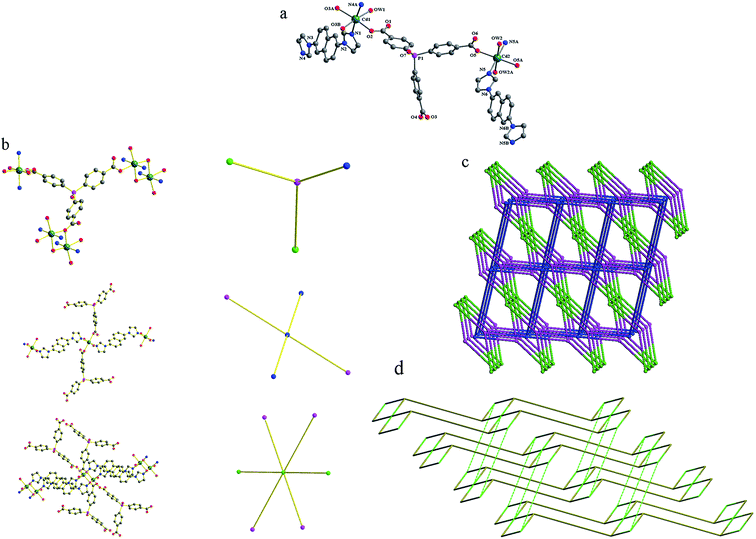
Compound 5 crystallizes in a triclinic space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) . The asymmetric unit contains one and a half Cd(II) atoms, one tpo3− anion, one and a half 2,6-bin, two coordination water molecules, one lattice water molecule and one lattice DMF molecule. As shown in Fig. 5a, there are two crystallographically Cd(II) atoms in compound 5. Both Cd1 and Cd2 are six-coordinated in distorted octahedral coordination geometries, but their coordination environments are entirely different. Cd1 is coordinated by two nitrogen atoms from two 2,6-bin ligands (Cd–N, 2.262(3)–2.299(3) Å), and four oxygen atoms from three tpo3− anions and one water molecule (Cd–O, 2.272(3)–2.448(3) Å). Cd2 is coordinated by two nitrogen atoms from one 2,6-bin ligands (Cd–N, 2.271(3) Å), and four oxygen atoms from two tpo3− anions and two water molecules (Cd–O, 2.285(3)–2.318(3) Å). The Cd(II) to O/N distances and bond angles are within the normal range (Table S5 in the ESI†). The tpo3− anion links four Cd(II) atoms by two monodentate and one monodentate-bridging carboxylates. Cd1 and its symmetry-related Cd1A are bridged by two μ2-O3 atoms from the monodentate-bridging carboxylates into a binuclear Cd2 unit, with a Cd1⋯Cd1A distance of 3.65 Å and a Cd1–O3–Cd1A angle of 99.68°. Each binuclear unit is surrounded by eight organic ligands (four 2,6-bin and four tpo3−), each Cd2 atom coordinates four organic ligands (two 2,6-bin and two tpo3−), and each tpo3− anion connects one Cd2 atom and two binuclear units. Topologically, the tpo3− anions can be considered as 3-connected nodes, Cd2 atoms can be viewed as 4-connected nodes, and the binuclear units can be reduced to 6-connected nodes (Fig. 5b). Therefore, the whole 3D framework can be simplified as a (3,4,6)-connected net of 3,4,6T206 topology (Fig. 5c). To the best of our knowledge, there is only one coordination polymer of this topology: {[Co3(L)(BPY)1.5]·H2O}n (CSD RefCode EWUHUB: H3L = 4-methyoxy-3,5-bis(4-carboxyphenyl)benzoic acid, BPY = 4,4′-bipyridine).6c The 3,4,6T206 net has a peculiar property of self-catenation. This property originates from mixing ligands of different types into one framework. The tricarboxilate tpo3− ligand forms 2D subnet (partial composition [Cd3(tpo)2]) of 3,4-c bex topology, which is very common for 2D coordination polymers.14 The 2D subnets pack parallel to each other, and 2,6-bin ligands steach them into 3D framework passing through strong 6-rings of the bex layers (Fig. 5d).
. The asymmetric unit contains one and a half Cd(II) atoms, one tpo3− anion, one and a half 2,6-bin, two coordination water molecules, one lattice water molecule and one lattice DMF molecule. As shown in Fig. 5a, there are two crystallographically Cd(II) atoms in compound 5. Both Cd1 and Cd2 are six-coordinated in distorted octahedral coordination geometries, but their coordination environments are entirely different. Cd1 is coordinated by two nitrogen atoms from two 2,6-bin ligands (Cd–N, 2.262(3)–2.299(3) Å), and four oxygen atoms from three tpo3− anions and one water molecule (Cd–O, 2.272(3)–2.448(3) Å). Cd2 is coordinated by two nitrogen atoms from one 2,6-bin ligands (Cd–N, 2.271(3) Å), and four oxygen atoms from two tpo3− anions and two water molecules (Cd–O, 2.285(3)–2.318(3) Å). The Cd(II) to O/N distances and bond angles are within the normal range (Table S5 in the ESI†). The tpo3− anion links four Cd(II) atoms by two monodentate and one monodentate-bridging carboxylates. Cd1 and its symmetry-related Cd1A are bridged by two μ2-O3 atoms from the monodentate-bridging carboxylates into a binuclear Cd2 unit, with a Cd1⋯Cd1A distance of 3.65 Å and a Cd1–O3–Cd1A angle of 99.68°. Each binuclear unit is surrounded by eight organic ligands (four 2,6-bin and four tpo3−), each Cd2 atom coordinates four organic ligands (two 2,6-bin and two tpo3−), and each tpo3− anion connects one Cd2 atom and two binuclear units. Topologically, the tpo3− anions can be considered as 3-connected nodes, Cd2 atoms can be viewed as 4-connected nodes, and the binuclear units can be reduced to 6-connected nodes (Fig. 5b). Therefore, the whole 3D framework can be simplified as a (3,4,6)-connected net of 3,4,6T206 topology (Fig. 5c). To the best of our knowledge, there is only one coordination polymer of this topology: {[Co3(L)(BPY)1.5]·H2O}n (CSD RefCode EWUHUB: H3L = 4-methyoxy-3,5-bis(4-carboxyphenyl)benzoic acid, BPY = 4,4′-bipyridine).6c The 3,4,6T206 net has a peculiar property of self-catenation. This property originates from mixing ligands of different types into one framework. The tricarboxilate tpo3− ligand forms 2D subnet (partial composition [Cd3(tpo)2]) of 3,4-c bex topology, which is very common for 2D coordination polymers.14 The 2D subnets pack parallel to each other, and 2,6-bin ligands steach them into 3D framework passing through strong 6-rings of the bex layers (Fig. 5d).
Structure description of 6
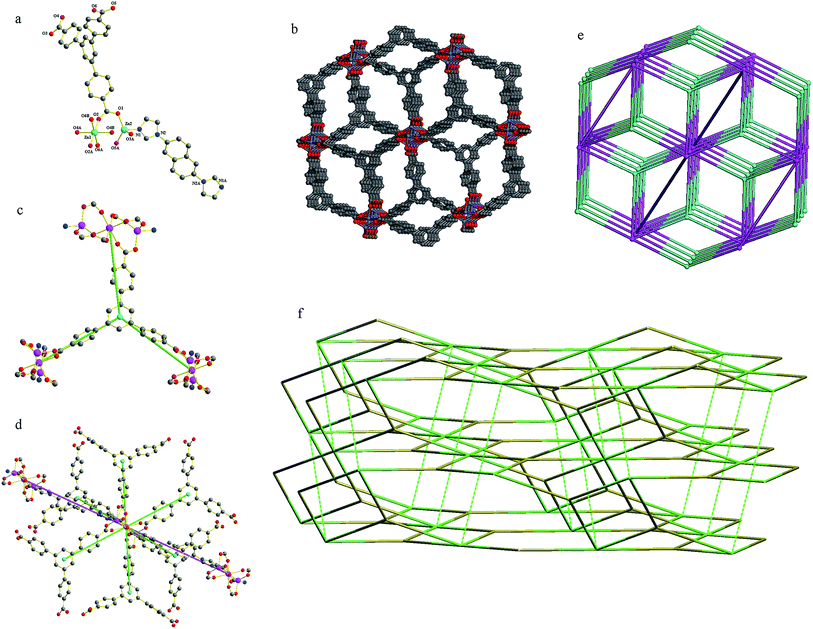
Compound 6 crystallizes in a triclinic space group C2/c. The asymmetric unit contains one and a half Zn(II) atoms, one btb3− anion and half a 2,6-bin ligand. As shown in Fig. 6a, there are two crystallographically different Zn(II) atoms in compound 6. Zn1 coordinates six oxygen atoms from six btb3− anions to give a distorted octahedral geometry (Zn–O, 2.067(2)–2.1019(19) Å). Zn2 is ligated by one nitrogen atom from one 2,6-bin ligand (Zn–N, 2.005(2) Å) and four oxygen atoms from four btb3− anions (Zn–O, 1.908(2)–1.962(2) Å) into a distorted square pyramid geometry. The Zn(II) to O/N distances and bond angles are within the normal range (Table S6 in the ESI†). The btb3− anion links six Zn(II) atoms by three chelate carboxylates. The Zn1 atom is situated at an inversion center and it is connected with two neighboring Zn2 atoms by six bis(monodentate) carboxylates to form a trinuclear [Zn3(CO2)6] cluster. Without considering the linear 2,6-bin ligands, these clusters are combined by the btb3− anions into a (3,6)-connected net of flu-3,6-C2/c topology. When the linear ligands are taken into account, topologically, the btb3− anions can be considered as 3-connected nodes (Fig. 6c), and the trinuclear [Zn3(CO2)6] cluster can be viewed as 8-connected nodes (Fig. 6d). Thus, the whole 3D framework can be simplified as a (3,8)-connected net of 3,8T72 topology (Fig. 6b and e). On the bases of automatic analysis by ToposPro, this is the second example of such topology, the first one was [Co3(L)2(DPDP)]n (CSD RefCode GUFQIJ: H3L = 4,4′,4′′-(nitrilotris-(methylene))tribenzoic acid, DPDP = 4,4′-(2,5-dibutoxy-1,4-phenylene)-dipyridine).15 The 3,8T72 net is also can be considered as self-catenated, where strong 4-rings (which are also strong rings of subnet flu-3,6-C2/c) are threaded by edges corresponded to 2,6-bin ligand (Fig. 6f).Adsorption capability towards methyl orange
The capture abilities of 1–6 towards to methyl orange (MO) were evaluated. The adsorption experiments were run as follows: 5 mg of host material was immersed to 10 mL of MO aqueous solutions (4 × 10−5 mol L−1), respectively. The clear solution after centrifuging was measured by UV/vis spectra to monitor the MO concentration at different time intervals. As shown in Fig. 7, for compounds 1–5, within 10–30 minutes, the adsorption efficiencies of MO reached 99.3% for 1, 97.9% for 3, 93.0% for 4 and almost 100% for 2 and 5. Obviously, these compounds exhibit highly efficient adsorption to MO. Comparatively, compound 6 presents interesting adsorption behavior to MO: at 20 minutes, the adsorption of MO reached a maximum of 60.3%; while, at 30 minutes, the value of adsorption decreased to 16.3%, which means the desorption of MO from compound 6; till to 60 minutes, the value of adsorption was stable at 55.4%.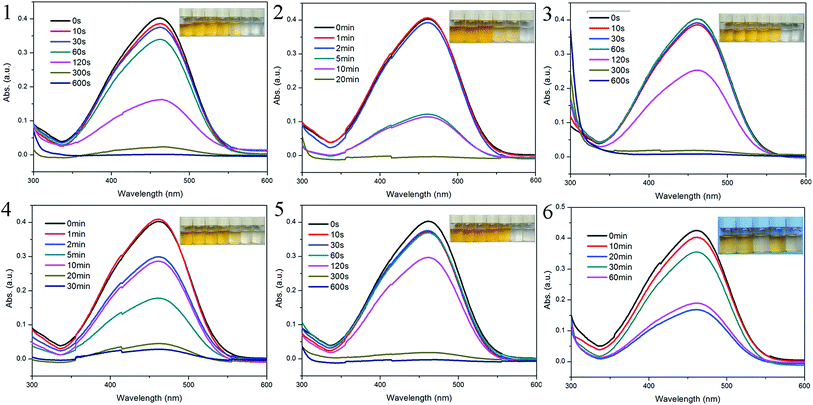 | ||
| Fig. 7 UV-vis spectra of MO with sample 1–6 at given intervals, respectively. Inset: photographic images of MO solutions for different period of time. | ||
From the structures above, we can find that only compounds 3 and 6 exhibit open frameworks, but the channels with minor window size are hard to trap MO molecules. Thus, the adsorption of 1–6 occurs mainly due to the surface interactions. In compound 6, the Zn3 cluster is surrounded by six bis(monodentate) carboxylates and the axial position of metal clusters are occupied by 2,6-bin ligands, which blockages the accessment of MO molecules. Thus, the adsorption is mainly ascribed by the π–π conjugations between benzene rings from host framework and MO molecules.16 Such weak interactions induce the spontaneous desorption of MO molecules in the process of adsorption. While, in compounds 1–5, besides the π–π conjugation effects, the bigger ionic radius of Cd(II) is favorable to the electrostatic interaction between central metal ions and anionic MO molecules.17 Such synergistic effect definitely improves the adsorption ability of host materials.
In order to confirm the mechanism of MO adsorption for 1–6, the adsorption–desorption cycle tests were evaluated. The release tests were carried out with methanol as the elution solvent. The MO-uptaked 1–6 were dispersed in 10 ml methanol by sonication for 5 min and centrifuged. After 5 times, the combined solution was measured by UV/vis spectra to monitor the MO concentration. The results revealed that the adsorbed MO molecules can be released almost completely (Fig. S9–S14, ESI†). Obviously, the weak surface interactions due to the π–π conjugation effects favor the release of MO molecules from host frameworks in methanol. The PXRD patterns of MO-released 1–6 indicate the stability of crystalline sample (Fig. S1–S6 in ESI†). Meanwhile, the MO-released 1–6 still show good reusability of MO adsorption (Fig. S9–S14, ESI†). The adsorption capacities of 1–6 towards cationic dyes methylene blue (MB) and rhodamine B (RhB) were also evaluated. As shown in Fig. S15–S20 (ESI),† the UV/vis adsorption spectra showed that these compounds almost do not exhibit adsorption of MB and RhB, in spite of their larger rigid aromatic cycle comparing to MO molecule. The results further demonstrated that the electrophilicity of central metal ions plays an important role for the adsorption of anionic dye.
Conclusions
In this study, based on the mixed-ligand strategy, six new MOFs have been synthesized an structurally characterized. Compound 1 exhibits a three-fold interpenetration of 4T25 network. Compound 2 presents a five-fold interpenetration of dia network. Compound 3 displays a 2D → 2D three-fold interpenetration of sql layers. Compound 4 features a 2D → 3D parallel polycatenation of sql layers. Compound 5 exhibits an unusual 3,4,6-connected self-catenated network of 3,4,6T206 topology. Compound 6 shows a rare 3,8-connected self-catenated network of 3,8T72 topology. The result of this study demonstrates that the rational selection of organic ligands with specific rigidity and length is an effective approach to the construction of entangled motifs. Almost all of these compounds exhibit selective adsorption to anionic methyl orange, which demonstrates that the surface physicochemical interactions including electrostatic interactions and π–π conjugation effects play a significant synergistic effect in the adsorbing dyes. These compounds not only fill the aesthetic diversity of coordinative network chemistry, but also may provide a potential way to the selective design of versatile network-based materials.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support for this work from the National Natural Science Foundation of China (No. 21601019), Jilin Natural Science Foundation of China (No. 20160414031GH) and Natural Science Foundation of Changchun Normal University (cscxy2017004, cxcxy2017028). EVA is gratefull to the Russian Science Foundation (Grant No. 16-13-10158).Notes and references
- (a) H. Xue, Q. H. Chen, F. L. Jiang, D. Q. Yuan, G. X. Lv, L. F. Liang, L. Y. Liu and M. C. Hong, Chem. Sci., 2016, 7, 5983 RSC; (b) V. Suba and G. Rathika, J. Adv. Phys., 2016, 5, 277 CrossRef; (c) A. V. Desai, B. Manna, A. Karmakar, A. Sahu and S. K. Ghosh, Angew. Chem., Int. Ed., 2016, 55, 7811 CrossRef CAS PubMed; (d) S. X. Duan, J. X. Li, X. Liu, Y. N. Wang, S. Y. Zeng, D. D. Shao and T. Hayat, ACS Sustainable Chem. Eng., 2016, 4, 3368 CrossRef CAS; (e) B. Wang, X. L. Lv, D. W. Feng, L. H. Xie, J. Zhang, M. Li, Y. B. Xie, J. R. Li and H. C. Zhou, J. Am. Chem. Soc., 2016, 138, 6204 CrossRef CAS PubMed.
- (a) V. K. Gupta and T. A. Saleh, Environ. Sci. Pollut. Res., 2013, 20, 2828 CrossRef CAS PubMed; (b) A. Dey, S. K. Konavarapu, H. S. Sasmal and K. Biradha, Cryst. Growth Des., 2016, 16, 5976 CrossRef CAS; (c) R. Bibi, L. F. Wei, Q. H. Shen, W. Tian, O. Oderinde, N. X. Li and J. C. Zhou, J. Chem. Eng. Data, 2017, 62, 1615 CrossRef CAS; (d) M. L. Gao, W. J. Wang, L. Liu, Z. B. Han, N. Wei, X. M. Cao and D. Q. Yuan, Inorg. Chem., 2017, 56, 511 CrossRef CAS PubMed.
- (a) L. Zhou, C. Gao and W. J. Xu, ACS Appl. Mater. Interfaces, 2010, 2, 1483 CrossRef CAS PubMed; (b) N. Peng, D. Y. Hu, J. Zeng, Y. Li, L. Liang and C. Y. Chang, ACS Sustainable Chem. Eng., 2016, 4, 7217 CrossRef CAS; (c) V. K. Gupta and A. Nayak, Chem. Eng. J., 2012, 180, 81 CrossRef CAS; (d) C. C. de Escobar, A. Fisch and J. H. Z. dos Santos, Ind. Eng. Chem. Res., 2015, 54, 254 CrossRef.
- (a) J. B. DeCoste and G. W. Peterson, Chem. Rev., 2014, 114, 5695 CrossRef CAS PubMed; (b) S. N. Zhao, X. Z. Song, M. Zhu, X. Meng, L. L. Wu, J. Feng, S. Y. Song and H. J. Zhang, Chem.–Eur. J., 2015, 21, 9748 CrossRef CAS PubMed; (c) Y. B. He, W. Zhou, G. D. Qian and B. L. Chen, Chem. Soc. Rev., 2014, 43, 5657 RSC; (d) J. N. Hao and B. Yan, Adv. Funct. Mater., 2017, 27, 1603856 CrossRef.
- (a) A. D. Pournara, S. Rapti, E. Skliri, G. S. Armatas, A. C. Tsipis and M. J. Manos, ChemPlusChem, 2017, 82, 1188 CrossRef CAS; (b) M. M. Guo, S. Y. Liu, H. D. Guo, Y. Y. Sun, X. M. Guo and R. P. Deng, Dalton Trans., 2017, 46, 14988 RSC.
- (a) Q. Chen, Q. Q. He, M. M. Lv, Y. L. Xu, H. B. Yang, X. T. Liu and F. Y. Wei, Appl. Surf. Sci., 2015, 327, 77 CrossRef CAS; (b) W. Yan, L. J. Han, H. L. Jia, K. Shen, T. Wang and H. G. Zheng, Inorg. Chem., 2016, 55, 8816 CrossRef CAS PubMed; (c) N. Zhao, F. X. Sun, N. Zhang and G. S. Zhu, Cryst. Growth Des., 2017, 17, 2453 CrossRef CAS.
- G. M. Sheldrick. SHELXS-97, Programs for X-ray Crystal Structure Solution, University of Göttingen, Göttingen, Germany, 1997 Search PubMed.
- G. M. Sheldrick. SHELXL-97, Programs for X-ray Crystal Structure Refinement, University of Göttingen, Göttingen, Germany, 1997 Search PubMed.
- A. Spek, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2009, 65, 148 CrossRef CAS PubMed.
- (a) E. V. Alexandrov, V. A. Blatov, A. V. Kochetkov and D. M. Proserpio, CrystEngComm, 2011, 13, 3947 RSC; (b) V. A. Blatov, A. P. Shevchenko and D. M. Proserpio, Cryst. Growth Des., 2014, 14, 3576 CrossRef CAS.
- L. Carlucci, G. Ciani and D. M. Proserpio, Coord. Chem. Rev., 2003, 246, 247 CrossRef CAS.
- B. Liu, J. Coord. Chem., 2015, 68, 1251 CrossRef CAS.
- E. V. Alexandrov, V. A. Blatov and D. M. Proserpio, CrystEngComm, 2017, 19, 1993 RSC.
- T. G. Mitina and V. A. Blatov, Cryst. Growth Des., 2013, 13, 1655 CAS.
- C. L. Zhang, Y. L. Li, T. Wang, Z. M. Ju, H. G. Zheng and J. Ma, Chem. Commun., 2015, 51, 8338 RSC.
- R. Bibi, L. F. Wei, Q. H. Shen, W. Tian, O. Oderinde, N. X. Li and J. C. Zhou, J. Chem. Eng. Data, 2017, 62, 1615 CrossRef CAS.
- (a) M. S. Bootharaju and T. Pradeep, Langmuir, 2013, 29, 8125 CrossRef CAS PubMed; (b) M. Dan-Hardi, C. Serre, T. Frot, L. Rozes, G. Maurin, C. Sanchez and G. Ferey, J. Am. Chem. Soc., 2009, 131, 10857 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: XRD, TGA. CCDC 1576790–1576795. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c7ra11754j |
| This journal is © The Royal Society of Chemistry 2018 |