Open Access Article
Open Access ArticleMC4R variant rs17782313 and manifestation of obese phenotype in Pakistani females
Sobia Rana *,
Soma Rahmani
*,
Soma Rahmani and
Saad Mirza
and
Saad Mirza
Molecular Biology and Human Genetics Laboratory, Dr. Panjwani Center for Molecular Medicine and Drug Research (PCMD), International Center for Chemical and Biological Sciences (ICCBS), University of Karachi, Karachi-75270, Pakistan. E-mail: molecularbiologist1@gmail.com; sobia.rana@iccs.edu; Tel: + 92 21 99261683
First published on 9th May 2018
Abstract
MC4R represents a key player involved in melanocortin-mediated control of energy balance. Recently identified near MC4R variant rs17782313 (T > C) can serve as a contributing factor for obese phenotype but its association with obesity has never been sought in a sample of the Pakistani population. The role of genetic variants as causal factors varies across populations. Association studies in a specific population can help us to distinguish global from local gene–gene and gene–environment interactions. This is the first study that investigated the association of rs17782313 with obesity and various obesity-linked anthropometric, metabolic, physical, and behavioural traits in Pakistani subjects including 306 OW/OB (overweight and obese) and 300 NW (normal weight) individuals. The comparison of various aforementioned obesity-linked continuous and categorical variables between OW/OB and NW subjects revealed that almost all variables were found significantly aberrant (p < 0.05) in OW/OB subjects as compared to their age- and gender-matched NW controls indicating greater risk of developing various cardio-metabolic disorders. The genotyping of rs17782313 showed significant association of this variant with obesity and obesity-linked anthropometric traits in females suggesting the gender-specific effect of this variant in our population. The minor allele C increased the risk of obesity by 1.55 times (95% CI = 1.1–2.18, p = 0.01) whereas homozygous CC genotype increased the risk by 2.43 times (95% CI = 1.19–4.96, p = 0.015) in females. However, no association of rs17782313 was observed with any of the obesity-linked metabolic, physical, and behavioural traits except random eating timings. In conclusion, the current study significantly contributes to the knowledge of the genetic proneness to obesity in Pakistani females. This could also be helpful for forthcoming meta-analysis studies elucidating which variants are truly associated with the susceptibility to develop an obese phenotype.
1. Introduction
Obesity can be viewed as a chronic destabilization of energy balance attributable to more energy intake (caloric consumption) and less energy expenditure (basal metabolism, adaptive thermogenesis, and physical activity) engendering an overall shift toward positive energy state ultimately reflected by excessive build-up of adipose tissue in the body and weight gain. It can have severe implications for longevity1 and health including type II diabetes, cardiovascular diseases, asthma, gall bladder disease, osteoarthritis, and many cancers.2The central melanocortin system regulates body weight and overall metabolic fitness by modulating the acute and long-term energetic states with appropriate behavioural and physiological output.3 These activities are dependent on the interoceptive function of two antagonistic populations of first-order neurons namely Agouti-related protein (AGRP)-producing neurons and proopiomelanocortin (POMC)-producing neurons in the arcuate nucleus of the hypothalamus (ARC). These two first-order neuronal populations represent a cellular interface between afferent indicators of physiological state and neural circuits governing response enactment.4 ARCAgRP neurons form the anabolic wing of the melanocortin pathway that is robustly stimulated by caloric insufficiency5 and is essential for driving energy intake, conserving energy expenditure, and promoting weight gain.6–8 Conversely, ARCPOMC neurons form the catabolic wing that is stimulated by caloric sufficiency and lead to satiety, increased energy expenditure, and weight loss.7,9,10 The proficiency of the melanocortin system to control both catabolic and anabolic processes of energy balance rests upon antagonistic involvement of second-order melanocortin-4 receptor (MC4R)-expressing neurons.
MC4R is a seven transmembrane G-protein coupled receptor critically involved in the central regulation of energy balance. It is a member of the melanocortin receptor family and expressed by multiple neuronal populations in the central nervous system.11 MC4R signalling is modulated by the first-order neuropeptides namely alpha-melanocyte stimulating hormone (α-MSH, a post-translational derivative of POMC) and AGRP. Both α-MSH and AGRP compete for binding to MC4R. The binding of α-MSH (an agonist for MC4R) stimulates MC4R activity whereas AGRP (an inverse agonist for MC4R) binding suppresses MC4R activity. Enhanced MC4R activity triggers an anorexigenic signal while diminished receptor activity triggers an orexigenic signal.12 Experimental evidence demonstrates a functional divergence in the melanocortinergic network such that the regulation of energy intake, energy expenditure, and glucose homeostasis proceeds through neuroanatomically distinct populations of MC4R-expressing neurons.13 MC4R-expressing glutamatergic neurons in the paraventricular nucleus of the hypothalamus (PVHMC4R) are the principal population for regulating energy intake but do not influence energy expenditure. MC4R-expressing PVH neurons are synaptically connected to neurons in the parabrachial nucleus (PBN), which relays visceral information to the forebrain, thus, lateral PBN serves as the site of functional outflow for melanocortin-regulated appetite.14 MC4R-expressing cholinergic preganglionic sympathetic neurons in the intermediolateral nucleus of the spinal cord (IMLMC4R) are the predominant population for regulating energy expenditure with no influence on energy intake. IMLMC4R neurons govern generalized sympathetic tone (sympatho-excitation) engendering increased energy expenditure, elevated blood pressure, and decreased plasma glucose. IMLMC4R neurons play a role in overall glucose homeostasis by increasing hepatic insulin action (sensitivity) including suppression of endogenous glucose production and stimulation of glucose disposal but do not directly influence insulin release. The MC4R-expressing preganglionic parasympathetic neurons in the dorsal motor nucleus of the vagus (DMVMC4R) are the primary population implicated in the glucose homeostasis with no influence on energy intake and expenditure. DMVMC4R neurons suppress parasympathetic tone (parasympatho-suppression) engendering tonic inhibition of pancreatic insulin release with no considerable effect on overall glycaemic state or insulin sensitivity.15–17
The human MC4R is a 332-amino acid protein encoded by a single exon gene localized on chromosome 18q22.18 Polymorphisms within the MC4R coding region or variants outside of the coding region that influence its expression can result in partial or complete dysfunction of MC4R leading to a clinical phenotype with lack of satiety, hyperphagia, a decline in energy expenditure, and consequently obesity.19,20 MC4R variants were originally identified as causing rare monogenic obesity but now known to be frequent enough to account for a considerable proportion of common obesity cases.21 Genome wide association studies (GWAS) conducted in Caucasians has identified new loci with variants associated with obesity. Among these variants, rs17782313 (T > C polymorphism) mapped at 188 kb downstream of the MC4R gene showed second strongest association with BMI.22 Energy intakes higher than estimated energy requirement23 and eating behaviour more specifically emotional eating and food cravings24 might be account for the association between rs17782313 and BMI.
The human genetic architecture differs across populations. The frequencies of risk alleles responsible for susceptibility to obesity differ among populations of changing geographic origin. To date, most of GWAS published reports have been executed in populations of western origin with non-western and multi-ethnic populations remain under-investigated so far. Thus, it is uncertain if the results of association studies in western world hold true in different non-western and multi-ethnic populations. Multi-ethnic study designs have great potential to rebuild the evolutionary history of genetic proneness to obesity, isolate disease-causing variants, and distinguish global from local gene–gene and gene–environment interactions. Association studies across different populations can help us to delineate more precisely which loci or variants could play a role in the obesity aetiology and help to understand the genetic and environmental factors contributing to obese phenotype. Therefore, association studies should be encouraged in non-western or isolated populations, especially in populations at low or high risk for obesity. According to a recent report by global burden of disease study, Pakistan has been placed at 9th position out of 188 countries in terms of overweight and obesity. One-third of adults in here are overweight and obese, and the gender gap in excess weight is widening with more women gaining weight than men.25 Such a high prevalence of overweight and obesity in Pakistan warrants investigations of various factors involved in manifestation of such phenotypes including genetic factors. To our knowledge, there is no study available that investigated the association of the MC4R variant rs17782313 with overweight and obese phenotypes in Pakistani population. Therefore, the current study has been carried out to investigate the association of this variant with the expression of obese phenotype and related traits in Pakistani population.
2. Materials and methods
2.1. Study design and sample population
The study was carried out at the International Center for Chemical and Biological Sciences (ICCBS), University of Karachi, Pakistan, after taking approval from Independent Ethics Committee (IEC) of the institute. The study was also approved by the University's Board of Advanced Studies and Research (BASR). Moreover, the study was conducted in accordance with the Helsinki Declaration as revised in 1989.A total of 606 human subjects of both genders between 12 and 62 years of age were recruited in the study. The total subjects constituting the sample population included 336 males (55.45%) and 270 females (44.55%). The mean age (mean ± SEM) of the sample population was 29.18 ± 0.37 years. Simple random sampling without replacement technique was used to recruit the subjects from general population of Karachi after obtaining written informed consents. However, all subjects were not permanent residents of the city. The recruited subjects were from diverse ethnic backgrounds including Urdu-speaking, Punjabi, Pashtun, Sindhi, Balochi, and others. Karachi is a cosmopolitan city and represents the people of diverse ethnic backgrounds from all over the Pakistan.
The study was based on a case-control design. A total sample population of 606 individuals included 306 overweight and obese subjects (cases) and their sex- and age-matched (±5 years) 300 normal-weight individuals (controls) with a calculated statistical power of 80.5. The sample size for the study population was estimated via Online Sample Size Estimator (OSSE) by substituting Minor Allele Frequency (MAF) in cases as 18.3% and in controls as 10.3%.26 The study population was further stratified on the basis of gender (males and females) and age (group1: subjects ≤20 years of age; group 2: subjects >20 years of age). The inclusion criteria for the case subjects with ages > 20 years was BMI ≥ 25 kg m−2 for overweight subjects and BMI ≥ 30 kg m−2 for obese subjects whereas the inclusion criteria for the control subjects (normal-weight) of the same age was BMI < 25 kg m−2 according to World Health Organization (WHO). On the other hand, the inclusion criteria for case subjects with ages ≤20 years was >85th – <95th percentile for overweight subjects and ≥95th percentile for obese subjects whereas the inclusion criteria for control subjects ≤20 years of age was 5th–85th percentile according to Center for Disease Control and Prevention (CDC) BMI for age growth charts. Individuals with the history of medication (tricyclic antidepressants, phenothiazine, anticonvulsants, and steroids) and history of endocrine disorders such as pituitary dysfunction, Cushing's syndrome and hypothyroidism were not included in the study.
2.2. Physical examination and behavioural features
Physical examination was performed to check the signs of hyperlipidaemia, acanthosis nigricans, and abdominal and axillary striae. Blood pressure (systolic and diastolic) was measured twice from the right arm of the subject in a sitting position using a standard mercury sphygmomanometer. Obesity-related behavioural information of each participant was obtained through questionnaire.2.3. Anthropometric parameters
Height in centimetres (to the nearest 0.1 cm) and weight in kilograms (to the nearest 0.1 kg) were measured by using a portable stadiometer (Seca 214, Germany) and a mechanical column scale (Seca 755, Germany), respectively. Both measurements were taken with light clothing and no shoes on. BMI was computed as weight (kg) divided by squared height (m2) from height and weight obtained during the in-person screen. Waist circumference (WC) was measured just above navel around the mid-point between the lower margin of the last rib and top of iliac crest by using a stretch-resistant measuring tape. Similarly, hip circumference was measured around the widest portion of the buttocks. The waist-to-hip (WHR) ratio was calculated by dividing the two measurements. Skin-fold thickness (subcutaneous fat) was measured in millimetres (mm) for biceps, triceps, suprailiac, abdomen, thigh, and sub-scapular by using skin-fold calliper (Slim Guide, MI, USA). Every measurement was done three times and the average value was taken. Skin-fold measurement of 4 sites (triceps, abdomen, supra-iliac, and thigh) was then used for estimation of percent body fat (% BF) by using gender-specific standard formulae27 as follows:| %BF in males = (0.29288)(4 skin-folds sum) − (0.0005)(4 skin-folds sum)2 + 0.15845(age) − 5.76377 |
| %BF in females = (0.29669)(4 skin-folds sum) − (0.00043)(4 skin-folds sum)2 + 0.02963(age) + 1.4072 |
2.4. Blood sample collection
Venous blood samples were drawn from each participant after an overnight fast of 8–12 hours. Fasting blood sample of 5 ml volume was drawn from each participant by using a 5 ml syringe (Cat no. 367863 BD, USA) via standard venepuncture technique. For each participant, 2 ml of fasting blood sample was collected in an EDTA (anticoagulant)-coated lavender top vacutainer tube (Cat no. 367841, BD, USA) whereas rest of 3 ml fasting blood sample was collected in a gold top vacutainer tube (Cat no. 367983, BD, USA) containing gel and clot activator. Blood sample from lavender top vacutainer tube was consumed for subsequent DNA extraction while blood sample from gold top vacutainer tube was used for subsequent serum isolation. Serum was isolated by centrifuging the gold top vacutainer tube containing 3 ml of fasting blood sample at 4000 rpm for 10 minutes. Isolated serum was used for the biochemical estimation of fasting insulin.2.5. Metabolic estimations
Fasting blood glucose (FBG) levels were estimated by using blood glucose monitoring system (Abbott, UK) while fasting serum insulin levels were determined by ELISA (enzyme linked immunosorbent assay) using DIAsource INS-EASIA Kit (Cat no. KAP1251, Belgium) according to manufacturer's instructions on Multiskan™ FC Microplate Photometer (ThermoFisher Scientific, MA, USA). The values of FBG and fasting serum insulin were utilized to calculate the homeostasis model assessment of insulin resistance (HOMA-IR) by the following formula: HOMA-IR = Fasting glucose (mmol L−1) × Fasting insulin (μIU ml−1)/22.5.282.6. DNA extraction and allelic discrimination assay
Genomic DNA was extracted from whole blood using EZ-10 spin column genomic DNA kit (Cat. no. BS684, Bio Basic, ON, Canada) according to the manufacturer's protocol. The variant rs17782313 (T → C transition in 188 kb downstream of the MC4R gene) was genotyped by real-time PCR and allelic discrimination using TaqMan® predesigned SNP genotyping assay (assay ID: C__32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 667
667![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 060_10, Cat no. 4
060_10, Cat no. 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 351376, ABI, Foster city, CA, USA) and TaqMan® genotyping master mix (Cat. no. 4381656, ABI, Foster City, CA, USA). The TaqMan genotyping reaction was amplified on an Applied Biosystems Inc. 7500 real-time PCR System (ABI, Foster City, CA, USA). The reaction components for each genotyping reaction include 50 ng of genomic DNA, 12.5 μL of TaqMan genotyping master mix (2X), 1.25 μL of assay mix (20X), and water up to a total volume of 25 μL. The thermocycler conditions employed were 95 °C for 10 minutes followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 1 minute. The reaction was then analysed for allelic discrimination by using Applied Biosystems Sequence Detection Software. Genotyping quality control was performed by duplicating 20% of the samples. Furthermore, two negative controls (NTC → No Template control) and a positive control (PC) for each genotype (TT, CT, and CC) were included in every batch.
351376, ABI, Foster city, CA, USA) and TaqMan® genotyping master mix (Cat. no. 4381656, ABI, Foster City, CA, USA). The TaqMan genotyping reaction was amplified on an Applied Biosystems Inc. 7500 real-time PCR System (ABI, Foster City, CA, USA). The reaction components for each genotyping reaction include 50 ng of genomic DNA, 12.5 μL of TaqMan genotyping master mix (2X), 1.25 μL of assay mix (20X), and water up to a total volume of 25 μL. The thermocycler conditions employed were 95 °C for 10 minutes followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 1 minute. The reaction was then analysed for allelic discrimination by using Applied Biosystems Sequence Detection Software. Genotyping quality control was performed by duplicating 20% of the samples. Furthermore, two negative controls (NTC → No Template control) and a positive control (PC) for each genotype (TT, CT, and CC) were included in every batch.
2.7. Statistical analysis
The statistical analysis was performed by using International Business Machines Statistical Package for Social Sciences software version 19.0 (IBM SPSS. Inc. Chicago, IL, USA). The normality of the data was examined by the Kolmogorov–Smirnov test. Continuous variables were expressed as mean ranks and categorical variables as percentages. Continuous variables were compared by Mann–Whitney U test and categorical variables by chi-square (χ2) test to observe the differences between OW/OB and NW subjects. The Hardy–Weinberg equation was applied to check whether the observed allelic and genotypic frequencies in overweight/obese and normal weight subjects were compatible with Hardy–Weinberg equilibrium (HWE) predictions. The whole data was stratified according to gender and age in subgroups. Allelic frequencies were calculated by gene counting. The differences in allelic and genotypic frequencies between OW/OB and NW subjects were determined by applying χ2 test assuming co-dominant (TT, CT, CC), dominant (TT vs. CT + CC) and recessive (CC vs. CT + TT) genetic models. Odd ratio (OR) and 95% confidence interval (CI) were calculated to find out the risk of obesity associated with the risk allele. Associations of obesity-related continuous variables (anthropometric and metabolic parameters) across genotypes of MC4R variant rs17782313 were assessed by applying Kruskal Wallis test and subsequent pair-wise associations were explored by using Dunn-Bonferroni post-hoc analysis. On the other hand, associations of obesity-related categorical variables across genotypes of MC4R variant rs17782313 were determined using χ2-test. A p-value < 0.05 was considered for significant differences.3. Results
3.1. Demographic data
The mean age (mean ± SEM) of overweight and obese subjects (cases) was 29.96 ± 0.538 years while the mean age of normal weight subjects (controls) was 28.39 ± 0.491 years. Among total population of overweight and obese subjects (306), 59.5% (182) reported the age of obesity onset as ≤20 years while rest of the 40.5% (124) reported the age of obesity onset as >20 years.3.2. Ethnic diversity
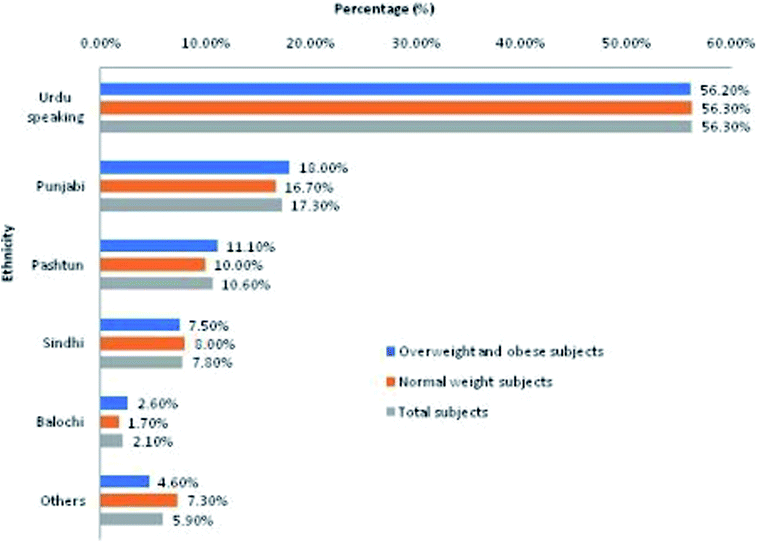
The sample population represented subjects with diverse ethnic backgrounds (Fig. 1). The order of ethnic representation in total study population (n = 606) from highest to lowest representation was Urdu-speaking (56.30%) > Punjabi (17.30%) > Pashtun (10.60%) > Sindhi (7.80%) > Balochi (2.10%) > others (5.90%; Balti, Kashmiri, Chitrali, etc.). Almost equal ethnic distribution was observed in overweight and obese (OW/OB) subjects (cases) and normal weight (NW) subjects (controls).3.3. Significantly aberrant continuous variables in overweight and obese cases
Comparison of continuous variables between OW/OB cases and NW controls is presented in Table 1. All the continuous variables including anthropometric measurements (weight, BMI, WC, HC, WHR, subcutaneous fat deposits, and % BF) except height, metabolic parameters (FBG, fasting insulin, HOMA-IR), and systolic (SBP) and diastolic (DBP) blood pressures were found significantly aberrant in total and gender-stratified OW/OB subjects as compared to their corresponding normal weight controls (p < 0.05) except the difference for SBP in males. The mean rank of SBP in male cases was higher than that of male controls but the p-value did not remain significant though it was at the very edge of significance (p = 0.052).| Total N = 606 | Females N = 270 (44.55%) | Males N = 336 (55.45%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameters | Groups | Mean | SD | Mean rank | p-Value | Mean | SD | Mean rank | p-Value | Mean | SD | Mean rank | p-Value |
| a Continuous variables are taken as mean rank and compared by Mann–Whitney U test between cases and controls. *A p-value < 0.05 was considered significant. Abbreviations; cases1: overweight and obese subjects, SD: standard deviation, BMI: body mass index, WC: waist circumference, HC: hip circumference, WHR: waist-to-hip ratio, SFT: skinfold thickness, % BF: percentage body fat, SBP: systolic blood pressure, DBP: diastolic blood pressure, FBG: fasting blood glucose, HOMA-IR: homeostasis model assessment of insulin resistance. | |||||||||||||
| Weight (kg) | Cases | 89.01 | 19.25 | 439.83 | <0.001* | 83.05 | 18.63 | 197.99 | <0.001* | 93.91 | 18.40 | 248.66 | <0.001* |
| Controls | 59.19 | 7.91 | 164.45 | 54.69 | 6.01 | 70.17 | 62.74 | 7.42 | 88.34 | ||||
| Height (cm) | Cases | 164.27 | 9.37 | 296.39 | 0.312 | 156.86 | 5.82 | 125.83 | 0.037* | 170.36 | 7.06 | 167.77 | 0.890 |
| Controls | 164.99 | 8.55 | 310.76 | 158.09 | 5.19 | 145.61 | 170.42 | 6.50 | 169.23 | ||||
| BMI (kg m−2) | Cases | 32.89 | 6.07 | 453.46 | <0.001* | 33.67 | 6.81 | 201.46 | <0.001* | 32.25 | 5.32 | 252.50 | <0.001* |
| Controls | 21.70 | 2.00 | 150.54 | 21.87 | 2 | 66.55 | 21.58 | 2.01 | 84.50 | ||||
| WC (cm) | Cases | 110.74 | 14.26 | 444.08 | <0.001* | 112.37 | 14.91 | 196.46 | <0.001* | 109.39 | 13.59 | 248.56 | <0.001* |
| Controls | 82.95 | 8.24 | 160.11 | 81.61 | 9.57 | 71.77 | 84 | 6.87 | 88.44 | ||||
| HC (cm) | Cases | 114.61 | 12.02 | 440.34 | <0.001* | 115.66 | 11.43 | 196.71 | <0.001* | 113.76 | 12.46 | 244.34 | <0.001* |
| Controls | 94.89 | 5.80 | 163.92 | 95.42 | 5.68 | 71.51 | 94.47 | 5.88 | 92.66 | ||||
| WHR | Cases | 0.97 | 0.06 | 405.91 | <0.001* | 0.97 | 0.07 | 183.17 | <0.001* | 0.96 | 0.06 | 224.70 | <0.001* |
| Controls | 0.87 | 0.07 | 199.04 | 0.85 | 0.08 | 85.66 | 0.89 | 0.06 | 112.30 | ||||
| Biceps SFT (mm) | Cases | 19.21 | 8.74 | 410.28 | <0.001* | 22.75 | 8.42 | 185.39 | <0.001* | 16.29 | 7.91 | 233.67 | <0.001* |
| Controls | 8.89 | 4.94 | 194.58 | 11.62 | 4.69 | 83.34 | 6.73 | 3.99 | 103.33 | ||||
| Triceps SFT (mm) | Cases | 31.36 | 9.94 | 430.44 | <0.001* | 31 | 8.01 | 188.40 | <0.001* | 31.66 | 11.29 | 243.39 | <0.001* |
| Controls | 14.89 | 6.74 | 174.02 | 18.55 | 5.61 | 80.19 | 12.01 | 6.13 | 93.69 | ||||
| Sub-scapular SFT (mm) | Cases | 33.19 | 11.18 | 425.03 | <0.001* | 34.96 | 10.66 | 192.60 | <0.001* | 31.75 | 11.42 | 233.66 | <0.001* |
| Controls | 17.26 | 6.18 | 178.12 | 17.98 | 5.93 | 75.80 | 16.69 | 6.33 | 101.95 | ||||
| Abdomen SFT (mm) | Cases | 47.25 | 15.39 | 428.19 | <0.001* | 42.98 | 12.58 | 190.38 | <0.001* | 50.77 | 16.59 | 239.34 | <0.001* |
| Controls | 24.28 | 9.36 | 176.32 | 24.65 | 7.11 | 78.13 | 23.99 | 10.81 | 97.66 | ||||
| Thigh SFT (mm) | Cases | 47.11 | 16.72 | 431.91 | <0.001* | 44.34 | 12.33 | 189.86 | <0.001* | 49.38 | 19.36 | 243.89 | <0.001* |
| Controls | 20.42 | 9.09 | 172.52 | 27.01 | 5.63 | 78.67 | 15.24 | 7.87 | 93.11 | ||||
| Supra-iliac SFT (mm) | Cases | 38.65 | 14.83 | 422.04 | <0.001* | 30.07 | 8.96 | 185.87 | <0.001* | 45.69 | 15.01 | 242.48 | <0.001* |
| Controls | 19.13 | 8.07 | 182.59 | 17.48 | 6.76 | 82.84 | 20.43 | 8.77 | 94.52 | ||||
| % BF | Cases | 35.05 | 5.86 | 438.08 | <0.001* | 35.84 | 6.57 | 190.72 | <0.001* | 34.39 | 5.13 | 248.18 | <0.001* |
| Controls | 20.34 | 6.97 | 166.23 | 22.66 | 6.87 | 77.77 | 18.52 | 6.49 | 88.82 | ||||
| SBP (mmHg) | Cases | 119.90 | 14.93 | 340.85 | <0.001* | 117.83 | 16.90 | 162.58 | <0.001* | 121.60 | 12.89 | 178.43 | 0.052 |
| Controls | 113.40 | 13.46 | 265.40 | 106.39 | 11.15 | 107.19 | 118.90 | 12.55 | 158.57 | ||||
| DBP (mmHg) | Cases | 79.80 | 11.05 | 342.52 | <0.001* | 77.36 | 11.48 | 151.32 | <0.001* | 81.81 | 10.29 | 192.01 | <0.001* |
| Controls | 74.51 | 9.52 | 263.70 | 72.21 | 8.79 | 118.96 | 76.31 | 9.70 | 144.99 | ||||
| FBG (mg dl−1) | Cases | 105.10 | 24.50 | 329.17 | <0.001* | 107.50 | 22.15 | 147.63 | 0.009* | 103.14 | 26.18 | 182.44 | 0.009* |
| Controls | 99.51 | 12.56 | 277.32 | 101.16 | 10.76 | 122.82 | 98.22 | 13.69 | 154.56 | ||||
| Insulin μl U ml−1 | Cases | 27.01 | 14.49 | 366.18 | <0.001* | 25.59 | 13.31 | 149.28 | 0.003* | 28.17 | 15.33 | 217.29 | <0.001* |
| Controls | 18.83 | 12.21 | 239.57 | 20.44 | 9.89 | 121.09 | 17.57 | 13.67 | 119.71 | ||||
| HOMA-IR | Cases | 6.99 | 3.95 | 368.39 | <0.001* | 6.86 | 4.02 | 151.37 | 0.001* | 7.09 | 3.89 | 217.75 | <0.001* |
| Controls | 4.77 | 3.97 | 237.31 | 5.23 | 2.89 | 118.91 | 4.40 | 4.63 | 119.25 | ||||
3.4. Significantly deviant categorical variables in overweight and obese cases
Comparison of categorical variables between OW/OB cases and NW controls is summarized in Table 2. Deviant life-style and behavioural traits (except diet unconsciousness) such as random eating timings, tendency towards fat dense food (TFDF), inadequate sleep, irregular sleep-wake timings, shift-work, and low physical activity (PA) were significantly more prevalent in OW/OB cases as compared to NW controls. However, after gender stratification, a significantly higher percentage of OW/OB males showed diet unconsciousness as compared to NW males while the difference disappeared for inadequate sleep in males, and for random eating timings and shift-work in females. Similarly, physical signs associated with metabolic disturbances such as acanthosis nigricans, axillary and abdominal striae, and hyperlipidaemia signs were significantly more frequent in OW/OB cases as compared to NW controls. However, the difference for signs of hyperlipidaemia vanished in males after gender stratification. In addition, co-morbidities associated with weight gain including hypertension, joint problems, depression and irregular menstrual cycle (in case of females) were significantly more common in OW/OB cases as compared to their NW controls. Furthermore, a significantly higher percentage of OW/OB cases had family history of obesity (FHO) as compared to their NW controls but this difference departed in both males and females after gender stratification. An interesting finding is that a significantly higher percentage of OW/OB subjects was found married as compared to normal weight subjects.| Categorical variables | Response | Cases | Controls | p-Value | Male cases | Male controls | p-Value | Female cases | Female controls | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| a Categorical variables are represented as counts (percentages in parenthesis) and compared by chi-square test between cases and controls. *A p-value < 0.05 was considered significant.b Consanguinity was taken as history of marriage with a first cousin.c Family history of obesity (FHO) included information about obesity in first-degree relatives (parents and full siblings).d Regular sleep/wake cycle was taken in terms of sleep timings between 8:00–12:00 pm and wake timings between 5:00–7.30 am. Irregular sleep/wake timings were taken as sleep and wake timings outside the range of regular sleep and wake up timings. Abbreviations; FHO: family history of obesity, TFDF: tendency towards fat dense food. | ||||||||||
| Marital status | Single | 175 (57.2%) | 218 (72.7%) | <0.001* | 102 (60.71%) | 120 (71.42%) | 0.038* | 73 (52.89%) | 98 (74.24%) | <0.001* |
| Married | 131 (42.8%) | 82 (27.3%) | 66 (39.28%) | 48 (28.57%) | 65 (47.10%) | 34 (25.75%) | ||||
| Parental consanguinityb | Yes | 74 (24.2%) | 67 (22.3%) | 0.590 | 45 (26.78%) | 38 (22.61%) | 0.376 | 29 (21%) | 29 (21.96%) | 0.848 |
| No | 232 (75.8%) | 233 (77.7%) | 123 (73.2%) | 130 (77.4%) | 109 (79%) | 103 (78%) | ||||
| FHOc | Yes | 158 (51.6%) | 129 (43%) | 0.033* | 84 (50%) | 72 (42.85%) | 0.189 | 74 (53.6%) | 57 (43.2%) | 0.086 |
| No | 148 (48.4%) | 171 (57%) | 84 (50%) | 96 (57.1%) | 64 (46.4%) | 75 (56.8%) | ||||
| Signs of hyperlipidaemia | Yes | 25 (8.2%) | 6 (2%) | 0.001* | 12 (7.14%) | 5 (2.97%) | 0.081 | 13 (9.4%) | 1 (0.8%) | 0.001* |
| No | 281 (91.8%) | 294 (98%) | 156 (92.9%) | 163 (97%) | 125 (90.6%) | 131 (99.2%) | ||||
| Acanthosis nigricans | Yes | 147 (48%) | 24 (8%) | <0.001* | 86 (51.19%) | 23 (13.69%) | <0.001* | 61 (44.2%) | 1 (0.8%) | <0.001* |
| No | 159 (52%) | 276 (92%) | 82 (48.8%) | 145 (86.3%) | 77 (55.8%) | 131 (99.2%) | ||||
| Axillary striae | Yes | 167 (54.6%) | 24 (8%) | <0.001* | 90 (53.57%) | 13 (7.74%) | <0.001* | 77 (55.79%) | 11 (8.33%) | <0.001* |
| No | 139 (45.4%) | 276 (92%) | 78 (46.4%) | 155 (92.3%) | 61 (44.2%) | 121 (91.7%) | ||||
| Abdominal striae | Yes | 161 (52.6%) | 22 (7.3%) | <0.001* | 82 (48.81%) | 10 (5.95%) | <0.001* | 79 (86.8%) | 12 (13.2%) | <0.001* |
| No | 145 (47.4%) | 278 (92.7%) | 86 (51.2%) | 158 (94%) | 59 (42.8%) | 120 (90.9%) | ||||
| Eating timings | Random | 200 (65.4%) | 159 (53%) | 0.002* | 111 (66.1%) | 86 (51.2%) | 0.006* | 89 (64.5%) | 73 (55.3%) | 0.123 |
| Specific | 106 (34.6%) | 141 (47%) | 57 (33.9%) | 82 (48.8%) | 49 (35.5%) | 59 (44.7%) | ||||
| Diet consciousness | Yes | 80 (26.1%) | 96 (32%) | 0.112 | 34 (20.23%) | 53 (31.54%) | 0.018* | 46 (33.3%) | 43 (32.6%) | 0.895 |
| No | 226 (73.9%) | 204 (68%) | 134 (79.8%) | 115 (68.5%) | 92 (66.7%) | 89 (67.4%) | ||||
| TFDF | High | 113 (36.9%) | 58 (19.3%) | <0.001* | 68 (40.47%) | 38 (22.61%) | 0.002* | 45 (32.6%) | 20 (15.2%) | 0.003* |
| Moderate | 91 (29.7%) | 122 (40.7%) | 37 (22%) | 53 (31.5%) | 54 (39.1%) | 69 (52.3%) | ||||
| Low | 102 (33.3%) | 120 (40%) | 63 (37.5%) | 77 (45.83%) | 39 (28.3%) | 43 (32.6%) | ||||
| Inadequate sleep (<7 hours) | Yes | 188 (61.4%) | 158 (52.7%) | 0.029* | 104 (61.9%) | 98 (58.3%) | 0.504 | 84 (60.9%) | 60 (45.4%) | 0.011* |
| No | 118 (38.6%) | 142 (47.3%) | 64 (38.1%) | 70 (41.7%) | 54 (39.1%) | 72 (54.5%) | ||||
| Sleep-wake timingsd | Irregular | 179 (58.5%) | 129 (43%) | <0.001* | 103 (61.3%) | 82 (48.8%) | 0.021* | 76 (55.1%) | 47 (35.6%) | 0.001* |
| Regular | 127 (41.5%) | 171 (57%) | 65 (38.7%) | 86 (51.2%) | 62 (44.92%) | 85 (64.4%) | ||||
| Shift workers | Yes | 38 (12.4%) | 18 (6%) | 0.006* | 33 (19.64%) | 15 (8.92%) | 0.005* | 5 (3.6%) | 3 (2.3%) | 0.513 |
| No | 268 (87.6%) | 282 (94%) | 135 (80.4%) | 153 (91.1%) | 133 (96.4%) | 129 (97.7%) | ||||
| Physical activity | High | 56 (18.3%) | 91 (30.3%) | <0.001* | 31 (18.452%) | 57 (33.92%) | <0.001* | 25 (18.1%) | 34 (25.8%) | 0.038* |
| Moderate | 110 (35.9%) | 125 (41.7%) | 46 (27.38%) | 56 (33.33%) | 64 (46.37%) | 69 (52.27%) | ||||
| Low | 140 (45.8%) | 84 (28%) | 91 (54.16%) | 55 (32.73%) | 49 (35.5%) | 29 (21.96%) | ||||
| Hypertension | Yes | 111 (36.3%) | 65 (21.7%) | <0.001* | 72 (42.9%) | 51 (30.4%) | 0.017* | 39 (28.3%) | 14 (10.6%) | <0.001* |
| No | 195 (63.7%) | 235 (78.3%) | 96 (57.1) | 117 (69.6%) | 99 (71.7%) | 118 (89.4%) | ||||
| Joint problems | Yes | 114 (37.3%) | 37 (12.3%) | <0.001* | 45 (26.78%) | 9 (5.36%) | <0.001* | 69 (50%) | 28 (21.2%) | <0.001* |
| No | 192 (62.7%) | 263 (87.7%) | 123 (73.2%) | 159 (94.6%) | 69 (50%) | 104 (78.8%) | ||||
| Depression | Yes | 114 (37.3%) | 38 (12.7%) | <0.001* | 42 (25%) | 13 (7.7%) | <0.001* | 72 (52.2%) | 25 (18.9%) | <0.001* |
| No | 192 (62.7%) | 262 (87.3%) | 126 (75%) | 155 (92.3%) | 66 (47.8%) | 107 (81.1%) | ||||
| Menstrual cycle | Regular | — | — | 93 (67.39%) | 119 (90.2%) | <0.001* | ||||
| Irregular | 40 (28.98%) | 13 (9.8%) | ||||||||
3.5. Association between stigma and obesity
Among total population of overweight and obese cases (n = 306), 44.77% reported that they were stigmatized for being overweight or obese (Table 2). However, after gender stratification of OW/OB subjects, significantly higher percentage (63.80%) of females were found to be stigmatized for being overweight and obese (χ2 test, p < 0.001) as compared to males (29.20%).3.6. Genotypic and allelic frequencies of MC4R variant rs17782313
The observed genotypic frequencies for the MC4R rs17782313 genotypes (TT, TC, and CC) were in Hardy–Weinberg equilibrium for both NW and OW/OB participants (χ2-test; p = 0.665 and 0.316, respectively). The observed minor allele frequency (MAF) of the MC4R risk ‘C’ allele was 42.1% (510/1212) in total study population. Similarly, the frequency of TT wild-type, TC heterozygote and CC homozygote carriers was 34% (206/606), 47.8% (290/606) and 18.2% (110/606), respectively. The comparison of genotypic and allelic frequencies of MC4R rs17782313 variant between OW/OB and NW subjects is mentioned in Table 3. MAF for the risk allele ‘C’ was 43.8% (268/612) and 40.3% (242/600) for OW/OB and NW subjects, respectively. No significant difference (p > 0.05) was observed in the distribution of genotypic and allelic frequencies of MC4R rs1778313 variant between OW/OB and NW subjects. However, after gender stratification of the total study population, significantly higher genotypic and allelic frequencies of MC4R rs1778313 variant were found in OW/OB females as compared to NW females (p < 0.05). When females were further stratified according to age in to >20 and ≤20 years' groups, this difference remained significant (p < 0.05) only in adult females (>20 years). However, it must be noted that the sample size reduced considerably in ≤20 years' females group. On the other hand, no significant difference was seen in genotypic and allelic frequencies between overall and age-stratified OW/OB males as compared to NW males (p > 0.05). Furthermore, no difference was found between OW/OB and NW subjects when study population was stratified on the basis of age alone (p > 0.05). Thus, gender and age related differences in genotypic and allelic frequencies of MC4R rs17782313 variant were found in our study population with higher genotypic and allelic frequencies of MC4R rs17782313 variant in OW/OB adult (>20 years) females as compared to their controls.| N | Co-dominant model | Dominant model | Recessive model | Alleles | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | TT | TC + CC | TT + TC | CC | T | C | ||
| a Genotypic and allelic frequencies are represented as counts (percentages in parentheses) and compared by chi-square test between cases and controls. Odd ratios (OR) calculated at 95% confidence interval (CI) to estimate overweight and obesity risk. *A p-value < 0.05 considered significant. Abbreviations; N: number of subjects, T: major allele, C: minor allele, TT: reference genotype. | ||||||||||
| Total subjects (N = 606) | ||||||||||
| Cases | (N = 306) | 101 (33%) | 142 (46.4%) | 63 (20.6%) | 101 (33%) | 205 (67%) | 243 (79.4%) | 63 (20.6%) | 344 (56.2%) | 268 (43.8%) |
| Controls | (N = 300) | 105 (35%) | 148 (49.3%) | 47 (15.7%) | 105 (35%) | 195 (65%) | 253 (84.3%) | 47 (15.7%) | 358 (59.7%) | 242 (40.3%) |
| OR (95% CI) | 0.997 (0.698–1.43) | 1.39 (0.874–2.22) | 1.09 (0.78–1.53) | 1.39 (0.92–2.12) | 1.15 (0.92–1.45) | |||||
| p-Value | — | 0.989 | 0.163 | 0.604 | 0.116 | 0.223 | ||||
| Female cases | (N = 138) | 38 (27.5%) | 68 (49.3%) | 32 (23.2%) | 38 (27.5%) | 100 (72.5%) | 106 (76.8%) | 32 (23.2%) | 144 (52.2%) | 132 (47.8%) |
| Female controls | (N = 132) | 52 (39.4%) | 62 (47%) | 18 (13.6%) | 52 (39.4%) | 80 (60.6%) | 114 (86.4%) | 18 (13.6%) | 166 (62.8%) | 98 (37.2%) |
| OR (95% CI) | 1.5 (0.873–2.58) | 2.43 (1.19–4.96) | 1.71 (1.03–2.85) | 1.91 (1.01–3.61) | 1.55 (1.1–2.18) | |||||
| p-Value | — | 0.142 | 0.015* | 0.039* | 0.043* | 0.01* | ||||
| Male cases | (N = 168) | 63 (37.5%) | 74 (44%) | 31 (18.5%) | 63 (37.5%) | 105 (62.5%) | 137 (81.5%) | 31 (18.5%) | 200 (59.5%) | 136 (40.5%) |
| Male controls | (N = 168) | 53 (31.5%) | 86 (51.2%) | 29 (17.3%) | 53 (31.5%) | 115 (68.5%) | 139 (82.7%) | 29 (17.3%) | 192 (57.1%) | 144 (42.9%) |
| OR (95% CI) | 0.72 (0.45–1.17) | 0.89 (0.48–1.68) | 0.77 (0.49–1.21) | 1.08 (0.62–1.89) | 0.91 (0.67–1.23) | |||||
| p-Value | — | 0.187 | 0.739 | 0.251 | 0.776 | 0.531 | ||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||||
| ≤20 years (N = 61) | ||||||||||
| ≤20 years cases | (N = 28) | 10 (35.7%) | 14 (50%) | 4 (14.3%) | 10 (35.7%) | 18 (64.3%) | 24 (85.7%) | 4 (14.3%) | 34 (60.7%) | 22 (39.3%) |
| ≤20 years controls | (N = 33) | 11 (33.3%) | 19 (57.6%) | 3 (9.1%) | 11 (33.3%) | 22 (66.7%) | 30 (90.9%) | 3 (9.1%) | 41 (62.1%) | 25 (37.9%) |
| OR (95% CI) | 0.811 (0.27–2.43) | 1.47 (0.26–8.22) | 0.9 (0.31–2.59) | 1.67 (0.34–8.17) | 1.06 (0.51–2.2) | |||||
| p-Value | — | 0.708 | 0.663 | 0.845 | 0.526 | 0.874 | ||||
| ≤20 years female cases | (N = 13) | 6 (46.2%) | 5 (38.5%) | 2 (15.4%) | 6 (46.2%) | 7 (53.8%) | 11 (84.6%) | 2(15.4%) | 17 (65.4%) | 9 (34.6%) |
| ≤20 years female controls | (N = 11) | 4 (36.4%) | 6 (54.5%) | 1 (9.1%) | 4 (36.4%) | 7 (63.6%) | 10 (90.9%) | 1 (9.1%) | 14 (63.6%) | 8 (36.4%) |
| OR (95% CI) | 0.55 (0.09–3.15) | 1.33 (0.08–20.11) | 0.67 (0.13–3.45) | 1.82 (0.142–23.25) | 0.93 (0.28–3.03) | |||||
| p-Value | — | 0.507 | 0.835 | 0.628 | 0.642 | 0.899 | ||||
| ≤20 years male cases | (N = 15) | 4 (26.7%) | 9 (60%) | 2 (13.3%) | 4 (26.7%) | 11 (73.3%) | 13 (86.7%) | 2 (13.3%) | 17 (56.7%) | 13 (43.3%) |
| ≤20 years male controls | (N = 22) | 7 (31.8%) | 13 (59.1%) | 2 (9.1%) | 7 (31.8%) | 15 (68.2%) | 20 (90.9%) | 2 (9.1%) | 27 (61.4%) | 17 (38.6%) |
| OR (95% CI) | 1.2 (0.27–5.39) | 1.75 (0.17–17.68) | 1.28 (0.30–5.49) | 1.54 (0.19–12.32) | 1.21 (0.47–3.12) | |||||
| p-Value | — | 0.80 | 0.635 | 0.736 | 0.683 | 0.686 | ||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||||
| >20 years (545) | ||||||||||
| >20 years cases | (N = 278) | 91 (32.7%) | 128 (46%) | 59 (21.2%) | 91 (32.7%) | 187 (67.3%) | 219 (78.8%) | 59 (21.2%) | 310 (55.7%) | 246 (44.3%) |
| >20 years controls | (N = 267) | 94 (35.2%) | 129 (48.3%) | 44 (16.5%) | 94 (35.2%) | 173 (64.8%) | 223 (83.5%) | 43 (16.5%) | 317 (59.4%) | 217 (40.6%) |
| OR (95% CI) | 1.025 (0.702–1.49) | 1.39 (0.85–2.25) | 1.12 (0.78–1.59) | 1.37 (0.89–2.1) | 1.16 (0.91–1.47) | |||||
| p-Value | — | 0.898 | 0.188 | 0.542 | 0.157 | 0.228 | ||||
| >20 years female cases | (N = 125) | 32 (25.6%) | 63 (50.4%) | 30 (24%) | 32 (25.6%) | 93 (74.4%) | 95 (76%) | 30 (24%) | 127 (50.8%) | 123 (49.2%) |
| >20 years female controls | (N = 121) | 48 (39.7%) | 56 (46.3%) | 17 (14%) | 48 (39.7%) | 73 (60.3%) | 104 (86%) | 17 (14%) | 152 (62.8%) | 90 (37.2%) |
| OR (95% CI) | 1.68 (0.95–2.99) | 2.65 (1.26–5.57) | 1.91 (1.11–3.29) | 1.93 (1–3.73) | 1.64 (1.14–2.34) | |||||
| p-Value | — | 0.074 | 0.01* | 0.019* | 0.047* | 0.007* | ||||
| >20 years male cases | (N = 153) | 59 (38.6%) | 65 (42.5%) | 29 (19%) | 59 (38.6%) | 94 (61.4%) | 124 (81%) | 29 (19%) | 183 (59.8%) | 123 (40.2%) |
| >20 years male controls | (N = 146) | 46 (31.5%) | 73 (50%) | 27 (18.5%) | 46 (31.5%) | 100 (68.5%) | 119 (81.5%) | 27 (18.5%) | 168 (56.9%) | 127 (43.1%) |
| OR (95% CI) | 0.69 (0.42–1.16) | 0.84 (0.44–1.61) | 0.73 (0.45–1.18) | 1.03 (0.57–1.84) | 0.89 (0.64–1.23) | |||||
| p-Value | — | 0.161 | 0.593 | 0.201 | 0.919 | 0.477 | ||||
3.7. Association of MC4R variant rs17782313 with obesity
Assuming co-dominant or general genetic model, only CC females (minor allele homozygotes) showed significant association with risk of overweight and obesity. Thus, CC genotype increases the risk of obesity by 2.43 times (95% CI: 1.19–4.96, p = 0.015) while the minor allele C increased the risk of overweight and obesity by 1.55 times (95% CI: 1.1–2.18, p = 0.01) in females. However, when females were further stratified according to age in to >20 and ≤20 years' groups, this difference remained significant only in adult females (>20 years) (Table 3).3.8. Association of MC4R variant rs17782313 with obesity-related continuous variables
Significant differences (p < 0.05) in anthropometric parameters namely weight, BMI, WC, HC, WHR, skin-fold thicknesses (biceps, subscapular, thigh) and % BF were observed across carriers of TT, TC and CC genotypes in females after gender stratification (Table 4). Subsequent multiple pair-wise comparisons (Table 5) revealed that female carriers of CT and CC showed significantly higher (p < 0.05) weight, BMI, WC, HC, and thigh SFT as compared to female carriers of TT genotype. Similarly, carriers of CC genotype showed significantly greater sub-scapular SFT as compared to female carriers of TT genotype while female carriers of CT genotype showed significantly greater WHR, biceps SFT, and % BF as compared to female carriers of TT genotype. Furthermore, the same associations of all the above-mentioned obesity-related anthropometric parameters across genotypes remained significant only in >20 years' females after age-based stratification of female population.| MC4R rs17782313 genotypes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TT (N = 90) | CT (N = 130) | CC (N = 50) | |||||||||
| Mean | SD | Mean ranks | Mean | SD | Mean ranks | Mean | SD | Mean ranks | Chi-square | p-Value | |
| a Continuous variables are taken as mean rank and compared for differences across MC4R rs17782313 genotypes by Kruskal Wallis test. *A p-value < 0.05 was considered significant. Abbreviation; BMI: body mass index, WC: waist circumference, HC: hip circumference; WHR: waist-to-hip ratio, SFT: skin-fold thickness, % BF: percentage body fat, SBP: systolic blood pressure, DBP: diastolic blood pressure, FBG: fasting blood glucose, HOMA-IR: homeostatic model assessment for insulin resistance, TT: wild-type genotype. | |||||||||||
| Weight (kg) | 63.52 | 15.63 | 114.36 | 71.54 | 21.12 | 142.82 | 73.29 | 21.58 | 154.52 | 10.71 | 0.005* |
| Height (cm) | 157.19 | 5.59 | 132.90 | 157.92 | 5.22 | 141.15 | 156.75 | 6.24 | 125.48 | 1.61 | 0.448 |
| BMI (kg m−2) | 25.76 | 6.42 | 114.10 | 28.65 | 8.20 | 141.71 | 29.80 | 8.15 | 157.87 | 11.69 | 0.003* |
| WC (cm) | 91.44 | 17.07 | 112.93 | 99.71 | 20.39 | 144.92 | 101.74 | 21.16 | 151.63 | 11.55 | 0.003* |
| HC (cm) | 101.94 | 11.56 | 113.89 | 107.05 | 14.07 | 142.78 | 109.30 | 14.43 | 155.45 | 11.29 | 0.004* |
| WHR | 0.89 | 0.092 | 115.64 | 0.93 | 0.095 | 145.33 | 0.93 | 0.1 | 145.69 | 8.73 | 0.013* |
| Bicep SFT (mm) | 15.56 | 8.44 | 118.89 | 18.31 | 8.92 | 144.96 | 17.86 | 8.92 | 140.81 | 6.22 | 0.045* |
| Triceps SFT (mm) | 23.21 | 8.59 | 122.27 | 25.38 | 9.90 | 137.89 | 26.79 | 8.68 | 153.10 | 5.25 | 0.072 |
| Subscapular SFT (mm) | 24.14 | 11.62 | 119.05 | 27.35 | 12.46 | 139.65 | 29.41 | 11.57 | 154.31 | 7.27 | 0.026* |
| Abdominal SFT (mm) | 31.73 | 12.97 | 123.08 | 34.75 | 14.07 | 139.01 | 36.24 | 14.07 | 148.74 | 3.98 | 0.137 |
| Thigh SFT (mm) | 33.44 | 14.22 | 115.04 | 37.25 | 12.85 | 144.72 | 36.63 | 10.24 | 148.36 | 9.35 | 0.009* |
| Supra-iliac SFT (mm) | 22.21 | 10.01 | 121.76 | 24.25 | 9.89 | 139.27 | 26.12 | 10.69 | 150.44 | 4.93 | 0.085 |
| % BF | 27.14 | 9.84 | 117.78 | 30.51 | 9.13 | 143.79 | 30.57 | 8.76 | 145.84 | 6.97 | 0.031* |
| SBP (mmHg) | 111.77 | 15.55 | 134.70 | 111.81 | 15.87 | 131.62 | 114.20 | 14.28 | 147.02 | 1.49 | 0.475 |
| DBP (mmHg) | 73.91 | 10.55 | 130.12 | 74.81 | 10.61 | 132.62 | 76.60 | 10.42 | 152.67 | 3.31 | 0.191 |
| FBG (mg dL−1) | 101.63 | 13.85 | 124.58 | 106.25 | 21.72 | 139.11 | 104.56 | 11.09 | 145.76 | 2.90 | 0.234 |
| Insulin (μIU ml−1) | 23.01 | 12.85 | 132.73 | 106.25 | 21.71 | 136.13 | 104.56 | 11.09 | 138.83 | 0.212 | 0.899 |
| HOMA IR | 5.92 | 3.71 | 130.34 | 6.03 | 3.49 | 137.55 | 6.39 | 3.73 | 139.46 | 0.611 | 0.737 |
| Genotypes | Test statistic | Std. error | Std. test statistic | p-Value | Adjusted p-value | |
|---|---|---|---|---|---|---|
| a Multiple pair-wise comparisons of MC4R rs17782313 genotypes were made by Dunn-Bonferroni post-hoc analysis for continuous variables which were found significantly different across MC4R rs17782313 genotypes by Kruskal Wallis test. *A p-value < 0.05 was considered significant. | ||||||
| Weight (kg) | CT vs. TT | 28.46 | 10.71 | 2.66 | 0.008 | 0.024* |
| CC vs. TT | 40.16 | 13.78 | 2.92 | 0.004 | 0.011* | |
| BMI (kg m−2) | CT vs. TT | 27.61 | 10.71 | 2.58 | 0.010 | 0.030* |
| CC vs. TT | 43.77 | 13.77 | 3.18 | 0.001 | 0.004* | |
| WC (cm) | CT vs. TT | 31.99 | 10.71 | 2.99 | 0.003 | 0.008* |
| CC vs. TT | 38.69 | 13.77 | 2.81 | 0.005 | 0.015* | |
| HC (cm) | CT vs. TT | 28.89 | 10.70 | 2.69 | 0.007 | 0.021* |
| CC vs. TT | 41.56 | 13.77 | 3.02 | 0.003 | 0.008* | |
| WHR | CT vs. TT | 29.68 | 10.71 | 2.77 | 0.006 | 0.017* |
| CC vs. TT | 30.05 | 13.77 | 2.18 | 0.029 | 0.087 | |
| Biceps SFT (mm) | CT vs. TT | 26.07 | 10.69 | 2.44 | 0.015 | 0.044* |
| CC vs. TT | 21.92 | 13.76 | 1.59 | 0.111 | 0.333 | |
| Thigh SFT (mm) | CT vs. TT | 29.67 | 10.70 | 2.77 | 0.006 | 0.017* |
| CC vs. TT | 33.32 | 13.77 | 2.42 | 0.016 | 0.047* | |
| Sub-scapular SFT (mm) | CT vs. TT | 20.60 | 10.70 | 1.93 | 0.054 | 0.163 |
| CC vs. TT | 35.26 | 13.77 | 2.56 | 0.010 | 0.031* | |
| % BF | CT vs. TT | 26.01 | 10.71 | 2.43 | 0.015 | 0.045* |
| CC vs. TT | 28.06 | 13.77 | 2.04 | 0.042 | 0.125 | |
3.9. No association of MC4R variant rs17782313 with obesity-related categorical variables except eating timings
No significant association (p > 0.05) of categorical variables including parental consanguinity, family history of obesity (FHO), diet consciousness, eating timings, tendency towards fat dense food (TFDF), physical activity, depression, sleep-wake timings, lack of sleep, and joint problems was observed across carriers of TT, TC and CC genotypes in overall and gender-stratified population (Table 6). The results remain same even after subsequent pair-wise analysis by logistic regression except for eating timings as CT genotype was found to be associated with random eating timings (p = 0.025) with and without gender adjustment in overall study population (data not shown).| Parameter | N | Total genotypes | Females genotypes | Males genotypes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT (N = 206) | CT (N = 290) | CC (N = 110) | p-Value | TT (N = 90) | CT (N = 130) | CC (N = 50) | p-Value | TT (N = 206) | CT (N = 290) | CC (N = 110) | p-Value | |||
| a Categorical variables are represented as counts (percentages in parenthesis) and compared for differences across genotypes between cases and controls using chi-square test. A p < 0.05 was considered significant. Abbreviations; FHO: family history of obesity, TFDF: tendency towards fat dense food. | ||||||||||||||
| Parental consanguinity | Yes | 141 | 53 (37.6%) | 58 (41.1%) | 30 (21.3%) | 0.181 | 23 (39.7%) | 23 (39.7%) | 12 (20.7%) | 0.336 | 30 (36.1%) | 35 (42.2%) | 18 (21.7%) | 0.432 |
| No | 465 | 153 (32.9%) | 232 (49.9%) | 80 (17.2%) | 67 (31.6%) | 107 (50.5%) | 38 (17.9%) | 86 (34%) | 125 (49.4%) | 42 (16.6%) | ||||
| FHO | Yes | 287 | 93 (32.4%) | 148 (51.6%) | 46 (16%) | 0.189 | 43 (32.8%) | 69 (52.7%) | 19 (14.5%) | 0.191 | 50 (32.1%) | 79 (50.6%) | 27 (17.3%) | 0.570 |
| No | 319 | 113 (35.4%) | 142 (44.5%) | 64 (20.1%) | 47 (33.8%) | 61 (43.9%) | 31 (22.3%) | 66 (36.7%) | 81 (45%) | 33 (18.3%) | ||||
| Diet consciousness | Yes | 176 | 59 (33.5%) | 79 (44.9%) | 38 (21.6%) | 0.352 | 27 (30.3%) | 44 (49.4%) | 18 (20.2%) | 0.736 | 32 (36.8%) | 35 (40.2%) | 20 (23%) | 0.197 |
| No | 430 | 147 (34.2%) | 211 (49.1%) | 72 (16.7%) | 63 (34.8%) | 86 (47.5%) | 32 (17.7%) | 84 (33.7%) | 125 (50.2%) | 40 (16.1%) | ||||
| Eating timings | Random | 359 | 110 (30.6%) | 184 (51.3%) | 65 (18.1%) | 0.080 | 48 (29.6%) | 84 (51.9%) | 30 (18.5%) | 0.244 | 62 (31.5%) | 100 (50.8%) | 35 (17.8%) | 0.321 |
| Specific | 247 | 96 (38.9%) | 106 (42.9%) | 45 (18.2%) | 42 (38.9%) | 46 (42.6%) | 20 (18.5%) | 54 (38.8%) | 60 (43.2%) | 25 (18%) | ||||
| TFDF | High | 171 | 54 (31.6%) | 88 (51.5%) | 29 (17%) | 0.843 | 16 (24.6%) | 35 (53.8%) | 14 (21.5%) | 0.469 | 38 (35.8%) | 53 (50%) | 15 (14.2%) | 0.776 |
| Moderate | 213 | 73 (34.3%) | 101 (47.4%) | 39 (18.3%) | 42 (34.1%) | 58 (47.2%) | 23 (18.7%) | 31 (34.4%) | 43 (47.8%) | 16 (17.8%) | ||||
| Low | 222 | 79 (35.6%) | 101 (45.6%) | 42 (18.9%) | 32 (39%) | 37 (45.1%) | 13 (15.9%) | 47 (33.6%) | 64 (45.7%) | 29 (20.7%) | ||||
| Physical activity | High | 147 | 45 (30.6%) | 70 (47.6%) | 32 (21.8%) | 0.296 | 18 (30.5%) | 30 (50.8%) | 11 (18.6%) | 0.437 | 27 (30.7%) | 40 (45.5%) | 21 (23.9%) | 0.348 |
| Moderate | 235 | 91 (38.7%) | 106 (45.1%) | 38 (16.2%) | 51 (38.3%) | 57 (42.9%) | 25 (18.8%) | 40 (39.2%) | 49 (48%) | 13 (12.7%) | ||||
| Low | 224 | 70 (31.2%) | 114 (50.9%) | 40 (17.9%) | 21 (26.9%) | 43 (55.1%) | 14 (17.9%) | 49 (33.6%) | 71 (48.6%) | 26 (17.8%) | ||||
| Depression | Yes | 152 | 50 (32.9%) | 69 (45.4%) | 33 (21.7%) | 0.418 | 28 (28.9%) | 50 (51.5%) | 19 (19.6%) | 0.506 | 22 (40%) | 19 (34.5%) | 14 (25.5%) | 0.080 |
| No | 454 | 156 (34.4%) | 221 (48.7%) | 77 (17%) | 62 (35.8%) | 80 (46.2%) | 31 (17.9%) | 94 (33.5%) | 141 (50.2%) | 46 (16.4%) | ||||
| Sleep-wake cycle | Irregular | 308 | 147 (47.7%) | 53 (17.2%) | 108 (35.1%) | 0.770 | 62 (50.4%) | 22 (17.9%) | 39 (31.7%) | 0.791 | 85 (45.9%) | 31 (16.7%) | 69 (37.3%) | 0.487 |
| Regular | 298 | 143 (48%) | 57 (19.1%) | 98 (32.9%) | 68 (46.3%) | 28 (19%) | 51 (34.7%) | 75 (49.7%) | 29 (19.2%) | 47 (31.1%) | ||||
| Lack of sleep | Yes | 346 | 115 (33.2%) | 168 (48.6%) | 63 (18.2%) | 0.896 | 44 (30.6%) | 74 (51.4%) | 26 (18.1%) | 0.491 | 71 (35.1%) | 94 (46.5%) | 37 (18.3%) | 0.886 |
| No | 260 | 91 (35%) | 122 (46.9%) | 47 (18.1%) | 46 (36.5%) | 56 (44.4%) | 24 (19%) | 45 (33.6%) | 66 (49.3%) | 23 (17.2%) | ||||
| Joint problems | Yes | 151 | 51 (33.8%) | 73 (48.3%) | 27 (17.9%) | 0.990 | 28 (28.9%) | 48 (49.5%) | 21 (21.6%) | 0.414 | 23 (42.6%) | 25 (46.3%) | 6 (11.1%) | 0.237 |
| No | 455 | 155 (34.1%) | 217 (47.7%) | 83 (18.2%) | 62 (35.8%) | 82 (47.4%) | 29 (16.8%) | 93 (33%) | 135 (47.9%) | 54 (19.1%) | ||||
4. Discussion
This is the first study that investigated the association of MC4R variant rs17782313 with obesity and various obesity-linked anthropometric, metabolic, physical, and behavioural traits in Pakistani subjects. The inclusion of such a considerable number of obesity-related traits at a time has never been done before. In particular, inclusion of categorical variables such as sleep duration, sleep-wake timings, shift work, joint problems, menstrual cycle, and eating timings are unique to our study. Obesity is an overtly evident yet most neglected public health problem that intimidates to beset both developed and developing countries. It is a medical disorder that may lead to a number of co-morbidities. The clinically identifiable risk factors for cardiovascular disease (CVD) and metabolic dysregulation are more rampant among OW/OB than among those of healthy weight subjects.29,30 Likewise, in our study, we also observed a significant association of risk factor variables for cardiovascular (increased weight, BMI, WC, HC, WHR, subcutaneous fat deposit, % BF, SBP, and DBP) and metabolic disorders (augmented FBG, fasting insulin, HOMA-IR, and also signs of hyperlipidaemia, acanthosis nigicans, axillary and abdominal striae) with overweight and obese phenotype. Similarly, obesity has been linked with the development or progression of a wide range of disabling musculoskeletal disorders including joint problems in adults accountable for impaired quality of life.31 Substantial evidence also exists for obesity giving rise to increased depression and less consistent data subsists for depression leading to obesity.32 Moreover, studies have provided evidence for the association between obesity and menstrual cycle irregularity in young adult women.33,34 In agreement, our study demonstrated a significantly higher percentage of OW/OB individuals experiencing joint problems, depression, and menstrual cycle irregularities (females) as compared to their corresponding normal weight controls. OW/OB subjects, especially women, are prone to pronounced stigmatization and discrimination because of their weight.35,36 In accord, a considerable percentage of OW/OB persons in our study reported about experiencing weight stigma with more OW/OB females were found to be stigmatized as compared to OW/OB males. The recent data in human subjects suggest that the timing of food consumption can substantially affect weight regulation with specific mealtimes have implications in preventing weight gain.37 Similarly, food health consciousness has been reported as one of the most important predictors of restrained eating38 and individuals with a lower ECI (Eating Choices Index that differentiates between healthy and unhealthy eating choices) score are more likely to be overweight or obese.39 Dietary fat persuades overconsumption and weight gain through its high caloric density and low satiety properties. OW/OB individuals demonstrate a tendency towards liking and selection of energy-dense foods that may contribute to expression and continuation of these conditions.40 In addition, physical activity plays an important role in the prevention of becoming OW/OB in childhood and adolescence, and reducing the risk of obesity in adulthood.41 In our study, along the same lines, a significantly higher number of OW/OB subjects reported random mealtimes, high TFDF, and low physical activity as compared to NW subjects whereas a significantly higher number of OW/OB males reported of not being diet conscious as compared to NW males. Research evidence largely suggests that inadequate sleep increases the risk of becoming obese by increasing the time available for eating and it does not permit the renewal of a hormonal profile facilitating appetite control.42,43 Moreover, irregular sleep-wake patterns including variability in nocturnal sleep duration and daytime napping have also been associated with obesity independent of mean sleep duration in both men and women.44,45 Recently, studies have also demonstrated that overweight and obesity are more common in shift-workers than day workers possibly as an outcome of physiological maladaptation to chronically sleeping and eating at abnormal circadian times.46,47 In the similar manner, our study also demonstrates significant association of inadequate sleep, irregular sleep-wake timings, and shift-work with obesity. Genetic studies have provided evidence that overweight and obesity can run in families and obesity risk is two to three times higher for a person with a family history as opposed to a person with no family history of obesity.48 Similarly, parental consanguinity can be considered as one of the possible risk factors for the occurrence of obesity.49 Further, married men and women are generally more likely to be OW/OB than never married persons.50,51 Likewise, a significant association of FHO and marital status (being married) with overweight and obesity has been observed in our study. The possible reason in our society for association between marital status and obesity may be that people do care about their physical fitness for marriage purposes but once they get married they do not care about their fitness and health anymore. However, no significant association between parental consanguinity and obese phenotype has been found in our study that conforms to the study by Saraswathi et al. (2012) in South India.52The MAF of MC4R rs17782313 observed in our study (42%) is substantially higher as compared to those seen in American,53,54 European,55–57 East Asian,23,58 and even Indian populations59,60 but comparable to that of Irani population.61 These disparities in MAF from different studies might be attributable to different regional, racial, and ethnic backgrounds. In context of the current study, the gender specific association of MC4R rs17782313 with overweight and obesity, and obesity-related anthropometric traits (weight, BMI, WC, HC, WHR, thigh SFT, sub-scapular SFT, biceps SFT, and % BF) observed in females of our population may partly explain widening gender gap in excess weight with 10% more women gaining weight than men in Pakistan.25 Sedentary life-style of most females and all times access to food while residing at home in our society can serve to facilitate the expression of this variant particularly in females. Moreover, the aforementioned association remained significant in only adult females (>20 years) upon age-based stratification. However, after this age-based stratification, sample size considerably reduced in ≤20 years of age group. Thus, further investigation of this association involving Pakistani children and adolescents (≤20 years) can be carried out to validate this observation. Congruent to our observation, some other studies also reported association of rs17782313 with obesity and/or obesity-related traits in adult females.53,58,62,63 Recently, it has been demonstrated that before bariatric surgery extremely obese women carrying MC4R variant rs17782313, are more unlikely to reach non-obesity BMI (<30 kg m−2) and tend to maintain a BMI > 35 kg m−2 during 60 months after surgery that characterize treatment failure.64 In contrast to our study, a number of studies reported association of rs17782313 with obesity and related anthropometric traits in both children and adults without any gender specific effect65,66 while some studies showed association in female adults and children only.67 In addition, some studies reported association in girls only.68 Furthermore, some studies reported association in adult males.69 This implies that this variant affects obesity or weight gain in terms of gender and age differently in different populations.
Many studies have reported association of rs17782313 with risk of developing metabolic disorders like diabetes70,71 and insulin resistance.72,73 In contrast to this, we found no association of rs17782313 with glucose-related metabolic disturbances (aberrant FBG, fasting insulin levels, and HOMA-IR) independent of obesity in whole and gender stratified population. This observation indicates involvement of rs17782313 in engendering susceptibility of our population to obesity but not to diabetes. However, it must be noted that our cases involved OW/OB subjects who never got checked themselves for diabetes whether they were diabetic or not. In agreement to our results, a number of other studies also did not find any association of this variant with diabetes and related metabolic disturbances.74–76 Our study also reports no association of this variant with blood pressure in our population. In agreement to our study, Timpson and colleagues did not find any association of this variant with blood pressure.77 On the other hand, significant association of this variant was reported with an increased nocturnal BP in Chinese Han population.78 We did not find any association of MC4R rs17782313 with eating behaviours like tendency towards fat dense food and diet consciousness. Similarly, Hasselbalch and colleagues reported no role of this variant in food intake and preference for specific food items.79 In contrast, many studies have reported association of this variant with disordered eating, and dietary fat and high energy intakes.53,61,80 However, association of MC4R rs17782313 with random eating timings was observed in the current study but we could not find any parallel study in literature that investigated the association of this variant with random eating timings like ours. We did not find any association of MC4R rs17782313 with PA levels, depression, sleep-wake timings, and sleep duration in our study population. Similarly, no association of MC4R rs17782313 with PA was found in Iranian population.61 In contrast, a recent study revealed that the carriers of C allele (MC4R rs17782313) were considerably less physically active than those with the TT genotype.81 Moreover, contrary to our observation, Yilmaz et al. reported that the MC4R rs17782313 variant has been associated with an increase in depressed mood but the effect of rs17782313 on BMI was not through depression.24 Another study reported that Korean adults with MC4R minor alleles had a higher risk of obesity in high stress states independent of other obesity related factors.23 We could not find any related study like ours that investigated the association of rs17782313 with sleep-wake timings and sleep duration though no such association was found in our study. In nutshell, the differences in genetic predisposition to obesity across different populations highlight the limitations of a ‘one size fits all’ approach and emphasize the importance of population-specific studies for association between genetic variants and risk for obesity and related traits as we move from large genetic data to precision medicine for all.
5. Conclusion
The current study significantly contributes to the knowledge of the genetic predisposition of obesity in Pakistani females and compare this with other populations. This could also be helpful for forthcoming meta-analysis studies elucidating which variants are truly associated with the susceptibility to develop an obese phenotype. In addition, the higher risk of developing various cardio-metabolic disorders observed for overweight and obese subjects of the current study advocates the need for effective community-based interventions for weight management in Pakistani population. Similarly, higher percentage of stigmatized overweight and obese subjects particularly females observed in our study point towards the need of immediate social and legal initiatives to combat weight discrimination.Conflicts of interest
The authors declare that they have no conflicts of interest.Acknowledgements
This work was supported by a recurring grant from the International Center for Chemical and Biological Sciences (ICCBS), University of Karachi, Pakistan.References
- S. J. Olshansky, D. J. Passaro, R. C. Hershow, J. Layden, B. A. Carnes, J. Brody, L. Hayflick, R. N. Butler, D. B. Allison and D. S. Ludwig, N. Engl. J. Med., 2005, 352, 1138–1145 CrossRef CAS PubMed.
- D. P. Guh, W. Zhang, N. Bansback, Z. Amarsi, C. L. Birmingham and A. H. Anis, BMC Public Health, 2009, 9, 88 CrossRef PubMed.
- M. J. Krashes, B. B. Lowell and A. S. Garfield, Nat. Neurosci., 2016, 19, 206–219 CrossRef CAS PubMed.
- Y. Yang, D. Atasoy, H. H. Su and S. M. Sternson, Cell, 2011, 146, 992–1003 CrossRef CAS PubMed.
- T. Liu, D. Kong, B. P. Shah, C. Ye, S. Koda, A. Saunders, J. B. Ding, Z. Yang, B. L. Sabatini and B. B. Lowell, Neuron, 2012, 73, 511–522 CrossRef CAS PubMed.
- Y. Aponte, D. Atasoy and S. M. Sternson, Nat. Neurosci., 2011, 14, 351–355 CrossRef CAS PubMed.
- D. Atasoy, J. N. Betley, H. H. Su and S. M. Sternson, Nature, 2012, 488, 172 CrossRef CAS PubMed.
- M. J. Krashes, S. Koda, C. Ye, S. C. Rogan, A. C. Adams, D. S. Cusher, E. Maratos-Flier, B. L. Roth and B. B. Lowell, J. Clin. Invest., 2011, 121, 1424–1428 CAS.
- C. Zhan, J. Zhou, Q. Feng, J. E. Zhang, S. Lin, J. Bao, P. Wu and M. Luo, J. Neurosci., 2013, 33, 3624–3632 CrossRef CAS PubMed.
- G. T. Dodd, S. Decherf, K. Loh, S. E. Simonds, F. Wiede, E. Balland, T. L. Merry, H. Munzberg, Z. Y. Zhang, B. B. Kahn, B. G. Neel, K. K. Bence, Z. B. Andrews, M. A. Cowley and T. Tiganis, Cell, 2015, 160, 88–104 CrossRef CAS PubMed.
- T. Kishi, C. J. Aschkenasi, C. E. Lee, K. G. Mountjoy, C. B. Saper and J. K. Elmquist, J. Comp. Neurol., 2003, 457, 213–235 CrossRef CAS PubMed.
- A. S. Garfield, D. D. Lam, O. J. Marston, M. J. Przydzial and L. K. Heisler, Trends Endocrinol. Metab., 2009, 20, 203–215 CrossRef CAS PubMed.
- N. Balthasar, L. T. Dalgaard, C. E. Lee, J. Yu, H. Funahashi, T. Williams, M. Ferreira, V. Tang, R. A. McGovern, C. D. Kenny, L. M. Christiansen, E. Edelstein, B. Choi, O. Boss, C. Aschkenasi, C. Y. Zhang, K. Mountjoy, T. Kishi, J. K. Elmquist and B. B. Lowell, Cell, 2005, 123, 493–505 CrossRef CAS PubMed.
- B. P. Shah, L. Vong, D. P. Olson, S. Koda, M. J. Krashes, C. Ye, Z. Yang, P. M. Fuller, J. K. Elmquist and B. B. Lowell, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 13193–13198 CrossRef CAS PubMed.
- J. Rossi, N. Balthasar, D. Olson, M. Scott, E. Berglund, C. E. Lee, M. J. Choi, D. Lauzon, B. B. Lowell and J. K. Elmquist, Cell Metab., 2011, 13, 195–204 CrossRef CAS PubMed.
- J. W. Sohn, L. E. Harris, E. D. Berglund, T. Liu, L. Vong, B. B. Lowell, N. Balthasar, K. W. Williams and J. K. Elmquist, Cell, 2013, 152, 612–619 CrossRef CAS PubMed.
- E. D. Berglund, T. Liu, X. Kong, J. W. Sohn, L. Vong, Z. Deng, C. E. Lee, S. Lee, K. W. Williams, D. P. Olson, P. E. Scherer, B. B. Lowell and J. K. Elmquist, Nat. Neurosci., 2014, 17, 911–913 CrossRef CAS PubMed.
- D. Sundaramurthy, D. A. Campbell, J. P. Leek, A. F. Markham and L. F. Pieri, Cytogenet. Cell Genet., 1998, 82, 97–98 CrossRef CAS PubMed.
- J. Hebebrand, A. L. Volckmar, N. Knoll and A. Hinney, Obes. Facts, 2010, 3, 294–303 CrossRef PubMed.
- D. S. Evans, M. A. Calton, M. J. Kim, P. Y. Kwok, I. Miljkovic, T. Harris, A. Koster, Y. Liu, G. J. Tranah, N. Ahituv, W. C. Hsueh and C. Vaisse, PloS one, 2014, 9, e96805 Search PubMed.
- A. J. Walley, J. E. Asher and P. Froguel, Nat. Rev. Genet., 2009, 10, 431–442 CrossRef CAS PubMed.
- R. J. Loos, C. M. Lindgren, S. Li, E. Wheeler, J. H. Zhao, I. Prokopenko, M. Inouye, R. M. Freathy, A. P. Attwood, J. S. Beckmann, S. I. Berndt, K. B. Jacobs, S. J. Chanock, R. B. Hayes, S. Bergmann, A. J. Bennett, S. A. Bingham, M. Bochud, M. Brown, S. Cauchi, J. M. Connell, C. Cooper, G. D. Smith, I. Day, C. Dina, S. De, E. T. Dermitzakis, A. S. Doney, K. S. Elliott, P. Elliott, D. M. Evans, I. Sadaf Farooqi, P. Froguel, J. Ghori, C. J. Groves, R. Gwilliam, D. Hadley, A. S. Hall, A. T. Hattersley, J. Hebebrand, I. M. Heid, C. Lamina, C. Gieger, T. Illig, T. Meitinger, H. E. Wichmann, B. Herrera, A. Hinney, S. E. Hunt, M. R. Jarvelin, T. Johnson, J. D. Jolley, F. Karpe, A. Keniry, K. T. Khaw, R. N. Luben, M. Mangino, J. Marchini, W. L. McArdle, R. McGinnis, D. Meyre, P. B. Munroe, A. D. Morris, A. R. Ness, M. J. Neville, A. C. Nica, K. K. Ong, S. O'Rahilly, K. R. Owen, C. N. Palmer, K. Papadakis, S. Potter, A. Pouta, L. Qi, J. C. Randall, N. W. Rayner, S. M. Ring, M. S. Sandhu, A. Scherag, M. A. Sims, K. Song, N. Soranzo, E. K. Speliotes, H. E. Syddall, S. A. Teichmann, N. J. Timpson, J. H. Tobias, M. Uda, C. I. Vogel, C. Wallace, D. M. Waterworth, M. N. Weedon, C. J. Willer, V. L. Wraight, X. Yuan, E. Zeggini, J. N. Hirschhorn, D. P. Strachan, W. H. Ouwehand, M. J. Caulfield, N. J. Samani, T. M. Frayling, P. Vollenweider, G. Waeber, V. Mooser, P. Deloukas, M. I. McCarthy, N. J. Wareham, I. Barroso, K. B. Jacobs, S. J. Chanock, R. B. Hayes, C. Lamina, C. Gieger, T. Illig, T. Meitinger, H. E. Wichmann, P. Kraft, S. E. Hankinson, D. J. Hunter, F. B. Hu, H. N. Lyon, B. F. Voight, M. Ridderstrale, L. Groop, P. Scheet, S. Sanna, G. R. Abecasis, G. Albai, R. Nagaraja, D. Schlessinger, A. U. Jackson, J. Tuomilehto, F. S. Collins, M. Boehnke and K. L. Mohlke, Nat. Genet., 2008, 40, 768–775 CrossRef CAS PubMed.
- S. Park, J. W. Daily, X. Zhang, H. S. Jin, H. J. Lee and Y. H. Lee, Nutr. Metab., 2016, 13, 38 Search PubMed.
- Z. Yilmaz, C. Davis, N. J. Loxton, A. S. Kaplan, R. D. Levitan, J. C. Carter and J. L. Kennedy, Int. J. Obes., 2005, 2015(39), 114–120 Search PubMed.
- M. Ng, T. Fleming, M. Robinson, B. Thomson, N. Graetz, C. Margono, E. C. Mullany, S. Biryukov, C. Abbafati, S. F. Abera, J. P. Abraham, N. M. Abu-Rmeileh, T. Achoki, F. S. AlBuhairan, Z. A. Alemu, R. Alfonso, M. K. Ali, R. Ali, N. A. Guzman, W. Ammar, P. Anwari, A. Banerjee, S. Barquera, S. Basu, D. A. Bennett, Z. Bhutta, J. Blore, N. Cabral, I. C. Nonato, J. C. Chang, R. Chowdhury, K. J. Courville, M. H. Criqui, D. K. Cundiff, K. C. Dabhadkar, L. Dandona, A. Davis, A. Dayama, S. D. Dharmaratne, E. L. Ding, A. M. Durrani, A. Esteghamati, F. Farzadfar, D. F. Fay, V. L. Feigin, A. Flaxman, M. H. Forouzanfar, A. Goto, M. A. Green, R. Gupta, N. Hafezi-Nejad, G. J. Hankey, H. C. Harewood, R. Havmoeller, S. Hay, L. Hernandez, A. Husseini, B. T. Idrisov, N. Ikeda, F. Islami, E. Jahangir, S. K. Jassal, S. H. Jee, M. Jeffreys, J. B. Jonas, E. K. Kabagambe, S. E. Khalifa, A. P. Kengne, Y. S. Khader, Y. H. Khang, D. Kim, R. W. Kimokoti, J. M. Kinge, Y. Kokubo, S. Kosen, G. Kwan, T. Lai, M. Leinsalu, Y. Li, X. Liang, S. Liu, G. Logroscino, P. A. Lotufo, Y. Lu, J. Ma, N. K. Mainoo, G. A. Mensah, T. R. Merriman, A. H. Mokdad, J. Moschandreas, M. Naghavi, A. Naheed, D. Nand, K. M. Narayan, E. L. Nelson, M. L. Neuhouser, M. I. Nisar, T. Ohkubo, S. O. Oti, A. Pedroza, D. Prabhakaran, N. Roy, U. Sampson, H. Seo, S. G. Sepanlou, K. Shibuya, R. Shiri, I. Shiue, G. M. Singh, J. A. Singh, V. Skirbekk, N. J. Stapelberg, L. Sturua, B. L. Sykes, M. Tobias, B. X. Tran, L. Trasande, H. Toyoshima, S. van de Vijver, T. J. Vasankari, J. L. Veerman, G. Velasquez-Melendez, V. V. Vlassov, S. E. Vollset, T. Vos, C. Wang, X. Wang, E. Weiderpass, A. Werdecker, J. L. Wright, Y. C. Yang, H. Yatsuya, J. Yoon, S. J. Yoon, Y. Zhao, M. Zhou, S. Zhu, A. D. Lopez, C. J. Murray and E. Gakidou, Lancet, 2014, 384, 766–781 CrossRef.
- A. Srivastava, B. Mittal, J. Prakash, V. S. Narain, S. M. Natu and N. Srivastava, Oman Med. J., 2014, 29, 114–118 CrossRef CAS PubMed.
- A. S. Jackson and M. L. Pollock, Physician Sportsmed., 1985, 13, 76–90 CrossRef CAS PubMed.
- D. R. Matthews, J. P. Hosker, A. S. Rudenski, B. A. Naylor, D. F. Treacher and R. C. Turner, Diabetologia, 1985, 28, 412–419 CrossRef CAS PubMed.
- X. Pi-Sunyer, Postgrad. Med., 2009, 121, 21–33 CrossRef PubMed.
- G. A. Head, Front Physiol., 2015, 6, 32 Search PubMed.
- A. Anandacoomarasamy, I. Caterson, P. Sambrook, M. Fransen and L. March, Int. J. Obes., 2005, 2008(32), 211–222 Search PubMed.
- M. S. Faith, M. Butryn, T. A. Wadden, A. Fabricatore, A. M. Nguyen and S. B. Heymsfield, Obes. Rev., 2011, 12, e438–453 CrossRef CAS PubMed.
- S. Wei, M. D. Schmidt, T. Dwyer, R. J. Norman and A. J. Venn, Obesity, Silver Spring, Md, 2009, 17, pp. 1070–1076 Search PubMed.
- A. N. Jung, J. H. Park, J. Kim, S. H. Kim, B. C. Jee, B. H. Cha, J. W. Sull and J. H. Jun, J. Womens Health, 2002, 2017(26), 83–90 Search PubMed.
- K. E. Giel, S. Zipfel, M. Alizadeh, N. Schaffeler, C. Zahn, D. Wessel, F. W. Hesse, S. Thiel and A. Thiel, BMC Public Health, 2012, 12, 525 CrossRef PubMed.
- S. Ambwani, K. M. Thomas, C. J. Hopwood, S. A. Moss and C. M. Grilo, Eat. Behav., 2014, 15, 366–370 CrossRef PubMed.
- S. Sofer, A. H. Stark and Z. Madar, Adv. Nutr., 2015, 6, 214–223 CrossRef CAS PubMed.
- C. Keller and K. van der Horst, Appetite, 2013, 62, 150–159 CrossRef PubMed.
- G. K. Pot, M. Richards, C. J. Prynne and A. M. Stephen, Public Health Nutr., 2014, 17, 2660–2666 CrossRef PubMed.
- D. J. Mela, Obes. Res., 2001, 9(Suppl. 4), 249s–255s CrossRef PubMed.
- J. M. Jakicic and K. K. Davis, Psychiatric Clinics of North America., 2011, 34, 829–840 CrossRef PubMed.
- G. Jean-Louis, N. J. Williams, D. Sarpong, A. Pandey, S. Youngstedt, F. Zizi and G. Ogedegbe, BMC Public Health, 2014, 14, 290 CrossRef PubMed.
- P. A. Vargas, Curr. Obes. Rep., 2016, 5, 38–50 CrossRef PubMed.
- S. R. Patel, A. L. Hayes, T. Blackwell, D. S. Evans, S. Ancoli-Israel, Y. K. Wing and K. L. Stone, Int. J. Obes., 2005, 2014(38), 1159–1164 Search PubMed.
- B. J. Taylor, K. A. Matthews, B. P. Hasler, K. A. Roecklein, C. E. Kline, D. J. Buysse, H. M. Kravitz, A. G. Tiani, S. D. Harlow and M. H. Hall, Sleep, 2016, 39, 457–465 CrossRef PubMed.
- A. W. McHill, E. L. Melanson, J. Higgins, E. Connick, T. M. Moehlman, E. R. Stothard and K. P. Wright Jr, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 17302–17307 CrossRef CAS PubMed.
- M. Y. Xiao, Z. Y. Wang, H. M. Fan, C. L. Che, Y. Lu, L. X. Cong, X. J. Gao, Y. J. Liu, J. X. Yuan, X. M. Li, B. Hu and Y. P. Chen, Zhonghua Liuxingbingxue Zazhi, 2016, 37, 1468–1472 CAS.
- R. J. Loos and C. Bouchard, J. Intern. Med., 2003, 254, 401–425 CrossRef CAS PubMed.
- S. Saeed, T. A. Butt, M. Anwer, M. Arslan and P. Froguel, Mol. Genet. Metab., 2012, 106, 121–126 CrossRef CAS PubMed.
- A. Lipowicz, S. Gronkiewicz and R. M. Malina, Am. J. Hum. Biol, 2002, 14, 468–475 CrossRef PubMed.
- B. Moghimi-Dehkordi, A. Safaee, M. Vahedi, A. Pourhoseingholi, M. Pourhoseingholi, S. Ashtari and M. Zali, Ann Med Health Sci Res., 2013, 3, 171–176 CrossRef CAS PubMed.
- Y. S. Saraswathi, M. Najafi, G. Sreenivasa, P. T. Chaithra and S. S. Malini, J. Pharm. Res., 2012, 5, 4642–4644 Search PubMed.
- L. Qi, P. Kraft, D. J. Hunter and F. B. Hu, Hum. Mol. Genet., 2008, 17, 3502–3508 CrossRef CAS PubMed.
- G. Liu, H. Zhu, Y. Dong, R. H. Podolsky, F. A. Treiber and H. Snieder, Eur. J. Epidemiol., 2011, 26, 463–473 CrossRef PubMed.
- F. Lauria, A. Siani, C. Pico, W. Ahrens, K. Bammann, S. De Henauw, R. Foraita, L. Iacoviello, Y. Kourides, S. Marild, D. Molnar, L. A. Moreno, Y. Pitsiladis, J. Sanchez, T. Veidebaum, G. Wang and P. Russo, J. Clin. Endocrinol. Metab., 2016, 101, 4229–4236 CrossRef CAS PubMed.
- J. a. Luan, B. Kerner, J.-H. Zhao, R. J. F. Loos, S. J. Sharp, B. O. Muthén and N. J. Wareham, BMC Proc., 2009, 3, S115 CrossRef PubMed.
- E. T. Liem, J. M. Vonk, P. J. Sauer, G. van der Steege, E. Oosterom, R. P. Stolk and H. Snieder, Am. J. Clin. Nutr., 2010, 91, 321–328 CrossRef CAS PubMed.
- J. Shi, J. Long, Y. T. Gao, W. Lu, Q. Cai, W. Wen, Y. Zheng, K. Yu, Y. B. Xiang, F. B. Hu, W. Zheng and X. O. Shu, Am. J. Epidemiol., 2010, 172, 244–254 CrossRef PubMed.
- S. K. Vasan, T. Fall, M. J. Neville, B. Antonisamy, C. H. Fall, F. S. Geethanjali, H. F. Gu, P. Raghupathy, P. Samuel, N. Thomas, K. Brismar, E. Ingelsson and F. Karpe, Obesity (Silver Spring, Md.), 2012, 20, 2268–2277 CrossRef CAS PubMed.
- A. E. Taylor, M. N. Sandeep, C. S. Janipalli, C. Giambartolomei, D. M. Evans, M. V. Kranthi Kumar, D. G. Vinay, P. Smitha, V. Gupta, M. Aruna, S. Kinra, R. M. Sullivan, L. Bowen, N. J. Timpson, G. Davey Smith, F. Dudbridge, D. Prabhakaran, Y. Ben-Shlomo, K. S. Reddy, S. Ebrahim and G. R. Chandak, J. Obes., 2011, 2011, 7 Search PubMed.
- A. Khalilitehrani, M. Qorbani, S. Hosseini and H. Pishva, Gene, 2015, 563, 125–129 CrossRef CAS PubMed.
- F. Bauer, C. C. Elbers, R. A. Adan, R. J. Loos, N. C. Onland-Moret, D. E. Grobbee, J. V. van Vliet-Ostaptchouk, C. Wijmenga and Y. T. van der Schouw, Am. J. Clin. Nutr., 2009, 90, 951–959 CrossRef CAS PubMed.
- D. C. Croteau-Chonka, A. F. Marvelle, E. M. Lange, N. R. Lee, L. S. Adair, L. A. Lange and K. L. Mohlke, Obesity (Silver Spring, Md.), 2011, 19, 1019–1027 CrossRef CAS PubMed.
- C. M. M. Resende, D. F. Durso, K. B. G. Borges, R. M. Pereira, G. K. D. Rodrigues, K. F. Rodrigues, J. L. P. Silva, E. C. Rodrigues, G. R. Franco and J. I. Alvarez-Leite, Clin. Nutr., 2017 DOI:10.1016/j.clnu.2017.05.018.
- C. I. Vogel, T. Boes, T. Reinehr, C. L. Roth, S. Scherag, A. Scherag, J. Hebebrand and A. Hinney, Obes. Facts, 2011, 4, 67–75 CrossRef PubMed.
- J. W. Sull, M. Lee and S. H. Jee, Endocrine, 2013, 44, 675–679 CrossRef CAS PubMed.
- K. Kvaloy, B. Kulle, P. Romundstad and T. L. Holmen, Int. J. Obes., 2005, 2013(37), 1221–1229 Search PubMed.
- G. Liu, H. Zhu, V. Lagou, B. Gutin, P. Barbeau, F. A. Treiber, Y. Dong and H. Snieder, J. Pediatr., 2010, 156, 598–605 CrossRef CAS PubMed.
- S. Cauchi, F. Stutzmann, C. Cavalcanti-Proenca, E. Durand, A. Pouta, A. L. Hartikainen, M. Marre, S. Vol, T. Tammelin, J. Laitinen, A. Gonzalez-Izquierdo, A. I. Blakemore, P. Elliott, D. Meyre, B. Balkau, M. R. Jarvelin and P. Froguel, J. Mol. Med., 2009, 87, 537–546 CrossRef CAS PubMed.
- O. P. Dwivedi, R. Tabassum, G. Chauhan, I. Kaur, S. Ghosh, R. K. Marwaha, N. Tandon and D. Bharadwaj, J. Hum. Genet., 2013, 58, 27–32 CrossRef PubMed.
- A. Marcadenti, F. D. Fuchs, U. Matte, F. Sperb, L. B. Moreira and S. C. Fuchs, Cardiovasc. Diabetol., 2013, 12, 103 CrossRef CAS PubMed.
- C. S. Janipalli, M. V. Kumar, D. G. Vinay, M. N. Sandeep, S. Bhaskar, S. R. Kulkarni, M. Aruna, C. V. Joglekar, S. Priyadharshini, N. Maheshwari, C. S. Yajnik and G. R. Chandak, Diabetic Med., 2012, 29, 121–127 CrossRef CAS PubMed.
- O. Tschritter, A. Haupt, H. Preissl, C. Ketterer, A. M. Hennige, T. Sartorius, F. Machicao, A. Fritsche and H. U. Haring, J. Obes., 2011, 2011, 283153 Search PubMed.
- W. Huang, Y. Sun and J. Sun, Endocrine, 2011, 39, 69–74 CrossRef CAS PubMed.
- M. Thomsen, M. Dahl, A. Tybjaerg-Hansen and B. G. Nordestgaard, J. Clin. Endocrinol. Metab., 2012, 97, E1074–1079 CrossRef CAS PubMed.
- M. D. Bazzi, F. A. Nasr, M. S. Alanazi, A. Alamri, A. A. Turjoman, A. S. Moustafa, A. A. Alfadda, A. A. Pathan and N. R. Parine, GMR, Genet. Mol. Res., 2014, 13, 10194–10203 CrossRef CAS PubMed.
- N. J. Timpson, R. Harbord, G. Davey Smith, J. Zacho, A. Tybjaerg-Hansen and B. G. Nordestgaard, Hypertension (Dallas, Tex. 1979), 2009, vol. 54, pp. 84–90 Search PubMed.
- Y. Sun, J. Sun, J. Wu and M. Yang, Cardiovasc. J. Afr, 2016, 27, 21–24 CrossRef PubMed.
- A. L. Hasselbalch, L. Angquist, L. Christiansen, B. L. Heitmann, K. O. Kyvik and T. I. Sorensen, J. Nutr., 2010, 140, 831–834 CrossRef CAS PubMed.
- J. A. Vega, G. Salazar, M. I. Hodgson, L. R. Cataldo, M. Valladares, A. M. Obregon and J. L. Santos, Ann. Nutr. Metab., 2016, 68, 35–41 CrossRef CAS PubMed.
- H. Lee, G. I. Ash, T. J. Angelopoulos, P. M. Gordon, N. M. Moyna, P. S. Visich, R. F. Zoeller, H. Gordish-Dressman, V. Deshpande, M. H. Chen, P. D. Thompson, E. P. Hoffman, J. M. Devaney and L. S. Pescatello, Sports Med., 2015, 1, 34 Search PubMed.
| This journal is © The Royal Society of Chemistry 2018 |