Open Access Article
Open Access ArticleMicrobial transformation of mestanolone by Macrophomina phaseolina and Cunninghamella blakesleeana and anticancer activities of the transformed products†
Rabia Farooqa,
Nusrat Hussaina,
Sammer Yousufa,
Atia-tul-Wahab*b,
Malik Shoaib Ahmadb,
Atta-ur-Rahmana and
M. Iqbal Choudhary *abc
*abc
aH. E. J. Research Institute of Chemistry, International Center for Chemical and Biological Sciences, University of Karachi, Karachi-75270, Pakistan. E-mail: iqbal.choudhary@iccs.edu
bDr. Panjwani Center for Molecular Medicine and Drug Research, International Center for Chemical and Biological Sciences, University of Karachi, Karachi-75270, Pakistan. E-mail: atia.tulwahab@iccs.edu
cDepartment of Biochemistry, Faculty of Science, King Abdulaziz University, Jeddah-21412, Saudi Arabia
First published on 14th June 2018
Abstract
The microbial transformation of anabolic androgenic steroid mestanolone (1) with Macrophomina phaseolina and Cunninghamella blakesleeana has afforded seven metabolites. The structures of these metabolites were characterized as 17β-hydroxy-17α-methyl-5α-androsta-1-ene-3,11-dione (2), 14α,17β-dihydroxy-17α-methyl-5α-androstan-3,11-dione (3), 17β-hydroxy-17α-methyl-5α-androstan-1,14-diene-3,11-dione (4), 17β-hydroxy-17α-methyl-5α-androstan-3,11-dione (5), 11β,17β-dihydroxy-17α-methyl-5α-androstan-1-ene-3-one (6), 9α,11β,17β-trihydroxy-17α-methyl-5α-androstan-3-one (7), and 1β,11α,17β-trihydroxy-17α-methyl-5α-androstan-3-one (8). All the metabolites, except 5 and 6, were identified as new compounds. Substrate 1 (IC50 = 27.6 ± 1.1 μM), and its metabolites 2 (IC50 = 19.2 ± 2.9 μM) and 6 (IC50 = 12.8 ± 0.6 μM) exhibited moderate cytotoxicity against the HeLa cancer cell line (human cervical carcinoma). All metabolites were noncytotoxic to 3T3 (mouse fibroblast) and H460 (human lung carcinoma) cell lines. The metabolites were also evaluated for immunomodulatory activity, and all were found to be inactive.
Introduction
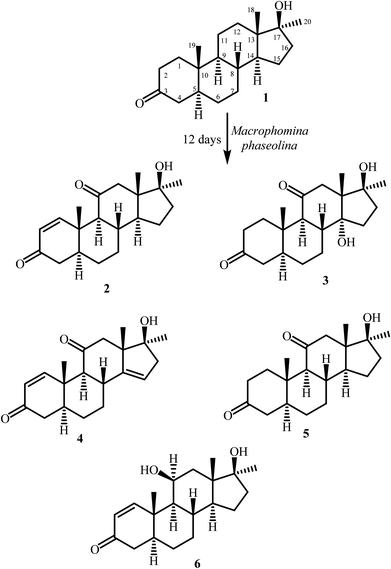
Biotransformation has been widely used in organic chemistry for stereoslective synthesis.1–6 Biotransformation reactions can be achieved by a variety of agents, such as enzymes, animals and plant cell cultures, and microorganisms; however, microorganisms are most effectively used for this purpose. Microbial enzyme systems can be used for reduction, oxidation, hydroxylation, and Michael addition. Regio- and stereo-selective oxidation and hydroxylation of steroids have been extensively achieved through microbial transformation. The cytochrome P450 monooxygenase system, present in microorganisms – particularly in fungi – is responsible for the stereoselective hydroxylation at various sites of the steroidal skeleton.7–9Mestanolone (1) (C20H18O2) is a member of the anabolic-androgenic class of steroids. It is weakly anabolic and strongly androgenic. Mestanolone was first synthesized by the oxidation of 17β-methylandrostan-3β,17β-diol. It is used as a starting material for the synthesis of other anabolic steroids, such as 17-methyl-1-testosterone, and oxandrolone.10,11 Compound 1 was earlier subjected to microbial transformation, and several new analogues were obtained.12
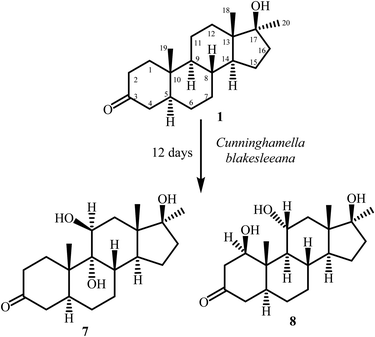
In continuation of our research on biotransformation of bioactive compounds, and drug molecules,13–15 mestanolone (1) was incubated with Macrophomina phaseolina, and Cunninghamella blakesleeana, which yielded metabolites 2–8 (Fig. 1 and 2).
Experimental
General
Mestanolone (1) was acquired from Hangzhu Dayangchem (Cat no. 541-11-9, China). Sabouraud dextrose agar (SDA) was purchased from Merck KGaA (Cat no. 146392, Germany). Silica gel precoated TLC plates (PF254, Merck KGaA, Germany) were used for thin layer chromatography; phosphomolybdic acid solution was used as a staining reagent for UV inactive compounds. Silica gel (70–230 mesh, Merck, Germany) was used for column chromatography. Final purification of the compounds was carried out by using recycling preparative HPLC-LC-908 (Japan), equipped with JAIGEL-ODS-L-80 column, with MeOH–H2O as the mobile phase. Melting points were recorded on Buchi M-560 (Buchi, Switzerland) apparatus. Optical rotations were measured on JASCO P-2000 (JASCO, Japan) polarimeter. UV Spectra were recorded on Hitachi U-3200 (Hitachi, Tokyo, Japan) spectrophotometer. IR Spectra were recorded as KBr discs on Bruker Vector 22 FT-IR (Bruker) spectrometer. Electron ionization (EI-MS) and high resolution electron ionization mass spectra (HREI-MS) were recorded on JEOL JMS600H mass spectrometer (JEOL, Japan). 1H-, 13C- and 2D-NMR spectra were recorded on Bruker Avance spectrometers (Bruker, Switzerland) in CD3OD. X-Ray diffraction data of the compound 6 was recorded on Bruker SMART APEX II single-crystal X-ray diffractometer (Germany).Microbial cultures and media preparation
Macrophomina phaseolina (KUCC730), and Cunninghamella blakesleeana (ATCC8688A) were acquired from the Karachi University Culture Collection (KUCC) and American Type Culture Collection (ATCC). Cultures were grown, and stored on Sabouraud dextrose agar (SDA) slant at 4 °C. Liquid media (5.0 L) was prepared to grow M. phaseolina (KUCC 730) in distilled H2O by using the following ingredients; glucose (50.0 g), glycerol (50.0 mL), peptone (25.0 g), yeast extract (25.0 g), KH2PO4 (25.0 g), and NaCl (25.0 g). Similarly media for C. blakesleeana was prepared by mixing peptone (20.0 g), glucose (40.0 g), yeast extract (20.0 g), KH2PO4 (20.0 g), NaCl (20.0 g), and glycerol (40.0 mL) in 4 L distilled water.General fermentation and extraction protocol
The media was prepared by using the aforementioned ingredients. The media was transferred to flasks and autoclaved at 120 °C. Seed flasks were prepared under sterilized conditions by transferring the spores of M. phaseolina from slants into the flasks, and then incubated at 25 ± 2 °C for 3 days on a rotary shaker (128 rpm). Similarly, seed flasks of C. blakesleeana were also prepared. The remaining flasks were inoculated by transferring the mycelia from the seed flasks and incubated on rotary shaker at 25 ± 2 °C. After appropriate fungal growth, compound 1 was dissolved in methanol and distributed evenly in all flasks. Fermentation was continued and the degree of transformation was analyzed by performing time course studies after different time intervals. In order to assess the fungal metabolites, and degradation of compound 1 in the aqueous media, two parallel control experiments were also conducted in order to assess the fungal metabolites a negative control (fungi + liquid media without compound) and, for degradation of compound 1 in the aqueous media, a positive control (liquid media + compound without fungi). After 12 days, the reaction was stopped by adding dichloromethane (CH2Cl2), and the broth was filtered to remove the mycelia. The broth was extracted with same solvent dichloromethane and dried over anhydrous sodium sulphate (Na2SO4). The organic layer was concentrated under reduced pressures to obtain a crude extract.Fermentation of mestanolone (1) with Macrophomina phaseolina
Mestanolone (1) (1 g) was dissolved in 25 mL of methanol, distributed evenly in 50 flasks cultured with M. phaseolina (KUCC 730), and incubated for 12 days on a rotary shaker at 25 ± 2 °C. After completion of 12 days, dichloromethane was added into each flask to stop the fermentation. The content of the flasks was filtered to remove the fungal biomass. The broth (aqueous filtrate) was then extracted with dichloromethane (3 × 6 L). The organic layer was dried over anhydrous sodium sulfate (Na2SO4), and concentrated on a rotary evaporator. A brown crude extract (1.3 g) was obtained and loaded on a silica gel column. The column was eluted with 5% gradient of hexanes and acetone. Four fractions (1–4) were obtained, which yielded metabolites 2–6 (Fig. 1) after purification through reverse phase recycling HPLC (methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water; 70
water; 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30). Fraction 1 resulted into compound 2 (RT = 38 min, 9 mg). Compound 3 (RT = 34 min, 16 mg) was obtained from fraction 2. Compounds 4 (RT = 22 min, 3 mg) and 5 (RT = 32 min 7.8 mg) were obtained from fraction 3. Fraction 4 yielded compound 6 (RT = 50 min, 56 mg).
30). Fraction 1 resulted into compound 2 (RT = 38 min, 9 mg). Compound 3 (RT = 34 min, 16 mg) was obtained from fraction 2. Compounds 4 (RT = 22 min, 3 mg) and 5 (RT = 32 min 7.8 mg) were obtained from fraction 3. Fraction 4 yielded compound 6 (RT = 50 min, 56 mg).
Fermentation of mestanolone (1) with Cunninghamella blakesleeana
Mestanolone (1) (500 mg) was dissolved in 20 mL of methanol, and dispensed equally in 40 flasks containing culture of C. blakesleeana (ATCC8688A). The flasks were kept on a shaker at 27 °C, and content was incubated for 12 days. The experiment was monitored through periodic TLC analysis. After 12 days of incubation, the substrate 1 seemed to be fully consumed. The contents of all flasks were collected, filtered off and then the aqueous media was extracted with CH2Cl2. The extract was then concentrated under reduced pressures. The crude extract (2.4 g) was then loaded on a silica gel column for fractionation. The mobile phase comprised gradients of hexane and acetone mixtures. Column chromatography yielded two fractions which were further purified by reverse phase recycling HPLC (methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water, 70
water, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30). Fraction 1 yielded metabolite 7 (RT = 32 min, 7 mg) on purification while metabolite 8 (9.2 mg, RT = 27 min) (Fig. 2) was purified from fraction 2.
30). Fraction 1 yielded metabolite 7 (RT = 32 min, 7 mg) on purification while metabolite 8 (9.2 mg, RT = 27 min) (Fig. 2) was purified from fraction 2.
17β-Hydroxy-17α-methyl-5α-androsta-1-ene-3,11-dione (2)
White solid: yield (percentage yield): 9 mg (0.9%); mp: 194–195 °C; [α]25D= −307 (c = 0.001, CH3OH); UV (MeOH) λmax 230 nm (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε 5.3); IR (KBr) νmax cm−1: 3490 (O–H stretching), 2956 (C–H stretching), 1701 (C
ε 5.3); IR (KBr) νmax cm−1: 3490 (O–H stretching), 2956 (C–H stretching), 1701 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1659 (C
O), 1659 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–C
C–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H-NMR (CD3OD, 500 MHz): Table 1; 13C-NMR (CD3OD, 125 MHz): Table 1; EI-MS m/z (rel. int., %): 316 [M+] (100), 298 (22), 283 (16), 259 (66), 240 (19), 121 (33); HREI-MS m/z 316.2039 [M+] (mol. formula C20H28O3, calcd value 316.2038).
O); 1H-NMR (CD3OD, 500 MHz): Table 1; 13C-NMR (CD3OD, 125 MHz): Table 1; EI-MS m/z (rel. int., %): 316 [M+] (100), 298 (22), 283 (16), 259 (66), 240 (19), 121 (33); HREI-MS m/z 316.2039 [M+] (mol. formula C20H28O3, calcd value 316.2038).
| Carbon | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 38.9 | 1.36 (overlap), 2.21 (m) | 162.3 | 7.57 (d, J1,2 = 10.5) | 38.5 | 1.27 (dd, J = 14.0, J = 5.2), 2.87 (ddd, J = 13.2, J = 6.4, J = 2.0) |
| 2 | 38.6 | 2.20 (m), 2.50 (td, J2a/2e, = 15.0, J2/1a,e = 7.0) | 127.7 | 5.78 (d, J2,1 = 10.5) | 38.7 | 2.17 (overlap), 2.50 (td, J2a/2e, =14.8, J2/1a,e = 6.8) |
| 3 | 214.6 | — | 202.2 | — | 214.4 | — |
| 4 | 45.2 | 2.01 (m), 2.35 (t, J4e/4a,5a = 14.1) | 41.4 | 2.16 (overlap), 2.42 (overlap) | 45.1 | 2.01 (dt, J = 14.8, J = 2.8), 2.36 (overlap) |
| 5 | 47.6 | 1.50 (overlap) | 45.5 | 1.91 (overlap) | 48.3 | 1.50 (overlap) |
| 6 | 32.6 | 0.94 (td), 1.51 (overlap) | 28.1 | 1.53 (m), 1.42 (m) | 29.2 | 1.37 (overlap), 1.43 (overlap) |
| 7 | 32.4 | 1.26 (overlap), 1.73 (m) | 32.6 | 1.24 (overlap), 1.84 (overlap) | 27.5 | 1.56 (m), 1.66 (overlap) |
| 8 | 38.6 | 1.54 (overlap) | 39.3 | 1.85 (overlap) | 43.4 | 2.16 (overlap) |
| 9 | 55.0 | 0.74 (td, J9a/8a = J9a/11a = 10.5, J9a/11e = 4.0) | 60.2 | 2.17 (overlap) | 59.1 | 2.32 (overlap) |
| 10 | 37.5 | — | 35.8 | — | 36.5 | — |
| 11 | 21.9 | 1.32 (overlap), 1.63 (overlap) | 213.3 | — | 214.6 | — |
| 12 | 29.7 | 1.29 (overlap), 1.36 (overlap) | 51.5 | 2.47 (overlap), 2.13 (overlap) | 47.6 | 1.88 (overlap), 2.93 (d, J12a/e = 12.0) |
| 13 | 46.8 | — | 50.9 | — | 55.4 | — |
| 14 | 51.6 | 1.22 (overlap) | 50.7 | 1.96 (overlap) | 83.4 | — |
| 15 | 24.0 | 1.28 (overlap), 1.61 (overlap) | 23.6 | 1.37 (m), 1.74 (overlap) | 32.6 | 1.83 (m), 1.70 (overlap) |
| 16 | 39.5 | 1.84 (m), 1.61 (overlap) | 39.2 | 1.82 (overlap), 1.90 (overlap) | 40.0 | 1.90 (overlap), 2.13 (overlap) |
| 17 | 82.1 | — | 80.8 | — | 81.5 | — |
| 18 | 14.4 | 0.85 (s) | 15.4 | 0.77 (s) | 20.8 | 0.85 (s) |
| 19 | 11.4 | 1.06 (s) | 13.9 | 1.26 (s) | 11.8 | 1.23 (s) |
| 20 | 26.0 | 1.17 (s) | 26.2 | 1.26 (s) | 29.4 | 1.47 (s) |
14α,17β-Dihydroxy-17α-methyl-5α-androstan-3,11-dione (3)
White solid: yield (percentage yield): 16 mg (1.6%); mp: 218–220 °C; [α]25D = −283 (c = 0.001, CH3OH); IR (KBr) νmax cm−1: 3523, 3490 (O–H stretching), 2942 (C–H stretching), 1692 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H-NMR (CD3OD, 400 MHz): Table 1; 13C-NMR (CD3OD, 75 MHz): Table 1; EI-MS m/z (rel. int., %): 334 (10), 316 (78), 283 (11), 258 (32), 159 (29), 147 (40), 121 (51), 91 (55); HREI-MS m/z 334.2140 [M+] (mol. formula C20H30O4, calcd value 334.2144).
O); 1H-NMR (CD3OD, 400 MHz): Table 1; 13C-NMR (CD3OD, 75 MHz): Table 1; EI-MS m/z (rel. int., %): 334 (10), 316 (78), 283 (11), 258 (32), 159 (29), 147 (40), 121 (51), 91 (55); HREI-MS m/z 334.2140 [M+] (mol. formula C20H30O4, calcd value 334.2144).
17β-Hydroxy-17α-methyl-5α-androsta-1,14-diene-3,11-dione (4)
White solid: yield (percentage yield): 3 mg (0.3%); mp: 180–182 °C; [α]25D = −82.8 (c = 0.0028, CH3OH); UV (MeOH) λmax 230 nm (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε 5.0); IR (KBr) νmax cm−1: 3460 (O–H stretching), 2930 (C–H stretching), 1708 (C
ε 5.0); IR (KBr) νmax cm−1: 3460 (O–H stretching), 2930 (C–H stretching), 1708 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1658 (C
O), 1658 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–C
C–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1H-NMR (CD3OD, 500 MHz): Table 2; 13C-NMR (CD3OD, 100 MHz): Table 2; EI-MS m/z (rel. int., %): 314 [M+] (100), 299 (22), 281 (10), 257 (26), 229 (11), 135 (56), 121 (75); HREI-MS m/z 314.1862 [M+] (mol. formula C20H26O3, calcd value 314.1876).
O), 1H-NMR (CD3OD, 500 MHz): Table 2; 13C-NMR (CD3OD, 100 MHz): Table 2; EI-MS m/z (rel. int., %): 314 [M+] (100), 299 (22), 281 (10), 257 (26), 229 (11), 135 (56), 121 (75); HREI-MS m/z 314.1862 [M+] (mol. formula C20H26O3, calcd value 314.1876).
| Carbon | 4 | 5 | 6 | |||
|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 161.6 | 7.45 (d, J1,2 = 10.0) | 38.6 | 2.17 (m), 2.52 (td, J = 15.0, J = 7.0) | 161.4 | 7.42 (d, J1,2 = 10.4) |
| 2 | 127.8 | 5.79 (d, J2,1 = 10.5) | 38.1 | 1.20 (overlap), 2.74 (ddd, J = 13.0, J = 6.5, J = 2.0) | 127.5 | 5.83 (d, J2,1 = 10.0) |
| 3 | 202.0 | — | 213.7 | — | 202.8 | — |
| 4 | 41.2 | 2.19 (overlap), 2.43 (overlap) | 45.0 | 2.01 (m), 2.36 (overlap) | 41.4 | 2.11 (dd, J4a/4e = 17.6, J4a/5a=3.6), 2.40 (dd, J4e/4a = 17.6, J4e/5α = 14.4) |
| 5 | 45.1 | 1.91 (m) | 48.1 | 1.52 (m) | 46.4 | 1.92 (overlap) |
| 6 | 27.9 | 1.53 (overlap), 1.59 (overlap) | 29.4 | 1.33 (overlap), 1.36 (overlap) | 28.3 | 1.54 (overlap), 1.40 (overlap) |
| 7 | 30.5 | 1.59 (overlap), 2.13 (m) | 33.0 | 1.18 (overlap), 1.84 (overlap) | 32.8 | 1.0 (overlap), 1.87 (overlap) |
| 8 | 36.9 | 2.60 (m) | 39.3 | 1.83 (overlap) | 33.9 | 1.96 (overlap) |
| 9 | 60.5 | 2.19 (overlap) | 64.4 | 1.90 (overlap) | 55.3 | 1.02 (overlap) |
| 10 | 39.4 | — | 36.4 | — | 40.7 | — |
| 11 | 212.6 | — | 214.3 | — | 68.0 | 4.52 (d, J11,9 α = 2.8) |
| 12 | 51.8 | 2.09 (d, J12a/e = 12), 2.56 (d, J12e/a = 12.0) | 51.7 | 2.06 (d, J = 11.5), 2.42 (overlap) | 41.3 | 1.48 (overlap), 1.73 (dd, J12 a/e=14.0, J12/11 = 2.4) |
| 13 | 55.6 | — | 48.1 | — | 46.1 | — |
| 14 | 150.0 | — | 50.9 | 1.91 (overlap) | 53.5 | 1.23 (m) |
| 15 | 119.7 | 5.45 (dd, J15/16a = 4.5, J15/16e = 2.0) | 23.7 | 1.36 (overlap), 1.71 (m) | 24.3 | 1.33 (overlap), 1.63 (overlap) |
| 16 | 46.9 | 2.38 (overlap), 2.52 (m) | 39.2 | 1.32 (overlap), 1.77 (overlap) | 39.1 | 1.62 (overlap), 1.89 (overlap) |
| 17 | 82.0 | — | 80.8 | — | 82.6 | — |
| 18 | 19.8 | 0.97 (s) | 15.3 | 0.76 (s) | 17.0 | 1.09 (s) |
| 19 | 13.5 | 1.31 (s) | 11.3 | 1.23 (s) | 15.5 | 1.28 (s) |
| 20 | 24.9 | 1.23 (s) | 26.1 | 1.25 (s) | 26.3 | 1.14 (s) |
7β-Hydroxy-17α-methyl-5α-androstan-3,11-dione (5)
White solid: yield (percentage yield): 7.8 mg (0.78%); mp: 150–152 °C; [α]25D = −95.7 (c = 0.007, CH3OH); IR (KBr) νmax cm−1: 3416 (O–H stretching), 1697 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H-NMR (CD3OD, 500 MHz): Table 2; 13C-NMR (CD3OD, 75 MHz): Table 2; EI-MS m/z (rel. int., %): 318 [M+] (79), 300 (13), 285 (12), 261 (100), 245 (19), 207 (21), 123 (25); HREI-MS m/z 318.221 [M+] (mol. formula C20H30O3, calcd value 318.2189).
O); 1H-NMR (CD3OD, 500 MHz): Table 2; 13C-NMR (CD3OD, 75 MHz): Table 2; EI-MS m/z (rel. int., %): 318 [M+] (79), 300 (13), 285 (12), 261 (100), 245 (19), 207 (21), 123 (25); HREI-MS m/z 318.221 [M+] (mol. formula C20H30O3, calcd value 318.2189).
11β,17β-Dihydroxy-17α-methyl-5α-androsta-1-ene-3-one (6)
White solid: yield (percentage yield): 56 mg (5.6%); mp: 234–236 °C; [α]25D = −49.2 (c = 0.0025, CH3OH); UV (MeOH) λmax 230 nm (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε 5.3); IR (KBr) νmax cm−1: 3414 (O–H stretching), 2942 (C–H stretching), 1662 (C
ε 5.3); IR (KBr) νmax cm−1: 3414 (O–H stretching), 2942 (C–H stretching), 1662 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–C
C–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1H-NMR (CD3OD, 400 MHz): Table 2; 13C-NMR (CD3OD, 100 MHz): Table 2; EI-MS m/z (rel. int., %): 318 [M+] (3), 300 (9), 267 (15), 242 (100), 227 (54), 215 (29), 161 (23); HREI-MS m/z 318.2238 (mol. formula C20H30O3, calcd value 318.2195). Single-crystal X-ray diffraction data: empirical formula = C20H30O3, crystal system: monoclinic; space group: P21; unit cell dimensions: a = 6.513(4) Å, b = 18.564(15) Å, c = 14.116(7) Å, α = γ = 90°, β = 92.67(6)°, volume 1704.7(19) Å3, Z = 2, ρ calc = 1.241 mg m−3, F(000): 696, crystal size: 0.41 × 0.18 × 0.17 mm, θ range for data collection: 3.13 to 50.02°. Total 6714 reflections were collected, out of which 3240 reflections were judged observed (Rint = 0.0605). Final R indices were R1 = 0.0641 for [I > 2siσ(I)], wR2 = 0.1595, R indices were R1 = 0.0799, wR2 = 0.1669 for all data largest difference peak and hole: 0.351, and −0.336 e. Å−3.
O); 1H-NMR (CD3OD, 400 MHz): Table 2; 13C-NMR (CD3OD, 100 MHz): Table 2; EI-MS m/z (rel. int., %): 318 [M+] (3), 300 (9), 267 (15), 242 (100), 227 (54), 215 (29), 161 (23); HREI-MS m/z 318.2238 (mol. formula C20H30O3, calcd value 318.2195). Single-crystal X-ray diffraction data: empirical formula = C20H30O3, crystal system: monoclinic; space group: P21; unit cell dimensions: a = 6.513(4) Å, b = 18.564(15) Å, c = 14.116(7) Å, α = γ = 90°, β = 92.67(6)°, volume 1704.7(19) Å3, Z = 2, ρ calc = 1.241 mg m−3, F(000): 696, crystal size: 0.41 × 0.18 × 0.17 mm, θ range for data collection: 3.13 to 50.02°. Total 6714 reflections were collected, out of which 3240 reflections were judged observed (Rint = 0.0605). Final R indices were R1 = 0.0641 for [I > 2siσ(I)], wR2 = 0.1595, R indices were R1 = 0.0799, wR2 = 0.1669 for all data largest difference peak and hole: 0.351, and −0.336 e. Å−3.
9α,11β,17β-Trihydroxy-17α-methyl-5α-androstan-3-one (7)
White solid; yield (percentage yield): 7.0 mg (0.7%); mp: 198–200 °C; [α]20D = −7.5 (c = 0.6, CHCl3); IR (KBr) νmax cm−1: 3377 (O–H stretching), 2935 (C–H stretching), 1701 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); IH-NMR (CD3OD, 600 MHz) Table 3; 13C-NMR (CD3OD, 125 MHz) Table 3; EI-MS m/z (rel. int) 336 [M+], 318 (57), 300 (39), 285 (37), 242 (30), 227 (26), 211 (43), 193 (48), 147 (43), 124 (58), 110 (77.6), 95 (62), 55 (65), 43 (100); HREI-MS m/z 336.2298 [M+] (mol. formula C20H32O4, calcd value 336.2301).
O); IH-NMR (CD3OD, 600 MHz) Table 3; 13C-NMR (CD3OD, 125 MHz) Table 3; EI-MS m/z (rel. int) 336 [M+], 318 (57), 300 (39), 285 (37), 242 (30), 227 (26), 211 (43), 193 (48), 147 (43), 124 (58), 110 (77.6), 95 (62), 55 (65), 43 (100); HREI-MS m/z 336.2298 [M+] (mol. formula C20H32O4, calcd value 336.2301).
| Carbon | 7 | 8 | ||
|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 32.8 | 2.01 (overlap), 1.70 (overlap) | 77.0 | 3.77 (dd, J1,2β = 10.8, J1,2α = 6.3) |
| 2 | 38.7 | 2.48 (overlap), 2.22 (overlap) | 47.4 | 2.52 (overlap) |
| 3 | 214.4 | — | 211.2 | — |
| 4 | 45.0 | 2.37 (t, J4α,4β/5 = 14.4), 2.01 (overlap) | 45.5 | 2.37 (t, J4α,4β/5 = 14.4), 1.92 (overlap) |
| 5 | 41.9 | 1.77 (dd, J5,4β = 12.0, J = 2.4) | 43.5 | 1.50 (overlap) |
| 6 | 28.2 | 1.70 (overlap) | 29.7 | 1.48 (overlap) |
| 7 | 27.8 | 1.50 (overlap), 1.30 (overlap) | 32.9 | 1.70 (overlap), 0.98 (overlap) |
| 8 | 34.9 | 2.48 (overlap) | 37.4 | 1.48 (overlap) |
| 9 | 79.3 | — | 61.7 | 0.98 (overlap) |
| 10 | 41.9 | — | 44.5 | — |
| 11 | 69.5 | 3.87, (d, J11,12α = 2.4) | 68.0 | 3.93 (m) |
| 12 | 39.2 | 1.87 (m), 1.70 (overlap) | 42.9 | 1.92 (overlap), 1.38 (overlap) |
| 13 | 46.8 | — | 47.6 | — |
| 14 | 40.7 | 2.22 (overlap) | 50.8 | 1.38 (overlap) |
| 15 | 23.5 | 1.70 (overlap), 1.30 (overlap) | 24.6 | 1.68 (overlap), 1.38 (overlap) |
| 16 | 37.9 | 1.61 (dt, J16α,16β = 13.8, J16α,15α = 2.4), 1.50 (overlap) | 39.2 | 1.92 (overlap), 1.70 (overlap) |
| 17 | 82.3 | — | 81.8 | — |
| 18 | 13.6 | 1.25 (s) | 15.1 | 0.85 (s) |
| 19 | 13.5 | 1.16 (s) | 7.3 | 1.11 (s) |
| 20 | 26.3 | 0.86 (s) | 25.9 | 1.18 (s) |
1β,11α,17β-Trihydroxy-17α-methyl-5α-androstan-3-one (8)
White solid; yield (percentage yield): 9.2 mg (0.92%); mp: 241–242 °C; [α]20D = −30.1 (c 0.024, CH3OH); IR (KBr) νmax cm−1: 3388 (O–H stretching), 2974 (C–H stretching), 1705 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); IH-NMR (CD3OD, 600 MHz) Table 3; 13C-NMR (CD3OD, 125 MHz) Table 3; EI-MS m/z (rel. int) 336 [M+], 318 (57), 300 (39), 285 (37), 242 (30), 227 (26), 180 (19), 155 (72), 137 (39), 121 (86), 109 (49), 95 (41), 55 (40), 43 (100); HREI-MS m/z 336.2296 [M+] (mol. formula C20H32O4, calcd value 336.2301).
O); IH-NMR (CD3OD, 600 MHz) Table 3; 13C-NMR (CD3OD, 125 MHz) Table 3; EI-MS m/z (rel. int) 336 [M+], 318 (57), 300 (39), 285 (37), 242 (30), 227 (26), 180 (19), 155 (72), 137 (39), 121 (86), 109 (49), 95 (41), 55 (40), 43 (100); HREI-MS m/z 336.2296 [M+] (mol. formula C20H32O4, calcd value 336.2301).
Immunomodulatory and cytotoxity assays
The compounds obtained by the microbial transformation of mestanolone (1) were evaluated for their immunomodulatory activity through oxidative burst inhibition assay but no significant activity was observed. The assay was performed according to standard protocol reported in the literature.16MTT assay protocol for cytotoxicity
MTT (3-(4,5-dimethyl thiazol-2yl)-2,5-diphenyl tetrazolium bromide) assay was employed to evaluate the anticancer activity of the compounds against HeLa (human cervical carcinoma ATCC CCl-2), NCl-H460 (human lung carcinoma ATCC HTB-177), and 3T3 (mouse fibroblast normal ATCC CRL-1658) cell lines. The cells were grown in DMEM (containing 100 μg mL−1 streptomycin, 5% of FBS, and 100 IU mL−1 penicillin), and incubated at 37 °C in a 5% CO2. The cells were plated into 96-well plates at a concentration of 1 × 105 cells mL−1, and allowed to stand for overnight. Medium was removed, 200 μL of fresh medium was added, and cells were treated for 24 h with various concentrations of the test compounds ranging between 30–0.2 μM. After treatment with test compounds the cells were incubated with MTT (0.5 mg mL−1) for 4 h at 37 °C, in a 5% CO2 incubator. MTT was removed and 0.1 mL of DMSO was added to each well and mixed by keeping on a stirrer. The presence of viable cells was visualized by the development of purple colour due to the reduction of MTT to formazan crystals. The absorbance was recorded by using micro plate reader (Spectra Max plus, Molecular Devices, CA, USA) at 545 and 570 nm for cancer, and normal cell lines. Cycloheximide was used as positive control for normal cell line, and doxorubicin for cancer cell lines, whereas DMSO was added in the negative control instead of compounds.17 The percent inhibition was calculated by using the following formula:| % inhibition = [100 − {(mean of O. D. of test compound − mean of O. D. of negative control)/(mean of O. D. of positive control − mean of O. D. of negative control) × 100}] |
Results and discussion
Structure elucidation
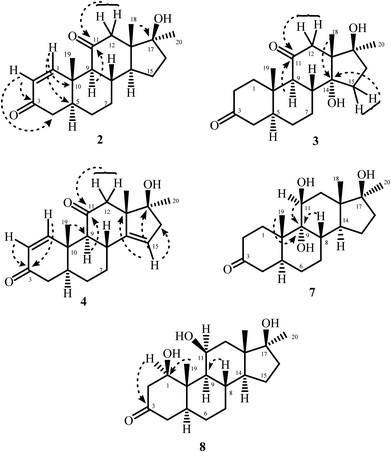
Microbial transformation of anabolic androgenic steroid, mestanolone [(5α,17β)-17-hydroxy-17-methylandrostan-3-one (C20H18O2)] (1), using M. phaseolina and C. blakesleeana is being reported here for the first time. Fermentation of 1 with M. phaseolina yielded five metabolites 2–6 (Fig. 1), while that with C. blakesleeana resulted into metabolites 7, and 8 (Fig. 2).Metabolite 2 showed the M+ at m/z 316.2039 in the HREI-MS, in agreement with the formula C20H28O3 (calcd 316.2033) with seven degrees of unsaturation. The presence of hydroxyl, and ketonic, and α,β-unsaturated carbonyl functionalities were inferred from the peaks at νmax (cm−1) 3490, 1701, and 1659, respectively, in the IR spectrum. The appearance of downfield doublets for the olefinic methine protons at δ 7.57 (J1,2 = 10.5 Hz) and 5.78 (J2,1 = 10.5 Hz) was observed in the 1H-NMR spectrum (Table 1). This suggested the presence of a double bond between C-1 and C-2, conjugated with C-3 ketonic carbonyl. Furthermore, the downfield shift of C-12 methylene protons at δ 2.47 (overlapped), and 2.13 (overlapped) suggested oxidation at C-11 position. In the 13C-NMR spectrum (Table 1) two new olefinic carbons appeared at δ 162.3 and 127.7, along with a new quaternary carbon at δ 213.3, which suggested the introduction of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and a ketonic carbon, respectively. The double bond was placed between C-1 and C-2 on the basis of HMBC correlations of H-1 (δ 7.45) with the ketonic C-3 (δ 202.0), C-5 (δ 45.5), and C-10 (δ 35.8). H-2 (δ 5.79) showed HMBC correlations with the C-3 ketonic carbon (δ 202.0), and C-4 (δ 41.4), which suggested an α,β-unsaturation. This was further supported by UV absorbance at λmax 230 nm. The position of new ketonic group (δ 213.3) was inferred from its HMBC correlations with H-9 (δ 2.17), H2-12 (δ 2.12, 2.47), and H-18 (δ 0.77) suggested a carbonyl group at C-11 (Fig. 3). NOESY correlations showed that the stereochemistry of metabolite 2 was retained as in substrate 1. Thus the structure of the new compound 2 was deduced as 17β-hydroxy-17α-methyl-5α-androsta-1-ene-3,11-dione.
C and a ketonic carbon, respectively. The double bond was placed between C-1 and C-2 on the basis of HMBC correlations of H-1 (δ 7.45) with the ketonic C-3 (δ 202.0), C-5 (δ 45.5), and C-10 (δ 35.8). H-2 (δ 5.79) showed HMBC correlations with the C-3 ketonic carbon (δ 202.0), and C-4 (δ 41.4), which suggested an α,β-unsaturation. This was further supported by UV absorbance at λmax 230 nm. The position of new ketonic group (δ 213.3) was inferred from its HMBC correlations with H-9 (δ 2.17), H2-12 (δ 2.12, 2.47), and H-18 (δ 0.77) suggested a carbonyl group at C-11 (Fig. 3). NOESY correlations showed that the stereochemistry of metabolite 2 was retained as in substrate 1. Thus the structure of the new compound 2 was deduced as 17β-hydroxy-17α-methyl-5α-androsta-1-ene-3,11-dione.
The HREI-MS of metabolite 3 showed the M+ at m/z 334.2140, was consistent with the formula C20H30O4 (calcd 334.2144). A 30 a.m.u. increase in mass as compared to the substrate 1 (m/z 334), suggested the addition of two oxygen atoms with the loss of two hydrogen atoms. The IR absorptions at νmax (cm−1) 3523 and 3490 were due to OH groups. The peak at 1692 cm−1 was due to the presence of a ketonic carbonyl group. In the 1H-NMR spectrum (Table 1), C-12 methylene protons appeared downfield at δ 1.88 (overlapped), and a doublet at δ 2.93 (d, J12a/e = 12.0 Hz), suggesting an oxidation at C-11. C-15 methylene protons appeared at δ 1.83 (multiplet) and 1.70 (overlapped), which suggested a change in the chemical environment at vicinal position. In the 13C-NMR spectrum (Table 1), two additional quaternary carbon signals appeared at δ 214.6 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), and 83.4 (C–OH). The HMBC correlations of H-9 (δ 2.32) and H2-12 (δ 1.88, 2.93) with the ketonic carbon (δ 214.6) indicated its presence at the C-11 position (Fig. 3). The position of new OH group was deduced through the HMBC correlations of H2-15 (δ 1.83, 1.70) and H-18 (δ 0.85) with the quaternary carbon (δ 83.4), which indicated hydroxylation at C-14 (Fig. 3). The OH-14 (δ 3.70) displayed NOESY correlation with the α-oriented H-9 (δ 2.32) (acetone-d6), suggesting its α-orientation (Fig. 4). NOESY correlations showed that the stereochemistry of metabolite 3 was retained as in substrate 1. The structure of the new metabolite 3 was thus deduced as 14α,17β-dihydroxy-17α-methyl-5α-androstan-3,11-dione.
O), and 83.4 (C–OH). The HMBC correlations of H-9 (δ 2.32) and H2-12 (δ 1.88, 2.93) with the ketonic carbon (δ 214.6) indicated its presence at the C-11 position (Fig. 3). The position of new OH group was deduced through the HMBC correlations of H2-15 (δ 1.83, 1.70) and H-18 (δ 0.85) with the quaternary carbon (δ 83.4), which indicated hydroxylation at C-14 (Fig. 3). The OH-14 (δ 3.70) displayed NOESY correlation with the α-oriented H-9 (δ 2.32) (acetone-d6), suggesting its α-orientation (Fig. 4). NOESY correlations showed that the stereochemistry of metabolite 3 was retained as in substrate 1. The structure of the new metabolite 3 was thus deduced as 14α,17β-dihydroxy-17α-methyl-5α-androstan-3,11-dione.
Metabolite 4 showed the M+ at m/z 314.1862 in the HREI-MS, supporting the formula C20H26O3 (calcd 314.1876), consistent with eight degrees of unsaturation. In the IR spectrum, the peaks at νmax (cm−1) 3460, 1708, and 1658 indicated the presence of hydroxyl, ketonic, and α,β-unsaturated carbonyl groups, respectively. Appearance of the olefinic doublets at δ 7.45 (J1,2 = 10.0 Hz) and 5.79 (J2,1 = 10.5 Hz) in the 1H-NMR spectrum suggested the presence of a double bond between C-1 and C-2 in ring-A conjugated with the C-3 ketonic carbonyl moiety. The 1H-NMR spectrum (Table 2) showed that C-12 methylene protons appeared as two sharp doublets at δ 2.09 (J12a/e = 12.0 Hz) and 2.56 (J12e/a = 12.0 Hz), which suggested an oxidation at C-11 position. The appearance of a double doublet at δ 5.45 (J15/16a = 4.5 Hz, J15/16e = 2.0 Hz) suggested a double bond between C-14 and C-15 in ring-D. The 13C-NMR spectrum (Table 2) also showed three olefinic signals at δ 161.6, 127.8, and 119.7, and two quaternary carbon signals at δ 150.0 and 212.6, indicating the presence of two double bonds, and a ketonic carbonyl group, respectively. COSY and HMBC techniques were used to deduce the final structure of metabolite 4. The presence of an α,β-unsaturated carbonyl was deduced from UV-visible spectrum (λmax 230 nm). Protons H-1 (δ 7.45) and H-2 (δ 5.79) showed HMBC correlations with the ketonic carbon (δ 202.0) and C-10 (δ 39.4) also indicating the presence of an α,β-unsaturated carbonyl moiety in ring-A. H-9 (δ 2.19) and H2-12 (δ 2.09, 2.56) showed HMBC correlations with the ketonic carbon (δ 212.6), and H-19 (δ 1.31) showed correlation with C-9 (δ 60.5). This indicated position of the carbonyl at C-11. The position of the other C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond between C-14 and C-15 was deduced on the basis of HMBC correlations of H-15 (δ 5.45) with C-13 (δ 55.6), C-16 (δ 46.9), and C-17 (δ 82.0 (Fig. 3). COSY correlations between C-16 methylene (δ 2.38, 2.52) and olefinic protons (δ 5.45) further supported a double bond between C-14 and C-15 in ring-D. Thus the new compound was identified as 17β-hydroxy-17α-methyl-5α-androsta-1,14-diene-3,11-dione.
C bond between C-14 and C-15 was deduced on the basis of HMBC correlations of H-15 (δ 5.45) with C-13 (δ 55.6), C-16 (δ 46.9), and C-17 (δ 82.0 (Fig. 3). COSY correlations between C-16 methylene (δ 2.38, 2.52) and olefinic protons (δ 5.45) further supported a double bond between C-14 and C-15 in ring-D. Thus the new compound was identified as 17β-hydroxy-17α-methyl-5α-androsta-1,14-diene-3,11-dione.
Metabolite 7 was obtained as a white solid. The molecular formula C20H32O4 was deduced through the HREI-MS, which displayed the M+ at m/z 336.2299 (calcd 336.2301). The 32 a.m.u. increase in molecular weight could be attributed to the addition of two oxygen atoms as hydroxyl groups. The presence of the hydroxyl groups was also inferred from the IR spectrum at νmax (cm−1) 3377, and 3350. A new methine proton signal at δ 3.87 (d, J11,12α = 2.4 Hz) (1H-NMR), and a methine carbon at δ 69.5, and a quaternary carbon signal at δ 79.3 (13C-NMR) suggested a dihydroxylation in mestanolone (1) (Table 3). COSY and HMBC correlations were used to deduce the positions of the newly introduced OH groups. H2-12 (δ 1.87, and 1.70) showed HMBC correlations with the newly formed methine carbon at δ 69.5 (C-11). COSY cross-peaks with the newly formed methine proton at δ 3.87 further supported the hydroxylation at C-11 (Fig. 3). The second OH was placed at C-9 on the basis of HMBC correlations of H3-19 (δ 1.16), and H-8 (δ 2.48) with C-9 (δ 79.3). H-11 (δ 3.87) also displayed HMBC correlations with C-9, which further supported an OH at C-9. H-11 was found to be α-oriented as it showed its NOESY correlation with H-1 (δ 2.22) (Fig. 4). This suggested that the geminal OH-11 has a β-orientation. H-11 appeared as a doublet (J11,12α = 2.4 Hz) which also supported a β-orientation (axial) of C-11 OH. The OH at C-9 was found to be α-oriented, as deduced from the NOESY correlations of OH-9 (δ 4.36) with H-14 (δ 2.23) (acetone-d6) (Fig. 4). Thus the new metabolite 7 was identified as 9α,11β,17β-trihydroxy-17α-methyl-5α-androstan-3-one.
The physical appearance of metabolite 8 was a white amorphous solid. The HREI-MS showed the [M+] at m/z 336.2299 (calcd 336.2301, C20H32O4). The 32 a.m.u. increase in mass than the substrate 1 suggested the addition of two oxygen atoms as OH. The IR absorptions at 3388, and 3370 cm−1 were due to OH groups. The 1H-NMR spectrum (Table 3) displayed signals for two new downfield methine protons at δ 3.93 (m), and 3.77 (dd, J1,2β = 10.8 Hz, J1,2α = 6.3 Hz, H-1) with their corresponding carbons at δ 68.0 (C-11), and 77.0 (C-1), respectively, in the 13C-NMR spectrum (Table 3). This further suggested dihydroxylation in substrate 1. The OH groups were placed on the basis of HMBC and COSY correlations. The C-12 methylene protons (δ 1.92, and 1.38) displayed COSY correlations with the newly formed methine proton at δ 3.93. These correlations indicated that one of the hydroxyl groups was at C-11. The H-9 (δ 0.98) showed HMBC correlations with C-11, and COSY correlations with H-11, thus placing a hydroxyl at C-11. The H-9 also showed HMBC correlations with the new downfield methine carbon at δ 77.0 (C-1). The CH3-19 (δ 1.11) displayed HMBC correlations with carbon at δ 77.0, suggested that the second OH was located at C-1 (Fig. 3). The newly appeared methine proton at δ 3.77 showed NOESY cross-peaks with H-5 (δ 1.50), and H-9 (δ 0.98). As H-5 and H-9 are α-oriented in substrate 1, therefore H-1 was also assumed to be α-oriented. Thus the geminal OH at C-1 was deduced to be β-oriented. H-11 (δ 3.93) showed NOESY correlations with H-8 (δ 1.48), H-18 (δ 0.85), and H-19 (δ 1.11). As these protons are β-oriented in the substrate mestanolone (1), therefore the resonance of α-OH was inferred at C-11. The structure of the new metabolite 8 was thus deduced as 1β,11α,17β-trihydroxy-17α-methyl-5α-androstan-3-one.
Metabolites 5, and 6 were characterized as known metabolites by comparing their spectroscopic data with the previously reported data in literature. These metabolites were identified as 17β-hydroxy-17α-methyl-5α-androstan-3,11-dione (5), and 11β,17β-dihydroxy-17α-methyl-5α-androsta-1-ene-3-one (6). Metabolite 5 was previously reported by Davitishvili et al. through chemical modification of 3α-hydroxy-5α-androst-9(11)-en-17-one.18 Metabolite 6 was also reported in the literature through chemical modification of androstan-17β-ol-3,11-dione.19
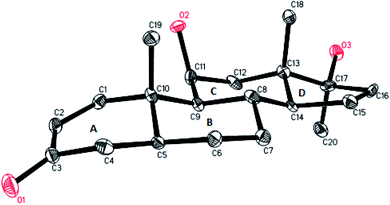
The structure of metabolite 6 was unambiguously deduced through the single-crystal X-ray diffraction techniques. X-ray diffraction studies showed that the molecule consists of four fused rings, rings A (C-1–C-5/C-10), B (C-5–C-10), C (C-8–C-9/C-11–C-14), and D (C-13–C-17). The six membered ring A exists in a pseudo chair conformation, while trans-fused rings B and C exist in chair conformation. trans-fused ring D exists in an envelope conformation. The hydroxyl group at C-11 was found to be β-oriented (Fig. 5). The single-crystal X-ray diffraction data was submitted to the Cambridge Crystallographic Data Collection with CCDC 1532897.
Cytotoxic activity
Mestanolone (1) and its metabolites 2–3, and 5–8 were evaluated for their cytotoxic activity against H460, and HeLa cancer, and 3T3 normal cell lines. Compounds 1, 2 and 6 showed moderate activity against HeLa cancer cell line (human cervical carcinoma) with IC50 values of 27.6 ± 1.1, 19.2 ± 2.9, and 12.8 ± 0.6 μM, respectively, as compared to the standard drug, doxorubicin (IC50 = 1.2 ± 0.4 μM). All test compounds were found to be non-cytotoxic against 3T3 (mouse fibroblast) (Table 4) and NCI-H460 cell line (human lung carcinoma) cell lines. Compounds 1, 2, and 6 showed selective cytotoxicity to HeLa cancer cell line. These results are significant, as the compounds only inhibited the growth of HeLa cell line, and were found to be non-cytotoxic to 3T3 normal cell line. The fundamental issue in all anti-cancer drugs is their cytotoxicity to normal cells. Therefore, the selective activity of these compounds to only cancer cells makes them as suitable leads for further studies.| Compounds | HeLa Cell line (Cancer cell line) IC50 ± SD [μM] | H460 Cell line (Cancer cell line) IC50 ± SD [μM] | 3T3 Cell line (Normal cell line) IC50 ± SD [μM] |
|---|---|---|---|
| 1 | 27.6 ± 1.1 | >30 | >30 |
| 2 | 19.2 ± 2.9 | >30 | >30 |
| 3 | >30 | >30 | >30 |
| 5 | >30 | >30 | >30 |
| 6 | 12.8 ± 0.6 | >30 | >30 |
| 7 | >30 | >30 | >30 |
| 8 | >30 | >30 | >30 |
| Standard drug, doxorubicin (chemotherapy medicine) | 1.2 ± 0.4 | 0.8 ± 0.03 | — |
| Standard, cycloheximide | — | — | 0.8 ± 0.2 |
Conclusion
In conclusion, biotransformation of mestanolone (1) with Macrophomina phaseolina and Cunninghamella blakesleeana, yielded five new metabolites 2–4, 7, and 8 along with two known metabolites 5–6. During these transformations, dehydrogenation at C-1/C-2, C-14/C-15, and oxidation at C-11, hydroxylations at C-1, C-9, C-11, and C-14 were observed. C-6, C-10, and C-11 were the sites for β-hydroxylation, whereas α-hydroxylation at C-9, C-11, and C-14 occurred. All the compounds were inactive in immunomodulatory assay. Substrate 1, and metabolites 2 and 6 were found to be active against HeLa cancer cell line, whereas all other compounds were found to be inactive. The tested compounds were found to be inactive against H460 cancer, and 3T3 normal cell lines.Conflicts of interest
Authors has no conflicts of interest.Acknowledgements
SY and MIC acknowledge the financial support of the OPCW (Organization for the Prohibition of Chemical Weapons) through a research project entitled, “Structure and biological studies on new anabolic steroids obtained by biotransformation”.References
- P. Zhang, H. H. Huang and D. F. Zhong, Acta Pharmacol. Sin., 2006, 27, 1097–1102 CrossRef PubMed.
- H. L. Holland and H. K. Weber, Curr. Opin. Biotechnol., 2000, 11, 547–553 CrossRef PubMed.
- H. L. Holland, Stereoselective hydroxylation reactions, Stereoselective biocatalysis, Marcel Dekker, New York, NY, 2000, pp. 131–152 Search PubMed.
- K. B. Borges, W. D. S. Borges, R. Duran-Patron, M. T. Pupo, P. S. Bonato and I. G. Collado, Tetrahedron: Asymmetry, 2009, 20, 385–397 CrossRef.
- S. M. Resnick, D. S. Torok, K. Lee, J. M. Brand and D. T. Gibson, Appl. Environ. Microbiol., 1994, 60, 3323–3328 Search PubMed.
- W. J. Jones, C. S. Mazur, J. F. Kenneke and A. W. Garrison, Environ. Sci. Technol., 2007, 41, 8301–8307 CrossRef PubMed.
- T. Dong, G. W. Wu, X. N. Wang, J. M. Gao, J. G. Chen and S. S. Lee, J. Mol. Catal. B: Enzym., 2010, 67, 251–256 CrossRef.
- Y. W. Wong and P. J. Davis, Pharm. Res., 1989, 6, 982–987 CrossRef.
- M. S. Ahmad, S. Zafar, M. Bibi, S. Bano, Atia-tul-Wahab, Atta-ur-Rahman and M. I. Choudhary, Steroids, 2014, 82, 53–59 CrossRef PubMed.
- R. Pappo and C. J. Jung, Tetrahedron Lett., 1962, 3, 365–371 CrossRef.
- S. K. Ginotra, B. S. Chhikara, M. Singh, R. Chandra and V. Tandon, Chem. Pharm. Bull., 2004, 52, 989–991 CrossRef PubMed.
- M. Y. Mohammad, S. G. Musharraf, A. M. Al-Majid, Atta-ur-Rahman and M. I. Choudhary, Biocatal. Biotransform., 2013, 31, 153–159 CrossRef.
- E. Baydoun, Atia-tul-Wahab, H. Mehmood, M. S. Ahmad, C. Smith and M. I. Choudhary, Steroids, 2016, 105, 121–127 CrossRef PubMed.
- E. Baydoun, M. Karam, Atia-tul-Wahab, M. S. A. Khan, M. S. Ahmad, Samreen, C. Smith, R. Abdel-Massih and M. I. Choudhary, Steroids, 2014, 88, 95–100 CrossRef PubMed.
- M. I. Choudhary, N. T. Khan, S. G. Musharraf and S. Anjum, Steroids, 2007, 72, 923–929 CrossRef PubMed.
- Z. Hussain, N. Dastagir, S. Hussain, A. Jabeen, S. Zafar, R. Malik, S. Bano, A. Wajid and M. I. Choudhary, Steroids, 2016, 112, 68–73 CrossRef PubMed.
- M. D. Fernando, L. C. R. Wijesundera, P. Soysa, D. De Silva and C. Nanayakkara, BMC Complementary Altern. Med., 2015, 15, 1–9 CrossRef PubMed.
- M. G. Davitishvili, R. G. Karpenko, O. F. Malyutina, L. N. Alekseeva and G. S. Grinenko, Khim.-Farm. Zh., 1988, 22, 1121–1125 Search PubMed.
- H. J. Ringold and R. George, Δ-Androsten-17β-ol-3-one having an 11β-hydroxy or an 11-oxo group as well as 17-esters of these derivatives and 17α-lower alkyl derivatives thereof, US Pat. no. 2888474A, 1959.
Footnote |
| † Electronic supplementary information (ESI) available: 1H, 13C NMR, HSQC, COSY, NOESY and MS data of all products. CCDC 1532897. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8ra01309h |
| This journal is © The Royal Society of Chemistry 2018 |