Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
DNA-induced synthesis of biomimetic enzyme for sensitive detection of superoxide anions released from live cell†
Ailing Ding,
Bin Wang *,
Xiaoqing Ma,
Jianglin Diao,
Jiushang Zheng,
Jiucun Chen and
Changming Li
*,
Xiaoqing Ma,
Jianglin Diao,
Jiushang Zheng,
Jiucun Chen and
Changming Li *
*
Institute for Clean Energy & Advanced Materials, Faculty of Materials and Energy, Southwest University, Chongqing 400715, P. R. China. E-mail: bwang@swu.edu.cn; ecmli@swu.edu.cn
First published on 3rd April 2018
Abstract
In this work, we successfully fabricate a rapid, sensitive sensor for the detection of superoxide anions O2˙− based on graphene/DNA/Mn3(PO4)2 biomimetic enzyme. In the design, graphene is served as excellent carrier to improve the catalysis of Mn3(PO4)2 nanoparticles; and DNA adsorbed on graphene acts as medium to assist the growth of Mn3(PO4)2 on graphene. The fabricated graphene/DNA/Mn3(PO4)2 composites exhibit excellently electrochemical activity, significantly decrease the response time and increase the sensitivity of the sensor towards O2˙−. The successful detection of O2˙− released from cancer cell demonstrated its potential applications in biology and medicine.
Introduction
Superoxide anions (O2˙−) are the primary type of reactive oxygen species (ROS). Under normal conditions, O2˙− is highly reactive and unstable and its metabolism is a rapid and spontaneous process. Overproduction of O2˙− can cause various disease, such as aging, asthma, ulcer disease, cancer, atherosclerosis, neurodegenerative diseases and other diseases.1–4 Thus the detection of O2˙− is very important in various biological systems. Many methods have been developed for the detection of O2˙− including chemiluminescent, spectrophotometric, fluorometric, and electrochemical technique, etc.5–8 Among all these techniques, electrochemical method receives most attention due to its advantages of real-time assay, high sensitivity and selectivity. And most of the reported electrochemical methods for O2˙− detection are based on enzyme catalysts such as cytochrome c and superoxide dismutase (SOD).2,9–11 Though enzyme improves the assay performance of sensors, but its high cost and poor long-term stability increase difficulty of direct monitoring of O2˙− in biological samples. Therefore, it is a serious challenge for us to establish a fast, reliable, and sensitive non-enzyme approach for O2˙− monitoring in physiological and pathological processes.As early as 1982, manganese was reported possessing an effective catalytic effect in vivo protection against superoxide toxicity.12 In recent years, further study found that manganese phosphate (Mn3(PO4)2) has the ability to catalyze the dismutation of O2˙− compared with free Mn2+ ion that only stoichiometrically reacts with O2˙−.13 Therefore Mn3(PO4)2 is usually selected as a substitute for enzyme to detect O2˙−.14 Mao et al. synthesize SiO2-Mn3(PO4)2 nanoparticles derived from phytic acid and utilize for O2˙− detection.15 Lan and co-workers deposit Mn3(PO4)2 nanoparticles on carbon nanomaterials directly for the fabrication of O2˙− sensor.16 These works verify the catalysis of Mn-superoxide dismutase mimics towards O2˙−. But the synthesis and property of nanoparticles need further study to improve the catalytic performance of sensor. In this work, we synthesize Mn3(PO4)2 nanoparticles by DNA induction, which results in even dispersion and excellent catalytic activity towards O2˙− disproportionation reaction.
Graphene is an increasingly important nanomaterials due to its excellent electronic conductivity, good stability, and promising catalytic performance in sensing.17–20 However, the two dimensional structure is easy to aggregate due to the single-atom-thickness, and hydrophobic aromatic structure. To solve this problem, we functionalize graphene with deoxyribonucleic acid (DNA) which can be adsorbed on graphene through π–π stacking and don't destroy the intact structure of the carbon materials.20 Under the induction of ssDNA, Mn3(PO4)2 nanoparticles was evenly deposited on the surface of graphene. The obtained graphene/DNA/Mn3(PO4)2 nanocomposites displayed significant biomimetic enzyme activity, rapid and sensitive response towards O2˙−. This approach holds a great promise for broad applications in biomedical research and clinical test.
Experimental section
Chemicals and materials
MCF-7 was bought from Cell bank of the representative culture preservation committee of the Chinese Academy of Sciences, China. Manganese sulfate (MnSO4), potassium phosphate tribasic (K3PO4) and Nafion were purchased from East Sichuan Chemical Industry (Group) Co., Ltd. (Chongqing, China) and used as received. Graphene was obtained from Sinocarbon Materials Technology Co., Ltd., China. Zymosan A (Zym, from Saccharomyces cerevisiae) SOD, DNA (low molecular weight extracted from salmon sperm), and potassium superoxide were purchased from Sigma-Aldrich and used without further purification. The O2˙− solutions were prepared by dissolving KO2 in PBS solution (pH 7.0, N2 saturated). The concentration of O2˙− was determined by the reduction of ferri cytochrome c spectrophotometrically.21 All the other solutions were prepared using deionized water (18 MΩ cm) and were degassed with high purity nitrogen before experiments. All electrochemical experiments were carried out at room temperature.Apparatus and instrumentations
Scanning electron microscopy (SEM) images were taken by JSM-6510LV, Japan. Field emission scanning electron microscopy (FESEM) images were taken by JSM-7800F, Japan. X-Ray diffraction (XRD) measurements were performed on a Shimadzu diffractometer (XRD-7000, Tokyo, Japan) operating in reflection mode with Cu Kα radiation at a step size of 0.06 per second. The nanosheets of graphene/DNA/Mn3(PO4)2 were characterized by transmission electron microscopy (TEM, JEM-2100F, Japan). Fourier transform infrared spectroscopy (FTIR) spectra were determined using a Thermo Nicolet 6700 FTIR spectrometer. Electrochemical performances were characterized using a CHI 660 system (Shanghai Chenhua, China). Graphene/DNA/Mn3(PO4)2 modified glassy carbon electrode (graphene/DNA/Mn3(PO4)2/GCE) served as working electrodes, platinum sheet as counter electrode and Ag/AgCl (in 3 M KCl) acted as reference electrode. All O2˙− measurements were performed in 10 mL 0.01 M PBS solution (pH = 7.4).Synthesis of graphene/DNA/Mn3(PO4)2 nanosheets
Graphene/DNA/Mn3(PO4)2 nanosheets were prepared by the previously reported method.22 Thirty milligrams of DNA was dissolved in 30 mL de-ionized water under stirring, after which the solution was annealed at 95 °C for 15 min to produce single-stranded DNA (ssDNA). Then the obtained ssDNA was mixed with 15 mL of 1 mg mL−1 graphene by mildly sonicated at 4 °C for 2 h, thereafter filtration and washing were performed to remove excess ssDNA. Subsequently, the mixture was dispersed into 0.1 M MnSO4 solution, then 0.1 M K3PO4 solution were dropwisely added under stirring and kept at room temperature for 30 min. Then graphene/DNA/Mn3(PO4)2 nanosheets were obtained by centrifugation at 8000 rpm for 10 minutes and washing with deionized water for 3 times.Glassy carbon electrode (GCE, d = 3 mm) was polished with alumina slurry to a smooth and bright surface, washed by sonication for 30 s and dried under nitrogen. Then 5 μL of the obtained graphene/DNA/Mn3(PO4)2 suspension was dropped on the electrode surface and dried at room temperature. Finally, 5% Nafion solution was coated to stabilize the fabricated electrode.
In situ detection of O2˙− released form living cells
MCF-7 cells were cultured in a humidified incubator (95% air with 5% CO2) at 37 °C. The cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) (Cellgro, USA) supplemented with 1 mol L−1 glutamine, 50 U mL−1 penicillin/streptomycin and 10% heat inactivated fetal bovine serum.14 For the detection of O2˙− released from living cell, the incubation solution was removed and washed with PBS solution (pH 7.4) for three times. Before electrochemical measurement, zymosan (Zym) was added to motivate cells generation of O2˙−. Amperometric response was recorded by CHI-660B electrochemical station at applied potential of 700 mV (versus Hg/Hg2Cl2).Results and discussion
Design of the modified graphene nanosheets
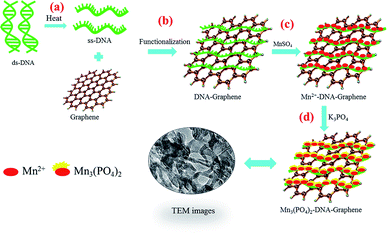
Scheme 1 illustrates the synthesis of graphene/DNA/Mn3(PO4)2 enzyme mimics by growth of Mn3(PO4)2 on graphene under the induction of DNA. As we all know, single-strand DNA (ssDNA) can be adsorbed on graphene by π–π interaction.23,24 Here ssDNA was prepared by annealing double-strand DNA (dsDNA) and applied for graphene modification. In the presence of Mn2+, the divalent cations accumulate along the ssDNA backbone by the electrostatic interaction, which facilitate the formation of Mn3(PO4)2 crystal nucleus on graphene and growth of Mn3(PO4)2 nanoparticles upon the addition of negatively charged PO43− groups.Characterization of graphene/DNA/Mn3(PO4)2 nanocomposites
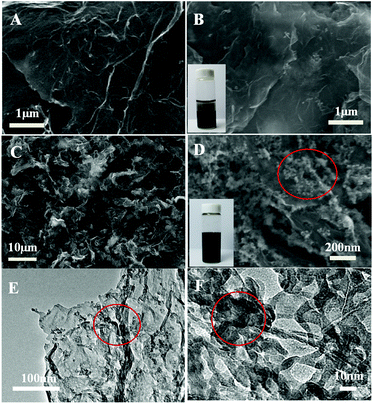
The morphology of the as-prepared graphene/DNA/Mn3(PO4)2 nanosheets is characterized with scanning electron microscope (SEM). Fig. 1A and B shows the SEM images of graphene before and after adding MnSO4 and K3PO4, typical wrinkle of graphene can be clearly observed and no evident Mn3(PO4)2 nanoparticles is found in Fig. 1B, suggesting that it is difficult to form Mn3(PO4)2 nanoparticles on graphene surface without DNA. However, when ssDNA is modified on graphene (Fig. 1C and D), even-distributed Mn3(PO4)2 nanoparticles can be evidently observed on the surface of graphene. The phenomena reveal the crucial role of ssDNA on the growth of Mn3(PO4)2 on graphene. The TEM images in Fig. 1E and F display different morphology of Mn3(PO4)2 nanoparticles due to the overlapping of particles. In order to verify the results, energy disperse spectrometer (EDS) was performed to explore chemical composition of the materials. The EDS results in Fig. S2† indicate that the major constituent elements of the wirelike nanostructure conclude O (68.7 at%), P (12.1 at%) and Mn (19.2 at%), and the atomic ratio of Mn and P is close to 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, suggesting the existence of Mn3(PO4)2 nanoparticles. The results further confirm the formation of well-defined thin-layer structure of the nanocomposites of Mn3(PO4)2 nanoparticles on graphene surface.
2, suggesting the existence of Mn3(PO4)2 nanoparticles. The results further confirm the formation of well-defined thin-layer structure of the nanocomposites of Mn3(PO4)2 nanoparticles on graphene surface.
Then FTIR spectra are determined to further verify the formation of graphene/DNA/Mn3(PO4)2 nanosheets. As shown in Fig. 2A, absorption peak at 1568.6 cm−1 can be assigned to the aliphatic carboxylic acid salts, the peaks at 996.0 cm−1 and 3196.3 cm−1 were attributed to inorganic phosphates. The results suggest that DNA and Mn3(PO4)2 have been successfully modified on the graphene. In addition, the X-ray diffraction pattern (XRD) of graphene/Mn3(PO4)2 in Fig. 2B shows that the diffraction peak of the nanomaterials is at around 28.2°, which is consistent with the results of Mn3(PO4)2. The results further confirm that Mn3(PO4)2 nanoparticles are successfully modified on the surface of graphene. Then the zeta potential at various stages of the synthesis were measured to further characterize the formation of the nanocomposites. The results in Fig. S1† indicate that the surface charge of graphene is about −0.203 mV, which decreases to −32.5 mV as DNA was adsorbed on its surface due to the negative charge of DNA backbone. When Mn3(PO4)2 was deposited, the zeta potential increase to −15.6 mV, indicating the formation of graphene/DNA/Mn3(PO4)2 nanocomposites.
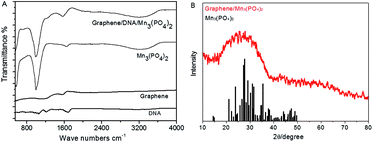 | ||
| Fig. 2 (A): FTIR image of different materials; (B): X-ray diffraction pattern of graphene/DNA/Mn3(PO4)2 nanosheets (red line) and the standard values of Mn3(PO4)2 (black line). | ||
Electrochemical properties of graphene/DNA/Mn3(PO4)2 nanosheets
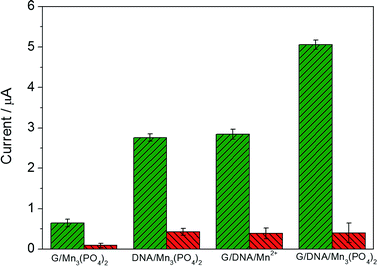
Fig. 3 exhibits the electrochemical response of different materials in the absence and presence of 1.0 μM O2˙− and the mixture of O2˙− and SOD (red column) in PBS. When graphene/DNA/Mn3(PO4)2 nanosheets were modified on GCE, the obtained modified electrode displayed evident redox peaks around 0.7 V and 0.5 V in the absence of O2˙− (as shown in Fig. S3†), which can be attributed to the electrochemical transformation between Mn2+ and Mn3+ species. However, no evident current response is observed when the mixture of O2˙− and SOD is added, suggesting the current response attributed to the catalysis of the graphene/DNA/Mn3(PO4)2 nanosheets towards O2˙−. These results verify the biomimetic enzyme activity of graphene/DNA/Mn3(PO4)2 to catalyze the dismutation of O2˙−.25In order to further study the electrochemical performance of graphene/DNA/Mn3(PO4)2 nanosheets, graphene/Mn3(PO4)2 were synthesized by direct depositing Mn3(PO4)2 nanoparticles on graphene in the absence of DNA, DNA/Mn3(PO4)2 nanoparticles were by prepared depositing Mn3(PO4)2 on dsDNA template and graphene/DNA/Mn2+ was synthesized by adsorbing DNA on graphene and subsequent depositing Mn2+ on DNA. Electrochemical performance of these materials were explored with electrodes modified with the above three nanomaterials (Fig. 3A–C). As shown in Fig. 3A, the electrode modified with graphene/Mn3(PO4)2 (graphene/Mn3(PO4)2/GCE) possesses excellent electrical conductivity but the current response towards O2˙− is weak, verifying the good conductivity of graphene and poor modification of Mn3(PO4)2 (Fig. 3A). Similarly, the electrodes modified with Mn3(PO4)2/DNA and graphene/DNA/Mn2+ display evident current response to O2˙−, but the current response is lower than that on graphene/DNA/Mn3(PO4)2/GCE (as shown in Fig. 3B and C). The phenomena further prove that DNA plays important roles in the synthesis of graphene/DNA/Mn3(PO4)2 nanosheets. In addition, in comparison with the Mn3(PO4)2/DNA/GCE (Fig. 3B), we can see that the graphene/DNA/Mn3(PO4)2/GCE (Fig. 3D) displays stronger current signal due to the excellent conductivity of graphene.
The reason for DNA facilitating the growth of Mn3(PO4)2 nanoparticles on graphene can be ascribed to two factors. First, the π–π interaction between DNA and graphene induces ordered assembly of DNA on the surface of graphene, which is a critical factor to the synthesis of Mn3(PO4)2 nanoparticles. Next, Mn2+ is adsorbed on DNA backbone to assist the nucleation of Mn3(PO4)2 nanoparticles. In addition, in comparison with other conventional method, the DNA induced synthesis is easier to control the morphology and dispersion of products by adjusting temperature, pH, or DNA concentration.
Electrochemical response of O2˙− at the present electrode
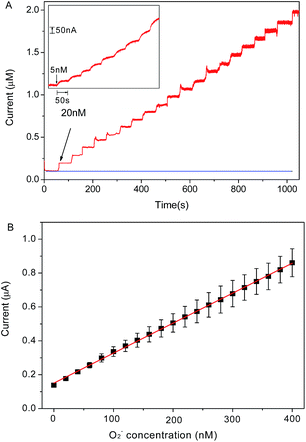
Chronoamperometry is utilized to investigate the electrochemical response of graphene/DNA/Mn3(PO4)2/GCE towards O2˙− in 10 mM PBS (pH = 7.4). Fig. 4A shows the stepwise current response of the present electrode with successive increasing O2˙− concentration from 5 nM to 400 nM. From the results in Fig. 4B we can see that the prepared sensor displays significant current response to the O2˙− concentrations in the range of 5 nM to 400 nM with a regression equation expressed as I (μA) = 0.00354 c (nM) + 0.1504 (R2 = 0.999). The detection limit of the assay is calculated to be about 1.67 nM (S/N = 3) with a sensitivity of 3.54 μA μM−1, which is much more superior to most of previous reports.14,23,26–28 The response time of the electrode to O2˙− is as short as 5 s, which is an advantage for the detection of O2˙− due to its short lifetime. These results demonstrate that the present sensor may meet the requirement of O2˙− detection in normal physiological conditions.29The selectivity and stability of the present electrode
The selectivity experience was carried out in the presence of various interfering species, such as K+, Na+, Cl−, NO3−, SO42−, hydrogen peroxide (H2O2), glucose (Glu), glutathione (GSH) uric acid (UA) and ascorbic acid (AA). Among these interferences, H2O2, UA and AA possess good electrochemical activity and perplex the detection of O2˙− due to their wide existence in biological systems. As shown in Fig. 5, in comparison to the electrochemical response of the as-present sensor to 100 nM O2˙−, the presence of 10 μM K+, Na+, Cl−, SO42−, NO3−, Glu. GSH, AA, UA and H2O2 did not cause any noticeable current response. The result suggests that the proposed sensor possesses excellent specificity to O2˙− detection in biological systems. In addition, the reproducibility and stability of the electrode were explored. The electrochemical response of the sensor was performed using five different electrodes fabricated at the same time, the standard derivations of current response was lower than 5.2%. When the electrode was stored at 4 °C for one month, the current response towards O2˙− decrease 6.8% of original value (as shown in Fig. S4†). These results indicate that the excellent reproducibility and stability of the sensor.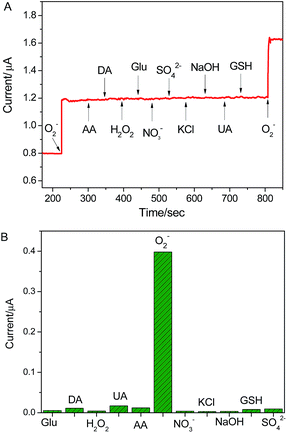 | ||
| Fig. 5 The selectivity of graphene/DNA/Mn3(PO4)2/GCE for the assaying of O2˙− against the different interfering species. The response obtained upon the addition of 10 μM interferes and 100 nM O2˙−. | ||
Detection of O2˙− released from cancer cell
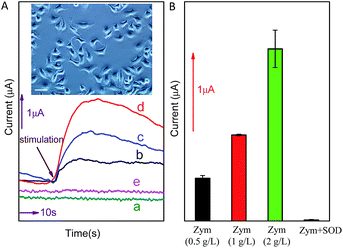
In order to further investigate the potential application of the method in biological systems, the detection of O2˙− released from MCF-7 cells was performed at ambient temperature. As shown in Fig. 6, upon the addition of different concentration (0.5 g L−1, 1.0 g L−1 or 2.0 g L−1) of zymosan (Zym, a drug was used to stimulate living cells to release O2˙−), a significant current response was observed (corresponding to O2˙− oxidation). According to the calibration curve in Fig. 5B, the concentration of O2˙− released from MCF-7 cells is calculated to be around 156.38 nM, 315.71 nM and 631.81 nM, respectively (the concentration of O2˙− released by per cell and total cells are calculated and listed on Table 1). The results indicate that current response of O2˙− released form cancer cell under Zym stimulation is a concentration-dependent behavior. In addition, the results of control experiments (curve d and e shown in Fig. 6) indicate that no current was obtained upon the addition of either Zym or SOD-Zym mixture, confirming that the current responses were attributed to the drug-induced O2˙− release from cells.| Stimulation | Response current (μA) | Concentration of O2˙− released by cells (nM) | O2˙− released by per cell (pM) |
|---|---|---|---|
| Zym 0.5 g L−1 | 0.704 | 156.38 | 0.782 |
| Zym 1 g L−1 | 1.268 | 315.71 | 1.579 |
| Zym 2 g L−1 | 2.387 | 631.81 | 3.159 |
Conclusions
A novel O2˙− biomimetic enzyme sensor based on graphene/DNA/Mn3(PO4)2 nanosheets is successfully developed. The synthesized nanosheets display strong electrocatalytic activity towards O2˙− with high sensitivity, excellent selectivity and fast current response. The successful determination of O2˙− released from cancer cells demonstrates the great potential for application in biological system. This work provides a new method for the fabrication of biomimetic enzyme sensor and a great promise for biosensing and biomedical application.Conflicts of interest
There are no conflicts to declareAcknowledgements
This work is financially supported by National Natural Science Foundation of China (21505108) and Technological and Developmental Grant (2015-09) from Beibei District Commission. The authors would also like to acknowledge the financial support of the Fundamental Research Funds (XDJK2016C132) for the Central Universities.Notes and references
- Y. W. Sheng, I. A. Abreu, D. E. Cabelli, M. J. Maroney, A. F. Miller, M. Teixeira and J. S. Valentine, Chem. Rev., 2014, 114, 3854 CrossRef CAS PubMed.
- Y. Tian, L. q. Mao and T. Ohsaka, Curr. Anal. Chem., 2006, 2, 51 CrossRef CAS.
- B. N. Ames, M. K. Shigenaga and T. M. Hagen, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 7915 CrossRef CAS.
- M. Ganesana, J. S. Erlichman and S. Andreescu, Free Radicals Biol. Med., 2012, 53, 2240 CrossRef CAS PubMed.
- Y. Ohara, T. E. Peterson and D. G. J. Harrison, J. Clin. Invest., 1993, 91, 2546 CrossRef CAS PubMed.
- J. L. Zweier, J. H. Flaherty and M. L. Weisfeldt, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 1404 CrossRef CAS.
- T. Ohyashiki, M. Nunomur and T. Katoh, Biochim. Biophys. Acta, 1999, 1421, 131 CrossRef CAS.
- Z. f. Deng, Q. Rui, X. Yin, H. Q. Liu and Y. Tian, Anal. Chem., 2008, 80, 5839 CrossRef CAS PubMed.
- V. Lvovich and A. Scheeline, Anal. Chem., 1997, 69, 454 CrossRef CAS PubMed.
- J. Chen, U. Wollenberger, F. Lisdat, B. X. Ge and F. W. Scheller, Sens. Actuators, B, 2000, 70, 115 CrossRef CAS.
- Z. Wang, D. Liu, H. Gu, A. Zhu, Y. Tian and G. Shi, Biosens. Bioelectron., 2013, 43, 101 CrossRef CAS PubMed.
- F. S. Archibald and I. Fridovich, Arch. Biochem. Biophys., 1982, 214, 452 CrossRef CAS PubMed.
- K. Barnese, E. B. Gralla, D. E. Cabelli and J. S. Valentine, J. Am. Chem. Soc., 2008, 130, 4604 CrossRef CAS PubMed.
- F. X. Hu, Y. J. Kang, F. Du, L. Zhu, Y. H. Xue, T. Chen, L. M. Dai and C. M. Li, Adv. Funct. Mater., 2015, 25, 5924–5932 CrossRef CAS.
- X. Shen, Q. Wang, Y. Liu, W. Xue, L. Ma, S. Feng, M. Wan, F. Wang and C. Mao, Sci. Rep., 2016, 6, 28989–28998 CrossRef CAS PubMed.
- X. Cai, L. Shi, W. Sun, H. Zhao, H. Li, H. He and M. Lan, Biosens. Bioelectron., 2018, 102, 171–178 CrossRef CAS PubMed.
- J. F. Ping, Y. X. Wang, K. Fan, J. Wu and Y. B. Ying, Biosens. Bioelectron., 2011, 28, 204 CrossRef CAS PubMed.
- T. Kuila, S. Bose, P. Khanra, A. K. Mishra, N. H. Kim and J. H. Lee, Biosens. Bioelectron., 2011, 26, 4637 CrossRef PubMed.
- B. Wang, Y. Wu, Y. Chen, B. Weng and C. Li, Sens. Actuators, B, 2017, 238, 802–808 CrossRef CAS.
- X. Zuo, S. He, D. Li, C. Peng, Q. Huang, S. Song and C. Fan, Langmuir, 2010, 26, 1936 CrossRef CAS PubMed.
- J. M. McCord and I. Fridovic, J. Biol. Chem., 1969, 244, 6049 CAS.
- C. X. Guo, L. Y. Zhang, J. w. Miao, J. Zhang and C. M. Li, Adv. Energy Mater., 2013, 3, 167 CrossRef CAS.
- X. Ma, W. Hu, C. Guo, L. Yu, L. Gao, J. Xie and C. M. Li, Adv. Funct. Mater., 2014, 24, 5897 CrossRef CAS.
- H. Zhang, H. Huang, Z. Lin and X. Su, Anal. Bioanal. Chem., 2014, 406, 6925 CrossRef CAS PubMed.
- K. Barnese, E. B. Gralla, D. E. Cabelli and J. S. Valentine, J. Am. Chem. Soc., 2008, 130, 4604 CrossRef CAS PubMed.
- B. P. Crulhas, L. C. Recco, F. K. Delella and V. A. Pedrosa, Electroanalysis, 2017, 29, 1252 CrossRef CAS.
- X. Liu, X. Liu, H. Wei, G. Song, H. Guo and X. Lu, Sens. Actuators, B, 2017, 252, 503 CrossRef CAS.
- R. B. Sadeghian, S. Ostrovidov, J. Han, S. Salehi, B. Bahraminejad, H. Bae, M. Chen and A. Khademhosseini, ACS Sens., 2016, 1(7), 921 CrossRef CAS.
- Y. Luo, Y. Tian and Q. Rui, Chem. Commun., 2009, 3014 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra12962a |
| This journal is © The Royal Society of Chemistry 2018 |