Open Access Article
Open Access ArticleRetracted Article: Linc00472 suppresses breast cancer progression and enhances doxorubicin sensitivity through regulation of miR-141 and programmed cell death 4
Pengwei Lu,
Xue Yang,
Yunqing Yang,
Fang Wang,
Lin Li and
Yuanting Gu *
*
Department of Breast Surgery, The First Affiliated Hospital of Zhengzhou University, No. 1 Jianshe East Road, Erqi District, Zhengzhou, 450000, China. E-mail: guyuantinggyt@163.com; Tel: +86-0371-67967171
First published on 23rd February 2018
Abstract
The existence of drug resistance strikingly hampers the therapy of many malignancies, including breast cancer. Long non-coding RNAs (LncRNAs) have been reported to participate in the regulation of various biological processes associated with cancer progression. Whereas, the role of linc00472 in breast cancer pathogenesis and doxorubicin (ADR) resistance have not been well elucidated. In the present study, it is found that linc00472 expression was decreased in breast cancer tissues and cells. Moreover, higher linc00472 expression was positively associated with favorable disease status and prognosis for breast cancer patients. Functional analyses revealed that linc00472 overexpression suppressed proliferation and invasion, facilitated apoptosis and enhanced ADR sensitivity in breast cancer cells. Mechanistic studies discovered that linc00472 acted as a competing endogenous RNA (ceRNA) of miR-141 to sequester miR-141 from its target mRNA PDCD4 (programmed cell death 4). Furthermore, the inhibition effect of linc00472 on breast cancer cell progression and ADR resistance could be partly abrogated by miR-141 up-regulation or PDCD4 knockdown. In vivo assays also demonstrated that linc00472 hindered tumor growth by suppressing miR-141 expression and enhancing PDCD4 expression. In conclusion, linc00472 blocked breast cancer progression and induced ADR sensitivity through regulation of miR-141 and PDCD4, highlighting a potential therapeutic strategy for breast cancer patients.
1. Introduction
Breast cancer is the most common malignancy among women, accounting for 29% of all new diagnosed cancers. Although much progress has been made in early detection and adjuvant treatment, breast cancer is still the second major cause of cancer-triggered death in women.1 Among breast cancer patients with surgery and radio-chemotherapy treatment, approximately 30–75% breast cancer will evolve into metastatic or recurrent disease.2,3 Moreover, most patients with metastatic breast cancer were incurable with a 5 year survival rate of 20–40%.4 Doxorubicin (ADR), also named as adriamycin, is a chemotherapy regimens widely used for the treatment of metastatic breast cancer patients.5 However, the existence of chemoresistance strikingly limited the curative effect of drugs including ADR in breast cancer.6,7 Hence, it is of great importance to gain insight into the molecular mechanisms underlying ADR resistance involved in breast cancer progression in order to figure out more effective treatment strategies for breast cancer.Long non-coding RNAs (lncRNAs), a class of transcripts longer than 200 nucleotides without protein coding potential, have been demonstrated to be critical mediators in various biology processes implicated in carcinogenesis and drug sensitivity/resistance.8 Also, a broad spectrum of lncRNAs are involved in breast cancer progression via modulating cell growth, apoptosis, migration and invasiveness at epigenetic, transcriptional and post-transcriptional levels.9 For instance, lncRNA NEAT1 contributed to cell growth and invasion via regulating miR-211/HMGA2 axis in breast cancer.10 Up-regulation of lncRNA HOTAIR promoted cell proliferation and conferred tamoxifen resistance in breast cancer.11 lncRNA-ATB overexpression induced epithelial–mesenchymal transition (EMT) and enhanced trastuzumab resistance in breast cancer through miR-200c-ZEB1/ZNF217 pathway.12 Linc00472, located on chromosome 6q13, has been discovered to be positively correlated with favorable disease status and prognostic value together with a better chemotherapy response in breast cancer.13,14 Moreover, high expression of linc00472 inhibited breast cancer progression such as proliferation and migration.13 However, the molecular mechanisms of linc00472 underlying breast cancer progression and ADR resistance have not been well elucidated till now.
In the present study, we firstly demonstrated that linc00472 expression was down-regulated in breast cancer tissues and cells, and linc00472 expression was positively related with favorable disease status and better prognosis of breast cancer patients. Function analysis manifested that linc00472 suppressed proliferation and invasion, enhanced ADR susceptibility, and facilitated apoptosis in breast cancer cells. Further mechanism study demonstrated that linc00472 exerted its anti-tumor effect by regulating miR-141/PDCD4 (programmed cell death 4) axis in breast cancer.
2. Materials and methods
2.1. Tissues specimens and cell culture
Breast cancer tissues specimens (n = 40) and adjacent normal tissues were obtained from the breast cancer patients suffering surgical resection or biopsy between 2014 and 2016 at our hospital. Our study got the approval of Research Ethics Committee of the First Affiliated Hospital of Zhengzhou University and every patient signed the informed consent before enrolling in the study. The TNM stage of breast cancer patients was determined according to the standard of the American Joint Committee on Cancer (AJCC). All cell lines were purchased from American Tissue Culture Collection (ATCC, Manassas, VA, USA). Human mammary epithelial cell line (MCF-10A) were grown in Clonetics MEGM™ mammary epithelial cells growth medium (Lonza Walkersville, Inc. Walkersville, MD, USA) containing 5% horse serum, cholera toxin (100 ng ml−1), hydrocortisone (0.5 μg ml−1), hEGF (20 ng ml−1) and insulin (10 μg ml−1). Human breast cancer cell line (MDA-MB-231, MDA-MB-453 and MCF-7) were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Invitrogen). Doxorubicin (ADR) was purchased from Sigma-Aldrich (St. Louis, MO, USA). All cells were maintained in humidified incubator containing 5% CO2 at 37 °C.2.2. Transfection
The full length sequences of linc00472 were amplified by PCR and constructed into pcDNA3.1 vector (Invitrogen) to form pcDNA-linc00472 overexpression plasmid. Then the putative binding sites in the pcDNA-linc00472 were mutant to generate pcDNA-linc00472-MUT plasmid. miR-141 mimic and its scramble control (miR-con) along with miR-141 inhibitor (anti-miR-141) and its scramble control (anti-miR-con) were purchased from Sigma-Aldrich (St. Louis). Linc00472 small interference RNA (si-linc00472) and its scramble control (si-con) together with the siRNA of PDCD4 (si-PDCD4) and its scramble control (si-con) were synthesized by GenePharma Co., Ltd (Shanghai, China). Then, 100 pmol oligonucleotides or 3 μg plasmids were transfected into cells using Lipofectamine 2000 reagent (Invitrogen) referring to the manufacturer's instructions. The amplification primers and endonucleases were listed in Table 1.| Primer sequences | Restriction enzyme cutting site | |
|---|---|---|
| pcDNA-linc00472 | BamHI and XhoI | |
| Forward | CGGGATCCATGCGAGGCTGGGGCCGGTTG | |
| Reverse | CCGCTCGAGTTTAGACTCCAAATGGCTG | |
| pmirGLO-linc00472-WT | SacI and XhoI | |
| Forward | CGAGCTCACCCTCTACCTTTCCAATC | |
| Reverse | CCGCTCGAGGTTACCTATCACTTCCTTTCG | |
| pmirGLO-PDCD4-WT | SacI and XhoI | |
| Forward | CGAGCTCATATAAGAACTCTTGCAGTC | |
| Reverse | CCGCTCGAGACCAGGTTCATTTTTCC | |
| pLenti-puro-linc00472 | BamHI and EcoRV | |
| Forward | CGCGGATCCATGCGAGGCTGGGGCCGGTTG | |
| Reverse | GCGCGATATCTTTAGACTCCAAATGGCTG | |
| pLenti-puro-pre-miR-141 | BamHI and EcoRV | |
| Forward | CGGGATCCGGGTCCATCTTCCAGTAC | |
| Reverse | GCGCGATATCGAGCCATCTTTACCAGAC | |
| miR-141 (reverse transcription primer), linc00472 (quantification primers) | GTCGTATCCAGTGCAGGGTCCGAGTATTCGCACTGGATACGACGCTGA | |
| Forward | CACTGGGCATTTTCTCTTCA | |
| Reverse | CCTATCCCTTTCCCTCTGCT | |
| PDCD4 (quantification primers) | ||
| Forward | GGCCTCCAAGGAGTAAGACC | |
| Reverse | AGGGGTCTACATGGCAACTG | |
| β-Actin (quantification primers) | ||
| Forward | GCATCCCCCAAAGTTCACAA | |
| Reverse | AGGACTGGGCCATTCTCCTT | |
| miR-141 (quantification primers) | ||
| Forward | GCCGCTGTAAACATCCTACACT | |
| Reverse | GTGCAGGGTCCGAGGT | |
| U6 snRNA (quantification primers) | ||
| Forward | CTCGCTTCGGCAGCACA | |
| Reverse | AACGCTTCACGAATTTGCGT |
2.3. RT-qPCR assays
Total RNA was isolated from breast tissues specimens or cells employing TRIzol Reagent (Invitrogen). Equal amounts of RNA was reversely transcribed into the first-strand cDNA using M-MLV reverse transcriptase (Invitrogen). Then SYBR Green Realtime PCR Master Mix (TOYOBO, Osaka, Japan) was used to detect the expression patterns of linc00472, miR-141 and PDCD4 mRNA. The reverse transcription (RT) primer of miR-141 and quantification primers of linc00472, miR-141, β-actin, U6 snRNA and PDCD4 were listed in Table 1. The relative gene expression was determined by 2−ΔΔCt method with U6 snRNA as an internal control of miR-141 and β-actin as an endogenous control of linc00472 and PDCD4.2.4. Western blot assays
Total protein was extracted from tissues and cells using pre-cold RIPA buffer (Thermo Scientific, Rockford, IL, USA). Then equal amounts of protein was separated on SDS-PAGE gel and transferred onto PVDF membranes (Millipore Corp. Bedford, MA, USA). Next, the membranes were blocked by 5% non-fat milk, followed by the incubation with primary antibodies against PDCD4 (sc-376430, mouse monoclonal antibody, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), ki-67 (sc-23900, mouse monoclonal antibody, 1
1000), ki-67 (sc-23900, mouse monoclonal antibody, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), β-actin (sc-58673, mouse monoclonal antibody, 1
1000), β-actin (sc-58673, mouse monoclonal antibody, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), activated caspase 3 (sc-22171, goat polyclonal antibody, 1
1000), activated caspase 3 (sc-22171, goat polyclonal antibody, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), pro-caspase 3 (sc-271759, mouse monoclonal antibody, 1
1000), pro-caspase 3 (sc-271759, mouse monoclonal antibody, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), GAPDH (sc-47724, mouse monoclonal antibody, 1
1000), GAPDH (sc-47724, mouse monoclonal antibody, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) overnight at 4 °C. All antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA. USA). Subsequently, the membranes were treated with HRP-conjugated secondary antibody (goat-anti-mouse polyclonal secondary antibody, ab6789, 1
1000) overnight at 4 °C. All antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA. USA). Subsequently, the membranes were treated with HRP-conjugated secondary antibody (goat-anti-mouse polyclonal secondary antibody, ab6789, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000; or rabbit-anti-goat polyclonal secondary antibody, ab6741, 1
5000; or rabbit-anti-goat polyclonal secondary antibody, ab6741, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, abcam, Cambridge, UK) for 1 h at room temperature. At last, specific protein signal was detected employing the BeyoECL Plus agent (Beyotime, Shanghai, China).
000, abcam, Cambridge, UK) for 1 h at room temperature. At last, specific protein signal was detected employing the BeyoECL Plus agent (Beyotime, Shanghai, China).
2.5. Luciferase reporter assays
The partial sequences of linc00472 and PDCD4 containing putative or mutant miR-141 binding sites were amplified by PCR and cloned into pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA), to generate LINC00472-WT, LINC00472-MUT, PDCD4-WT (WT), PDCD4-MUT (MUT). Then the constructed luciferase vectors were respectively transfected into breast cancer cells together with miRNAs (miR-con, miR-141), miRNA inhibitors (anti-miR-con, anti-miR-141), overexpression plasmids (pcDNA3.1 vector, pcDNA-lnc00472) or siRNAs (si-con, si-lnc00472). At 48 h after transfection, the luciferase activity was determined using Dual-Luciferase Reporter Assay System (Promega) following the manufacturer's instructions.2.6. EdU (5-ethynyl-2′-deoxyuridine) immunofluorescence and flow cytometry assays
Cell proliferation patterns were also measured by EdU (5-ethynyl-2′-deoxyuridine) immunofluorescence assays using Cell-Light TM EdU kit (RiboBio) referring the manufacturer's instructions. For immunofluorescence assays, transfected cells (1 × 105 per well, 96-well plate) were labeled with complete medium containing 50 μΜ EdU for 2 h, and then fixed using 4% paraformaldehyde for 30 min at room temperature, followed by the addition of 2 mg ml−1 glycine for 5 min. Next, 100 μl 1× Apollo solution was added into each well. After incubation for 30 min in dark, Apollo solution was discarded and 100 μl of 0.5% TritonX-100 was added into each well for 10 min. After 3 times of repetition, cells were stained with DAPI solution (1 μg ml−1, Beyotime, Shanghai, China) for 20 min, and then imaged using a confocal microscopy. For flow cytometry assays, transfected cells were labeled with complete medium containing 50 μΜ EdU for 2 h, followed by the digestion with 0.25% trypsin for 1 min and fixation with 4% paraformaldehyde for 15 min. Then cells were stained using Apollo and DAPI solutions as described above. At last, cells were collected at 1000 rpm and re-suspended in 300 μl PBS prior to subjecting to flow cytometer (BD Biosciences, San Jose, CA, USA).2.7. Colony formation assay
Briefly, transfected MCF-7 and MDA-MB-453 cells were seeded in 6-well plates and incubated for 15 days. Next, the treated cells were fixed using acetic acid/methanol 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 (v/v) and stained with 0.5% crystal violet solution (Sigma-Aldrich). As last, cell colonies were counted under an inverted microscope.
7 (v/v) and stained with 0.5% crystal violet solution (Sigma-Aldrich). As last, cell colonies were counted under an inverted microscope.
2.8. Cell cycle and apoptosis analyses
At 48 h after transfection, MCF-7 and MDA-MB-453 cells were treated for further analyses of cell cycle and apoptosis. For cell cycle analysis, cells were fixed using 80% ethanol at −20 °C overnight and then treated with DNase-free RNase A (Thermo Scientific, 50 μg ml−1) and propidium iodide (PI, Sigma-Aldrich, 50 μg ml−1) for 1 h at room temperature in the dark, followed by the analysis on flow cytometer (BD Biosciences). Cell apoptosis rate was determined by Annexin V-FITC/PI Apoptosis Detection kit (Beyotime) referring to the instructions of manufacturer.2.9. Cell viability assays and IC50 value determination
Cell Proliferation Kit I (MTT) (Sigma-Aldrich) was used to assess cell viability and IC50 value (defined as the half maximal inhibitory concentration of drugs) of ADR. At the indicated time points (0, 24, 48, 72 h) after transfection, MTT assays were performed to detect cell viability. Additionally, transfected cells were inoculated into 96-well plates and incubated for 48 h, followed by the addition of gradient concentrations of ADR (0.001, 0.01, 0.1, 1, 10, 100 μM) for another 24 h and IC50 value determination using MTT assays. MTT assays were carried out as follows. Briefly, MTT solution was added into the 96-well plates and incubated for 4 h. Then cell medium containing MTT was removed and 150 μl dimethyl sulfoxide (DMSO) was added into each well to dissolve formed formazan crystals. At last, the absorbance was measured at the wavelength of 570 nm.2.10. RNA immunoprecipitation (RIP) assays
RIP assays were carried out employing the EZ-Magna RIP kit (Millipore, Billerica, MA, USA) according to the manufacturer's protocol. Briefly, MCF-7 and MDA-MB-453 cells were lysed with pre-cold RIPA buffer and then incubated with magnetic beads and primary antibody against Ago2 (Millipore) or IgG. Then the complex was treated with RNase-free DNase I (Promega, 10U) for 30 min at 37 °C and digested with proteinase K (1 mg ml−1) for 20 min at 45 °C to remove the superfluous DNA and protein. Finally, RT-qPCR assays were used to detect the expression patterns of specific RNAs in purified RNA.2.11. Subcellular fractionation
Cytoplasmic and Nuclear RNA Purification Kit (Norgen, Thorold, ON, Canada) was employed to isolate the nuclear and cytoplasm fractions. Then the expression levels of GAPDH, U6 and linc00472 in nuclear and cytoplasm fractions of MCF-7 and MDA-MB-453 cells were detected using RT-qPCR assays.2.12. Transwell invasion assays
Transwell assays were performed to assess cell invasion capability. Briefly, approximately 1 × 105 MCF-7 and MDA-MB-453 cells were inoculated into the upper chamber containing matrigel (BD Biosciences). Cells in the upper chamber were maintained in serum-free RPMI-1640 medium, while complete medium with 5% FBS was added to the lower chamber. After incubation for 48 h, the cells adhered to lower surface were fixed in the 100% pre-cold methanol, staining with 0.1% crystal violet solution (Sigma-Aldrich) and then counted by microscope.2.13. Lentivirus production and infection
pLenti-puro was a gift from Ie-Ming Shih (Addgene, Cambridge, Massachusetts, USA). The sequences of miR-141 precursor and linc00472 were amplified by PCR from genomic DNA and subcloned into pLenti-puro vector to generate the overexpression plasmid of miR-141 and linc00472, named as lenti-pre-miR-141 and lenti-linc00472. Then the lentiviral plasmids were respectively transfected into HEK293T cells along with psPAX2 and pMD2.G plasmids (Addgene). After transfection of 72 h, cell medium containing lentivirus were collected and filtered using 0.45 mm pore filter. Then, MCF-7 cells were infected with treated lentivirus, followed by the screening with puromycin (Sigma-Aldrich) for about 1 week so as to get stable lentivirus-transfected cell lines.2.14. Tumor formation in nude mice
All experiment processes were performed according to the national standard of the care and use of laboratory animals and got the approval of Institutional Committee for Animal Research. BALB/c athymic nude mice were obtained from Hubei Research Center of Laboratory Animal (Wuhan, China). Approximately 5 × 106 MCF-7 cells stably transfected with lenti-pre-miR-141, lenti-linc00472, lenti-linc00472+lenti-pre-miR-141 were subcutaneously injected into the armpit of nude mice. At 7 days post-inoculation, tumor volumes were detected using a caliper once a week for 6 weeks. Tumors were photographed and tumor weights were measured at 42 days after injection. Also the expression levels of miR-141, linc00472 and ki-67 in tumors tissues were analyzed by RT-qPCR and western blot assays.2.15. Statistical analysis
All assays were repeated at least three times with the results displayed as mean ± standard deviation (S.D.). Overall survival (OS) was calculated from the first surgery until death or the last follow-up investigation and was analyzed by Kaplan–Meier analysis. The correlation studies were performed using Spearman's correlation analysis. Student's t-test and one-way ANOVA were used to compare the significant difference of different groups. IC50 value was measured through SPSS software employing probit regression analysis. P < 0.05 meant the difference was statistically significant.3. Results
3.1. Linc00472 expression was down-regulated in breast cancer tissues and cells
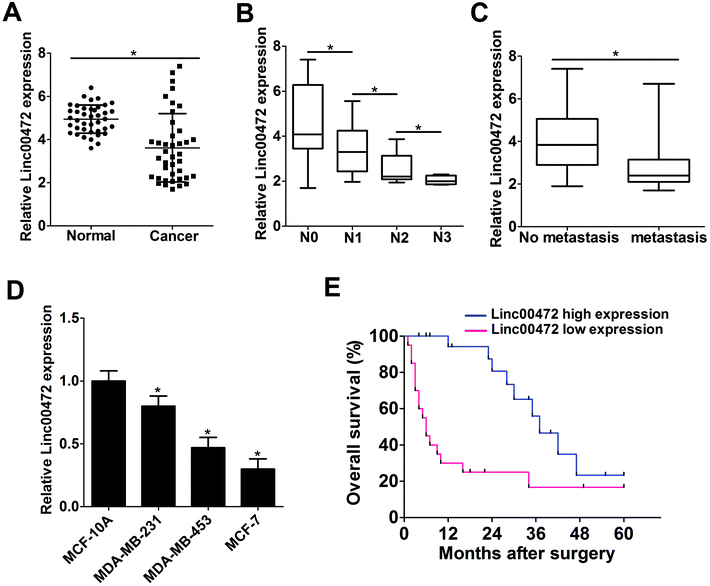
Firstly, RT-qPCR assays were performed to detect the expression pattern of linc00472 in breast cancer tumor tissues. As shown in Fig. 1A, linc00472 expression level was remarkably decreased in 40 pairs of breast cancer tissues (Cancer) compared with adjacent normal tissues (Normal) (Fig. 1A). We also found that linc00472 level was notably down-regulated with the development of lymph-node metastasis in breast cancer (Fig. 1B). In addition, low-level expression of linc00472 might be related with distant metastasis of breast cancer (Fig. 1C). Subsequently, we measured linc00472 expression in several breast cancer cell lines (MDA-MB-231, MDA-MB-453, MCF-7). The results showed that linc00472 expression was markedly reduced in breast cancer cell lines compared with human normal mammary epithelial cell line MCF-10A (Fig. 1D). Kaplan–Meier survival analysis indicated that patients with low linc00472 expression displayed a poorer overall survival when compared to that with high linc00472 expression (P = 0.0014, log-rank test) (Fig. 1E). All these data indicated that dysregulation of linc00472 might be associated with the development and prognosis of breast cancer.3.2. Ectopic expression of linc00472 suppressed proliferation and invasion of breast cancer cells
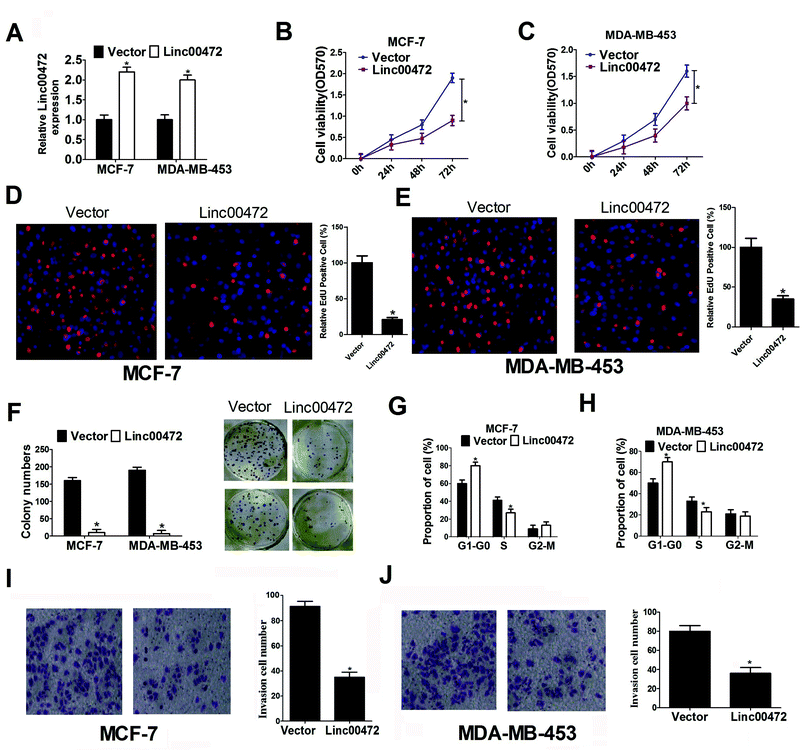
To delineate the role of linc00472 in breast cancer progression, pcDNA-linc00472 overexpression plasmid (linc00472) was constructed and transfected into breast cancer cells (MCF-7 and MDA-MB-453). RT-qPCR results showed that transfection of pcDNA-linc00472 strikingly enhanced linc00472 expression in MCF-7 and MDA-MB-453 cells compared with empty vector (Fig. 2A), indicating that pcDNA-linc00472 could be used for the following gain-of-function experiments. Then we proceeded to explore the effect of linc00472 on proliferation of breast cancer cells. MTT assays manifested that enforced expression of linc00472 markedly suppressed proliferation of MCF-7 and MDA-MB-453 cells (Fig. 2B and C). EdU assays also showed that overexpression of linc00472 greatly decreased the number of EdU-positive cells compared with control vector (Fig. 2D and E). Moreover, colony formation assays disclosed that the number of formed colonies was notably lower in linc00472-overexpressing group than that in control vector group (Fig. 2F). Also, cell cycle distribution exhibited that cell proportion in G0/G1 phase was increased and cell percentage in S-phase population was reduced in MCF-7 and MDA-MB-453 cells transfected with linc00472 compared with mock cells (Fig. 2G and H). All these data revealed that linc00472 up-regulation hampered proliferation capability of breast cancer cells. Then, we went on to explore the effect of linc00472 on invasion of breast cancer cells. As presented in Fig. 2I and J, transwell invasion assays revealed that the invasion ability of MCF-7 and MDA-MB-453 cells was weakened after introduction of linc00472-overexpression plasmid. Taken together, these data suggested that linc00472 overexpression blocked proliferation and invasion of breast cancer cells.3.3. Linc00472 overexpression improved ADR chemosensitivity and enhanced ADR-mediated apoptosis in breast cancer cells
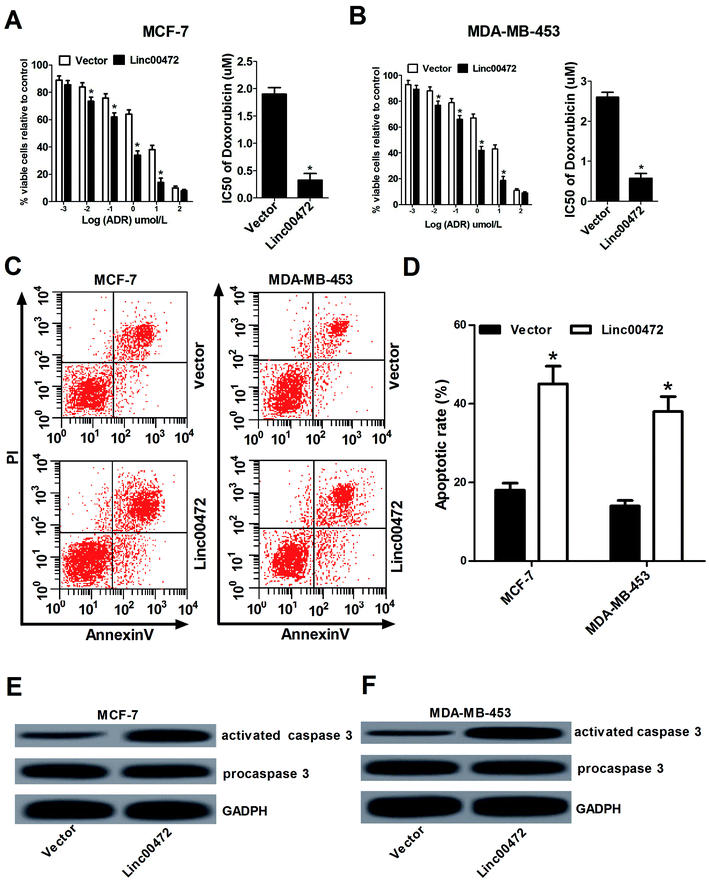
Then we proceeded to investigate the effect of linc00472 on ADR chemoresistance in breast cancer cells. To determine half maximal inhibitory concentration (IC50) of ADR in breast cancer cells, MCF-7 cells and MDA-MB-453 cells transfected with pcDNA3.1 vector or pcDNA-linc00472 were treated with different concentration of ADR (0.001, 0.01, 0.1, 1, 10, 100 μM) for 48 h. Then, MTT assays were performed to determine cell survival rate and IC50 values of ADR. As shown in Fig. 3A and B, introduction of linc00472 resulted in a significant reduction of breast cancer cell viability compared with empty vector at ADR treatment concentration ranging from 0.001 μM to 10 μM. Moreover, we demonstrated that the IC50 values of ADR were considerably decreased in linc00472-treated MCF-7 and MDA-MB-453 cells compared with control cells (vector). In a word, up-regulation of linc00472 expression increased ADR chemosensitivity of breast cancer cells. Next, to assess whether linc00472-induced sensitivity to ADR was associated with apoptosis, MCF-7 cells and MDA-MB-453 cells transfected with pcDNA3.1 vector or pcDNA-linc00472 were treated with 0.1 μM ADR for 48 h. Flow cytometry analysis demonstrated that linc00472 overexpression enhanced ADR-induced apoptosis in MCF-7 cells and MDA-MB-453 (Fig. 3C and D). Moreover, western blot assays revealed that the level of activated caspase 3 was markedly elevated in linc00472-overexpressing MCF-7 and MDA-MB-453 cells compared with control cells (vector) (Fig. 3E and F). In a word, these results revealed that linc00472 overexpression enhanced ADR chemosensitivity and promoted ADR-mediated apoptosis in breast cancer cells.3.4. Linc00472 acted as a decoy of miR-141
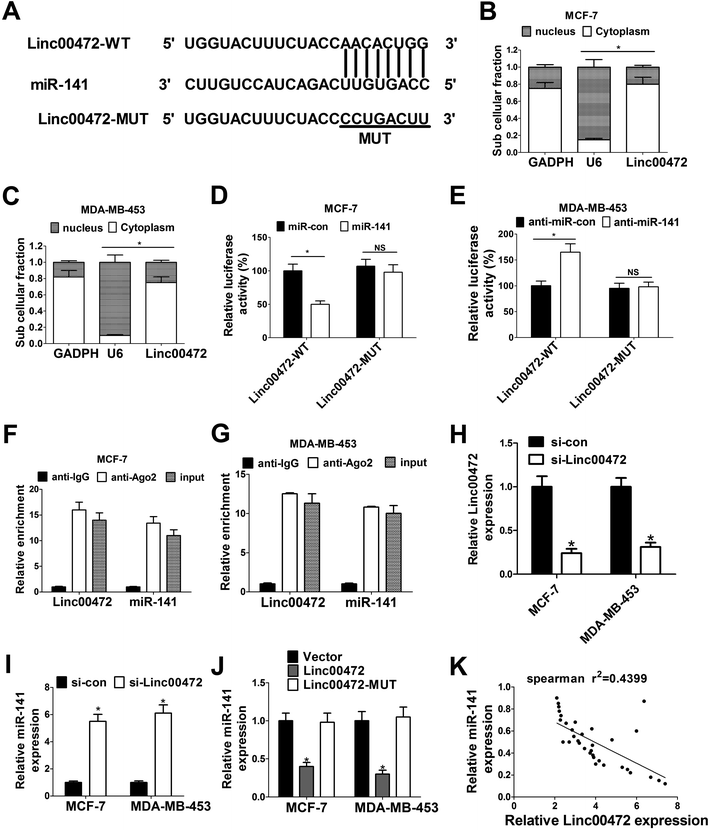
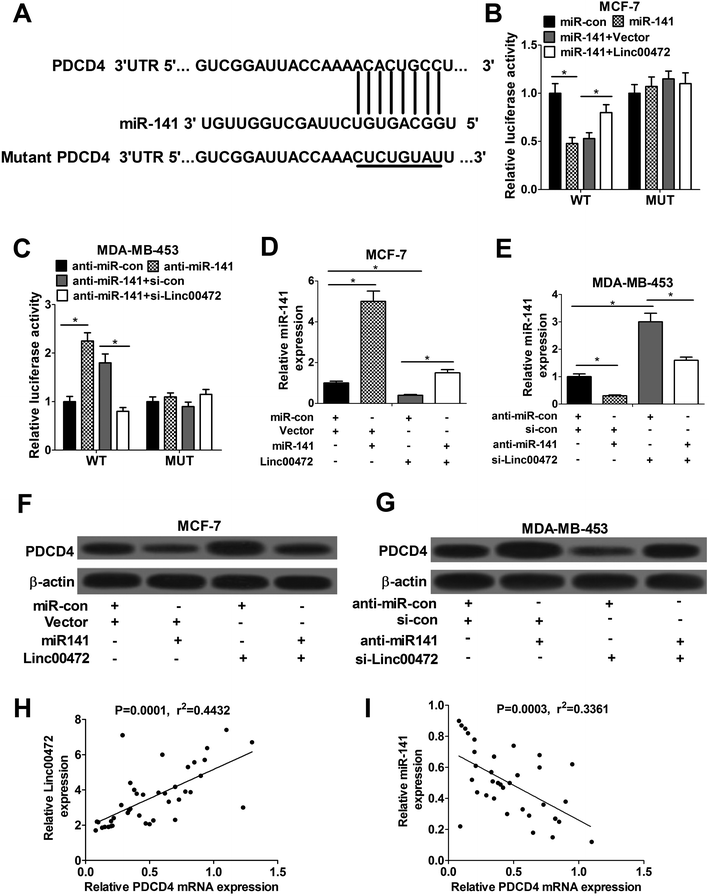
To further investigate the molecular mechanism of linc00472 in breast cancer progression, miRcode online website was used to search for the potential target miRNAs of linc00472. The results showed there existed some complementary sites between linc00472 and miR-141 (Fig. 4A). To explore the potential interaction between linc00472 and miR-141, cellular fractionation assays were performed to identify the subcellular location of linc00472 in MCF-7 and MDA-MB-453 cells. As displayed in Fig. 4B and C, linc00472 was substantially enriched in the cytoplasmic fraction, indicating that linc00472 had a chance to interact with cytoplasmic miRNAs. Next, to further confirm the direct interaction between linc00472 and miR-141, the wide-type sequence of linc00472 complementary with miR-141 or its mutant sequence was subcloned into luciferase reporter vector to generate linc00472-WT or linc00472-MUT luciferase reporter. Subsequent luciferase assays revealed that the relative luciferase activity of linc00472-WT systems was markedly decreased by miR-141 overexpression, but was notably increased following miR-141 depletion (Fig. 4D and E). However, the upregulation or downregulation of miR-141 had no effect on luciferase activity of linc00472-MUT systems compared with their corresponding counterparts (Fig. 4D and E). RIP assays also manifested that linc00472 and miR-141 was substantially enriched by Argonaute 2 (Ago2) antibody in MCF-7 and MDA-MB-453 cells when compared to IgG group (Fig. 4F and G). To observe the actual influence of linc00472 on miR-141, si-linc00472 was transfected into MCF-7 and MDA-MB-453 cells. At first, the knockdown efficiency of si-linc00472 was measured with the results showing that si-linc00472 transfection effectively decreased linc00472 expression in MCF-7 and MDA-MB-453 cells (Fig. 4H). As expected, knockdown of linc00472 gave rise to a significant increase of miR-141 expression in MCF-7 and MDA-MB-453 cells (Fig. 4I). Moreover, transfection of linc00472-overexpression plasmid containing miR-141 binding sites markedly suppressed miR-141 expression in MCF-7 and MDA-MB-453 cells, while introduction of linc00472-overexpression plasmid with mutant miR-141 binding sites (linc00472-MUT) had no effect on miR-141 level (Fig. 4J). Additionally, Spearman's correlation analysis also found that miR-141 expression level was inversely associated with linc00472 level in 40 cases of breast cancer tissues (Fig. 4K). Collectively, these results proved that linc00472 suppressed miR-141 expression by direct interaction.3.5. Linc00472 up-regulated PDCD4 expression by sponging miR-141
Accumulating evidence shows that miRNA exerts function by hindering target mRNAs expression. Hence, TargetScan website was employed to search for the potential target mRNAs of miR-141. Prediction results showed that there existed some complementary sites between PDCD4 and miR-141 (Fig. 5A). To further validate the interaction between linc00472, miR-141 and PDCD4, wide-type or mutant-type PDCD4-3′UTR luciferase reporter vector was constructed, followed by co-transfected with (miR-con, miR-141, miR-141+pcNDA3.1 vector, or miR-141+pcNDA-linc00472) or (anti-miR-con, anti-miR-141, anti-miR-141+si-con, or anti-miR-141+si-linc00472) into MCF-7 or MDA-MB-453 cells. As shown in Fig. 5B, introduction of miR-141 mimic strikingly decreased luciferase activity of wide-type PDCD4-3′UTR systems, while linc00472 overexpression notably abolished this effect of miR-141. Moreover, the luciferase activity of WT-PDCD4-3′UTR systems was enhanced by miR-141 inhibitor, while this effect of anti-miR-141 was substantially abrogated by linc00472 knockdown. However, change of miR-141 or linc00472 expression had no influence on luciferase activity of mutant-type PDCD4-3′UTR systems (Fig. 5B and C). In a word, miR-141 directly interacted with PDCD4-3′UTR region, while this interaction was attenuated by linc00472. Then, we continued to demonstrate the actual effect of miR-141 or linc00472 on PDCD4 expression. The results unveiled that miR-141 expression level was markedly enhanced by miR-141 mimic in MCF-7 cells, but was notably suppressed by pcDNA-linc00472 overexpression plasmid (Fig. 5D). Moreover, linc00472-induced decrease in miR-141 expression was reversed by transfection with miR-141 mimic (Fig. 5D). Conversely, anti-miR-141 curbed miR-141 expression and knockdown of linc00472 facilitated miR-141 expression in MDA-MB-453 cells (Fig. 5E). Moreover, miR-141 inhibition attenuated si-linc00472-mediated increase on miR-141 expression (Fig. 5E). We further demonstrated that PDCD4 expression was suppressed by miR-141, but was increased by linc00472 overexpression in MCF-7 cells. In addition, miR-141 overexpression abated pcDNA-linc00472-mediated enhancement effect on PDCD4 expression (Fig. 5F). Conversely, miR-141 inhibitor contributed to PDCD4 expression and linc00472 knockdown suppressed PDCD4 expression in MDA-MB-453 cells. Also, introduction of miR-141 inhibitor abrogated si-linc00472-mediated inhibition effect on PDCD4 expression (Fig. 5G). Spearman's correlation analyses revealed a positive relevance between linc00472 and PDCD4 expression (Fig. 5H), as well as an inverse correlation between miR-141 and PDCD4 (Fig. 5I) expression in 40 cases of breast cancer tissues. All these data suggested that linc00472 could sponge miR-141 to affect PDCD4 expression.3.6. miR-141 overexpression or PDCD4 knockdown abolished the inhibition effect of linc00472 on breast cancer cell progression
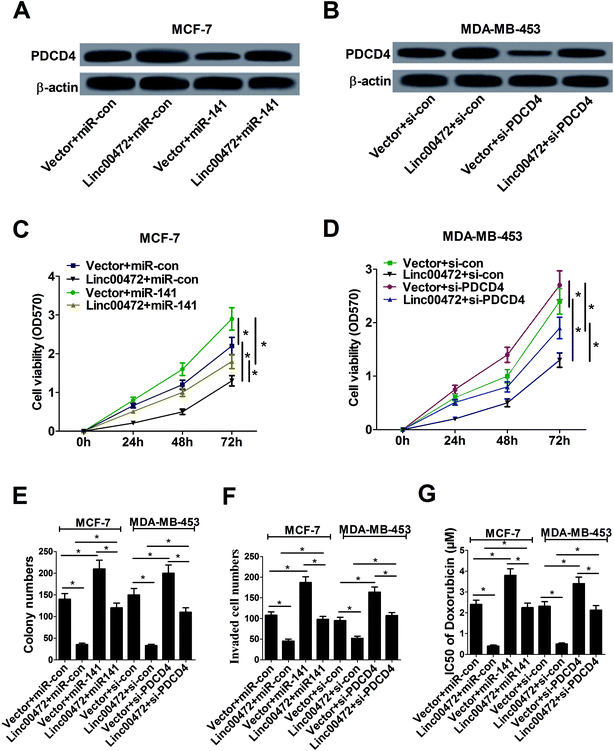
Plentiful studies demonstrated that lncRNAs acted as ceRNAs of miRNAs to sequester them from target mRNAs and enhance mRNA expression. As mentioned above, linc00472, miR-141 and PDCD4 was closely associated with each other, hence, we proceeded to investigate their function relevance in MCF-7 cells. As displayed in Fig. 6A, overexpression of linc00472 abated the inhibition effect of miR-141 on PDCD4 expression, and up-regulating miR-141 also weakened linc00472-mediated enhancement on PDCD4 expression. Moreover, we further validated that depletion of PDCD4 attenuated linc00472-induced promotion effect on PDCD4 expression, and linc00472 overexpression could also abate si-PDCD4-triggered diminishment in PDCD4 expression (Fig. 6B). Function analysis revealed that the inhibitory effects of linc00472 overexpression on cell proliferation, invasion and ADR resistance were markedly abrogated by miR-141 up-regulation or PDCD4 knockdown, and the promotive effects of miR-141 overexpression or PDCD4 knockdown on cell proliferation, invasion and ADR resistance were significantly overturned following restoration of linc00472 expression (Fig. 6C–G). Collectively, these results manifested that linc00472 suppressed breast cancer progression by regulating miR-141 and PDCD4.3.7. Linc00472 blocked tumor growth in breast cancer in vivo
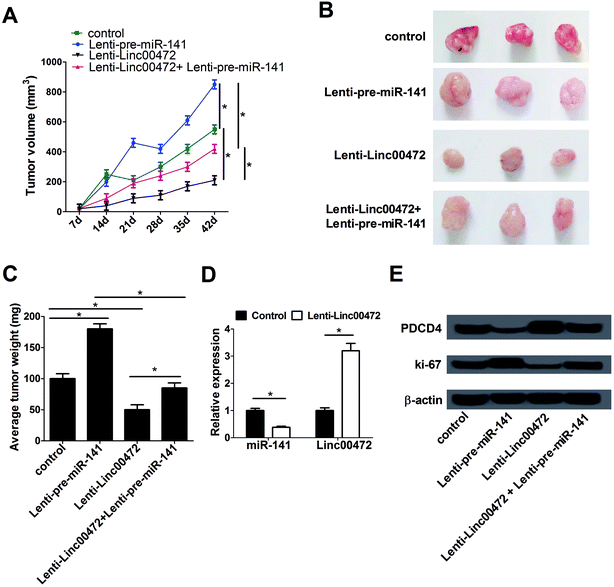
To further investigate the effect of miR-141 and linc00472 on tumor growth, MCF-7 cells stably transfected with lenti-con (control), lenti-pre-miR-141, lenti-linc00472 or lenti-linc00472+lenti-pre-miR-141 were subcutaneously injected into the armpit of nude mice. Tumor volume was detected once every 7 days for 42 days using a caliper. Tumor weight was measured at 42 days after implantation. As presented in Fig. 7A–C, overexpression of miR-141 notably accelerated tumor growth, while the tumor promotion effect of miR-141 was substantially reversed by linc00472 up-regulation. Moreover, enforced expression of linc00472 inhibited tumor growth, whereas linc00472-mediated anti-tumor effect was greatly counteracted following enhanced miR-141 expression. We also compared the expression patterns of miR-141 and linc00472 in linc00472-overexpression and control tumor tissues. As expected, transfection of lenti-linc00472 enhanced linc00472 expression and suppressed miR-141 expression (Fig. 7D). Furthermore, miR-141 up-regulation resulted in a decrease of PDCD4 expression and an increase of ki-67 (a proliferation marker) expression level, and linc00472 overexpression resulted in an elevation of PDCD4 expression and a decline of ki-67 expression in resected tumor tissues (Fig. 7E). Moreover, lenti-linc00472-mediated alteration of PDCD4 and ki-67 expression was greatly reversed following the regaining of miR-141 expression (Fig. 7E). These data linc00472 inhibited tumor growth partly through regulating miR-141 and PDCD4 expressions in breast cancer mice models.4. Discussion
LncRNAs and miRNAs have been reported to be implicated in the progression and drug resistance/sensitivity of various cancers including breast cancer.15 For instance, downregulation of lncRNA GAS5 enhanced trastuzumab resistance and facilitated proliferation by regulating miR-21/PTEN in breast cancer.16 Overexpression of lncRNA BCAR4 promoted proliferation and enhanced endocrine resistance in breast cancer cells.17 miR-451 could enhance ADR sensitivity of ADR-resistant breast cancer cells.18 Here, our study was performed to elucidate the role and mechanism of linc00472 in breast cancer progression.In the present study, we firstly demonstrated that linc00472 expression level was down-regulated in breast cancer tissues and cells, and was positively related with favorable disease status and prognosis of breast cancer patients (Fig. 1), which is in accordance with previous studies.13,14 Gain-of-function analysis showed that linc00472 overexpression suppressed proliferation and invasion in breast cancer cells (Fig. 2). Moreover, linc00472 overexpression enhanced the susceptibility of breast cancer cells to ADR (Fig. 3), presented by decreased IC50 value of ADR (Fig. 3A and B), increased apoptotic rate (Fig. 3C and D) and enhanced expression of activated caspase 3 (Fig. 3E and F) in breast cancer cells following linc00472 upregulation.
Accumulating evidence indicate that lncRNAs exert functions by acting as ceRNAs of miRNAs, therefore, miRcode website was employed to search for the potential target miRNAs of linc00472. The prediction results showed that miR-141 might interact with linc00472 (Fig. 4A), which was further validated by the following subcellular fractionation (Fig. 4B and C), luciferase (Fig. 4D and E) and RIP (Fig. 4F and G) assays. Furthermore, linc00472 knockdown strikingly promoted miR-141 expression, while linc00472 overexpression induced a notable downregulation of miR-141 level in breast cancer cells (Fig. 4I and J). Additionally, spearman's correlation analysis revealed that miR-141 expression was negatively associated with linc00472 expression in breast cancer tissues (Fig. 4K).
miR-141, a member of miR-200 family, has been validated to be involved in progression and drugs resistance of some cancers by acting as a tumor suppressor or an oncogene. For example, miR-141 expression was down-regulated in gastric cancer and overexpression of miR-141 suppressed the proliferation of gastric cancer cells.19 However, some findings revealed that miR-141 was highly expressed in ovarian cancer and high expression of miR-141 enhanced cisplatin resistance of ovarian cancer cells.20,21 Previous studies also showed that all members of miR-200 family could regulate EGF-driven invasion and miR-200a/141 cluster could lead to G1 arrest in breast cancer cells, hinting the association between miR-141 and tumorigenesis of breast cancer.22 Moreover, miR-141 expression was found to be strikingly up-regulated in ADR and verapamil resistance breast cancer cell line (MCF-7/AdrVp) compared with parental MCF-7 cell line, indicating a link between miR-141 and ADR resistance of breast cancer cells.23 In the present study, we aimed to further explore the role and molecular mechanism of miR-141 in the progression and drug resistance of breast cancer.
Previous studies showed that miRNAs exerted their roles through regulating target mRNAs expression. Hence, TargetScan software was used to figure out the candidate target mRNAs of miR-141 with the results showing that there existed some complementary sites between PDCD4 and miR-141 (Fig. 5A). Then, luciferase (Fig. 5B and C), RT-qPCR (Fig. 5D and E) and western blot (Fig. 5F and G) assays further demonstrated that linc00472 performed as a ceRNA of miR-141 to enhance target gene PDCD4 expression. Moreover, spearman's correlation analysis revealed that PDCD4 expression was positively related to linc00472 expression (Fig. 5H), but was inversely associated with miR-141 expression (Fig. 5I).
PDCD4, localized to chromosome 10q24, has been demonstrated to be associated with the development of cancers by regulating various proteins implicated in tumor progression, cell differentiation and cell cycle.24,25 PDCD4 also has been identified as a tumor suppressor in multiple cancers with a low-level expression.25,26 For example, overexpression of PDCD4 suppressed ovarian cancer cell proliferation, migration and invasion.27 Also, previous studies manifested that PDCD4 inhibited migration and invasion as well as induced apoptosis in breast cancer cells.28,29 Moreover, PDCD4 participated in mediating the sensitivity of cancer cells to chemotherapy drugs such as docetaxel, paclitaxel and cisplatin.30,31 Taken together, these data promoted us to suppose that linc00472 might affect tumorigenesis and ADR resistance by regulating miR-141 and PDCD4 in breast cancer.
Furthermore, we demonstrated that increased miR-141 or depleted PDCD4 could weaken linc00472-mediated promotive effect on PDCD4 expression in breast cancer cells (Fig. 6A and B). Besides, miR-141 overexpression facilitated proliferation and invasion as well as improved ADR resistance in breast cancer cells (Fig. 6C–G). Restoration assays further unveiled that miR-141 overexpression or PDCD4 knockdown abolished the inhibitory effects of linc00472 on proliferation, invasion and ADR resistance of breast cancer cells (Fig. 6C–G). Furthermore, we noticed a significant decrease in tumor volume and weight, a notable inhibition of miR-141 and ki-67 expression, as well as a striking increase of PDCD4 expression in linc00472-overexpressing mice models. On the contrary, overexpression of miR-141 promoted tumor growth, presented by the increase of tumor volume and weight in mouse xenografts following the upregulation of miR-141. Also, linc00472-mediated anti-tumor effect was partly weakened by miR-141 overexpression and miR-141-induced promotive effect on tumor growth was attenuated following linc00472 up-regulation (Fig. 7A and B). Moreover, PDCD4 expression was decreased in miR-141-overexpression tumor tissues, while it was increased in linc00472-overexpression tumor tissues (Fig. 7D and E). All these data indicated that linc00472 suppressed breast cancer progression in vitro and in vivo.
5. Conclusions
In a word, our study demonstrated that linc00472 hindered proliferation and invasion, induced apoptosis, and enhanced ADR sensitivity by miR-141/PDCD4 axis in breast cancer cell lines. Our findings contributed to a better understanding of the mechanism of linc00472 involved in breast progression and ADR chemosensitivity, providing a potential biomarker and therapy target for breast cancer.Author contributions
Pengwei Lu, Lin Li and Yuanting Gu designed and performed the experiments. Pengwei Lv, Yunqing Yang and Fang Wang analyzed the data and wrote the manuscript. XueYang supervised the study and reviewed the manuscript.Conflicts of interest
The authors have no conflict of interest to declare.Acknowledgements
Not applicable.References
- R. L. Siegel, K. D. Miller and A. Jemal, Ca-Cancer J. Clin., 2016, 66, 7–30 CrossRef PubMed.
- F. Cardoso, M. Castiglione and E. G. W. Group, Ann. Oncol., 2009, 22, 15–18 Search PubMed.
- T. Gamucci, A. M. D'Ottavio, E. Magnolfi, M. Barduagni, A. Vaccaro, I. Sperduti, L. Moscetti, F. Belli and L. Meliffi, Br. J. Cancer, 2007, 97, 1040–1045 CrossRef CAS PubMed.
- C. E. Desantis, S. A. Fedewa, S. A. Goding, J. L. Kramer, R. A. Smith and A. Jemal, Ca-Cancer J. Clin., 2015, 66, 31–42 CrossRef PubMed.
- G. W. Sledge, D. Neuberg, P. Bernardo, J. N. Ingle, S. Martino, E. K. Rowinsky and W. C. Wood, J. Clin. Oncol., 2003, 21, 588–592 CrossRef PubMed.
- C. Marquette and L. Nabell, Curr. Treat. Options Oncol., 2012, 13, 263–275 CrossRef PubMed.
- A. Faversani, V. Vaira, G. P. Moro, D. Tosi, A. Lopergolo, D. C. Schultz, D. Rivadeneira, D. C. Altieri and S. Bosari, Breast Cancer Res., 2014, 16, 1–13 CrossRef PubMed.
- M. Majidinia and B. Yousefi, DNA Repair, 2016, 45, 25–33 CrossRef CAS PubMed.
- Y. Liu, S. Sharma and K. Watabe, Front. Biosci., 2015, 7, 94 CrossRef.
- X. Li, S. Wang, Z. Li, X. Long, Z. Guo, G. Zhang, J. Zu, Y. Chen and L. Wen, Int. J. Biol. Macromol., 2017, 105, 346–353 CrossRef CAS PubMed.
- X. Xue, Y. A. Yang, A. Zhang, K. W. Fong, J. Kim, B. Song, S. Li, J. C. Zhao and J. Yu, Oncogene, 2016, 35, 2746–2755 CrossRef CAS PubMed.
- S. J. Shi, L. J. Wang, B. Yu, Y. H. Li, Y. Jin and X. Z. Bai, Oncotarget, 2015, 6, 11652–11663 Search PubMed.
- Y. Shen, D. Katsaros, L. W. Loo, B. Y. Hernandez, C. Chong, E. M. Canuto, N. Biglia, L. Lu, H. Risch and W. M. Chu, Oncotarget, 2015, 6, 8579–8592 Search PubMed.
- Y. Shen, Z. Wang, L. W. Loo, Y. Ni, W. Jia, P. Fei, H. A. Risch, D. Katsaros and H. Yu, Breast Cancer Res. Treat., 2015, 154, 473–482 CrossRef CAS PubMed.
- D. Ayers and J. Vandesompele, Genes, 2017, 8, E95 CrossRef PubMed.
- W. Li, L. Zhai, H. Wang, C. Liu, J. Zhang, W. Chen and Q. Wei, Oncotarget, 2016, 7, 27778–27786 Search PubMed.
- M. Godinho, D. Meijer, B. Setyono-Han, L. C. Dorssers and T. V. Agthoven, J. Cell. Physiol., 2011, 226, 1741–1749 CrossRef CAS PubMed.
- O. Kovalchuk, J. Filkowski, J. Meservy, Y. Ilnytskyy, V. P. Tryndyak, V. F. Chekhun and I. P. Pogribny, Mol. Cancer Ther., 2008, 7, 2152–2159 CrossRef CAS PubMed.
- D. Ying, Y. Xu, D. Ling, H. Yao, Y. Hong, T. Zhou and J. Si, J. Gastroenterol., 2009, 44, 556–561 CrossRef PubMed.
- Y. C. Gao and J. Wu, Tumor Biol., 2015, 36, 4843–4850 CrossRef CAS PubMed.
- M. T. van Jaarsveld, J. Helleman, A. W. Boersma, P. F. van Kuijk, W. F. van Ijcken, E. Despierre, I. Vergote, R. H. Mathijssen, E. M. Berns and J. Verweij, Oncogene, 2013, 32, 4284–4293 CrossRef CAS PubMed.
- S. Uhlmann, J. D. Zhang, A. Schwäger, H. Mannsperger, Y. Riazalhosseini, S. Burmester, A. Ward, U. Korf, S. Wiemann and O. Sahin, Oncogene, 2010, 29, 4297–4306 CrossRef CAS PubMed.
- G. Q. Chen, Z. W. Zhao, H. Y. Zhou, Y. J. Liu and H. J. Yang, Med. Oncol., 2010, 27, 406–415 CrossRef CAS PubMed.
- A. P. Jansen, C. E. Camalier and N. H. Colburn, Cancer Res., 2005, 65, 6034–6041 CrossRef CAS PubMed.
- B. Lankatbuttgereit and R. Göke, Biol. Cell., 2009, 101, 309–317 CrossRef CAS PubMed.
- X. Liu, Z. Zhang, L. Sun, N. Chai, S. Tang, J. Jin, H. Hu, Y. Nie, X. Wang and K. Wu, Carcinogenesis, 2011, 32, 1798–1805 CrossRef CAS PubMed.
- N. Wei, S. S. Liu, K. K. Chan and H. Y. Ngan, PLoS One, 2012, 7, e30311 CAS.
- O. Afonja, D. Juste, S. Das, S. Matsuhashi and H. H. Samuels, Oncogene, 2004, 23, 8135–8145 CrossRef CAS PubMed.
- A. N. Santhanam, A. R. Baker, G. Hegamyer, D. A. Kirschmann and N. H. Colburn, Oncogene, 2010, 29, 3921–3932 CrossRef CAS PubMed.
- G. H. Shi, D. W. Ye, X. D. Yao, S. L. Zhang, B. Dai, H. L. Zhang, Y. J. Shen, Y. Zhu, Y. P. Zhu and W. J. Xiao, Acta Pharmacol. Sin., 2010, 31, 867–873 CrossRef CAS PubMed.
- M. Shiota, H. Izumi, A. Tanimoto, M. Takahashi, N. Miyamoto, E. Kashiwagi, A. Kidani, G. Hirano, D. Masubuchi and Y. Fukunaka, Cancer Res., 2009, 69, 3148–3156 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |