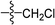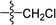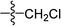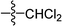Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Design, microwave-assisted synthesis, bioactivity and SAR of novel substituted 2-phenyl-2-cyclohexanedione enol ester derivatives†
Fei Ye,
Peng Ma,
Yue Zhai,
Fei Yang,
Shuang Gao,
Li-Xia Zhao and
Ying Fu *
*
Department of Applied Chemistry, College of Science, Northeast Agricultural University, Harbin, 150030, P. R. China. E-mail: fuying@neau.edu.cn
First published on 29th May 2018
Abstract
Based on the structure–activity relationship and active substructure combination, a novel class of substituted 2-phenyl-2-cyclohexanedione enol ester derivatives was designed for use as potential herbicide safeners. A microwave-assisted synthetic route was developed for the substituted 2-phenyl-2-cyclohexenone enol ester derivatives via coupling and acylation reactions. In the modified protocol, the reactions were performed under microwave irradiation, resulting in significant improvements in the yields and reaction times. All of the structures were characterized using IR, 1H NMR, 13C NMR and HRMS spectroscopies. The bioassay results demonstrated that most of these compounds could alleviate clethodim injury to maize. Molecular docking modeling showed that the potential antagonism between compound 3(S24) and clethodim plays a key role in the metabolism of herbicides. This paper presents a new safener candidate for maize protection.
Introduction
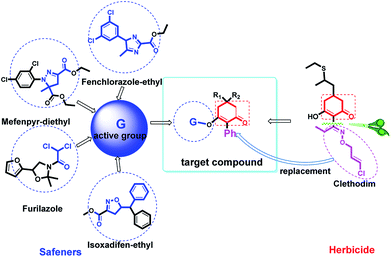
Clethodim, an acetyl CoA carboxylase (ACCase) inhibitor that belongs to the chemical family of cyclohexenone herbicides, specifically targets the carboxylase-transferase (CT) domain of the plastidic ACCase, which inhibits fatty acid biosynthesis and ultimately causes crop death.1,2 Cyclohexenone herbicides are toxic to non-target organisms that also contain this enzyme.3,4 Clethodim has been shown to cause albinism in canola via the inhibition of fatty acid biosynthesis, which not only causes dramatic crop yield losses but also die-off. As a consequence, the continuous development of new safeners to protect crops is urgently required to expand the application and increase the safety of herbicides.It is widely accepted that broad-spectrum herbicides can be combined with a safener for crop protection and efficient weed management. The safener induces the degradation of the herbicides only in the crops, not in the weeds.5 This effect is connected with the increased expression of gene coding for the enzymes responsible for herbicide degradation in crops, such as the cytochrome P450 monooxygenases (CYP), glutathione S-transferases (GST) and ATP-binding cassette (ABC) transporters.6–9 For example, isoxadifen-ethyl exhibited safening activity to protect corn against injury caused by the herbicide foramsulfuron by inducing CYP or glycosyl transferase activity.10–12 Several studies have reported that the safener isoxadifen-ethyl enhances GST activity and herbicide tolerance.13,14 A new era in safener research began with the discovery of 1,2,4-triazolcarboxylates and fenchlorazole-ethyl was developed as a post-emergence safener for wheat against the ACCase inhibitor fenoxaprop-ethyl.15 Similarly, the dihydropyrazol dicarboxylate mefenpyr-diethyl has been used against ACCase inhibitors, including fenoxaprop herbicides.16
The structure–activity relationship (SAR), an important measure for novel agrochemical discovery, has been introduced into many research works focused on the search for bioactive compounds.17 Stephenson et al. showed that compounds that share structural similarities to the thiocarbamate herbicides were found to be highly active antidotes for those herbicides in corn.18 Sulfamide compounds have been used as safeners to protect plants from the injury caused by sulfonylurea herbicides. Cyprosulfamide, designed based on the SAR, could protect plants from the injury caused by thiencarbazone-methyl.19 Safeners, having structural features closely resembling those of herbicides, may interfere in the metabolism of herbicides that protect crops.20
In recent years, the active substructure combination has become an effective method for improving the hit of compounds. The safener benzhydryloxy-acetic acid combined with the potential safener 5-phenyl-4,5-dihydroisoxazole-3-ethyl ester led to the production of the strong rice safener isoxadifen-ethyl.21 A class of new isoquinolinium-like compounds and novel chiral succinate dehydrogenase inhibitors was also designed by utilizing the active substructure combination theory, and they exhibited excellent and broad spectrum antifungal activity.22,23 In connection with the facts mentioned above and our earlier work on the design and synthesis of N-containing heterocyclic safeners,24–26 herein substituted 2-phenyl-2-cyclohexanedione enol ester derivatives were designed based on the SAR and active substructure combination (Scheme 1). Greenhouse experiments demonstrated that some of them exhibited promising safener activity for protecting crops from injury by clethodim.
Experimental
Chemicals and instruments
Infrared (IR) spectra were recorded on FTIR-8400S and ALPHA-T infrared spectrophotometers as KBr pellets. The nuclear magnetic resonance (NMR) spectra were recorded on Bruker AVANVE 400 MHz and Bruker AVANVE 600 MHz spectrometers, using CDCl3 as the solvent and TMS as the internal standard. The melting points were determined on a Beijing Taike melting point apparatus (X-4) and were uncorrected. The mass spectra were obtained by a Xevo TQ spectrometer. Microwave activation was carried out using an XH-100A focused microwave (2450 MHz, 1000 W, 40 L, normal pressure, Beijing Xiang Hu Sci. and Tech., Beijing, NC, China). All reagents used were of analytical grade (purchased from Aladdin Industrial Inc., Shanghai, China).General procedure for the preparation of substituted 3-hydroxy-2-phenylcyclohex-2-en-1-one derivatives, 2
To dimethylsulfoxide (DMSO, 50 ml) kept at 90 °C under microwave irradiation (800 W), iodobenzene (1.02 g, 5 mmol), L-proline (0.12 g, 1 mmol) and CuI (0.10 g, 0.5 mmol) were added. Substituted 1,3-cyclohexanedione 1 (15 mmol) was added via syringe. After 40 min, the organic phase was dried over anhydrous Na2SO4 and the solvent was removed by vacuum distillation. The pure products were obtained by column chromatography on silica gel eluted with petroleum ether and ethyl acetate (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1554 (C
O), 1554 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1176 (C–O).
C), 1176 (C–O).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1557 (C
O), 1557 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1137 (C–O).
C), 1137 (C–O).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1581 (C
O), 1581 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1133 (C–O).
C), 1133 (C–O).General procedure for the preparation of substituted 2-phenyl-2-cyclohexanedione enol ester derivatives, 3
To CH2Cl2 (20 ml) maintained at 0 °C, intermediate 2(a–c) and Et3N (1.5 mmol) were added. After five minutes of stirring, acyl chloride (1.2 mmol) was added via syringe. The solution was warmed to room temperature over 1 hour. The reaction mixture was extracted with CH2Cl2 and filtered. The organic layer was dried over anhydrous Na2SO4 and the CH2Cl2 was evaporated under vacuum. The pure products were obtained by recrystallization or column chromatography on silica gel eluting with petroleum ether and ethyl acetate (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–9
1–9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1587 (C
O), 1587 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1234 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic 329.1256, found 329.1148.
C), 1234 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic 329.1256, found 329.1148.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1599 (C
O), 1599 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1257 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 321.1412, found 321.1485.
C), 1257 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 321.1412, found 321.1485.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1572 (C
O), 1572 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1256 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 383.0320, found 383.0212.
C), 1256 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 383.0320, found 383.0212.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1571 (C
O), 1571 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1256 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 397.0476, found 397.0369.
C), 1256 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 397.0476, found 397.0369.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1571 (C
O), 1571 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1256 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 411.0633, found 411.0525.
C), 1256 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 411.0633, found 411.0525.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1598 (C
O), 1598 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1232 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 265.7042, found 265.0626.
C), 1232 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 265.7042, found 265.0626.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1645 (C
O), 1645 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1126 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 301.0710, found 301.0602.
C), 1126 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 301.0710, found 301.0602.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1598 (C
O), 1598 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1141 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 293.0866, found 293.0939.
C), 1141 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 293.0866, found 293.0939.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1576 (C
O), 1576 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1257 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 327.0476, found 327.0549.
C), 1257 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 327.0476, found 327.0549.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH2–O), 2.73–2.77 (t, 2H, J = 6.2 Hz, CH2), 2.63–2.66 (t, 2H, J = 6.9 Hz, CH2), 2.17–2.24 (m, 2H, CH2); 13C NMR (100 MHz, CDCl3): 197.5, 165.9, 164.2, 157.3, 131.1, 131.0, 129.7 (2C), 129.6 (2C), 128.1 (2C), 128.0, 122.0, 114.4 (2C), 64.8, 37.5, 28.9, 20.6; IR (KBr, cm−1): 3034–2852 (C–H), 1767, 1653 (C
C–CH2–O), 2.73–2.77 (t, 2H, J = 6.2 Hz, CH2), 2.63–2.66 (t, 2H, J = 6.9 Hz, CH2), 2.17–2.24 (m, 2H, CH2); 13C NMR (100 MHz, CDCl3): 197.5, 165.9, 164.2, 157.3, 131.1, 131.0, 129.7 (2C), 129.6 (2C), 128.1 (2C), 128.0, 122.0, 114.4 (2C), 64.8, 37.5, 28.9, 20.6; IR (KBr, cm−1): 3034–2852 (C–H), 1767, 1653 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1586 (C
O), 1586 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1124 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 345.1405, found 345.1097.
C), 1124 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 345.1405, found 345.1097.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH2–O), 2.71–2.73 (m, 1H, CH2), 2.67–2.69 (m, 1H, CH2), 2.53–2.60 (m, 1H, CH2), 2.47–2.51 (m, 1H, CH), 2.33–2.40 (m, 1H, CH2), 1.19–1.20 (d, 3H, J = 9.6 Hz, CH3). 13C NMR (150 MHz, CDCl3): 197.5, 166.0, 163.6, 157.4, 131.0, 130.5, 129.7 (2C), 129.6 (2C), 128.1 (2C), 128.0, 122.0, 114.4 (2C), 64.8, 45.7, 36.9, 28.3, 20.8; IR (KBr, cm−1): 3035–2850 (C–H), 1770, 1665 (C
C–CH2–O), 2.71–2.73 (m, 1H, CH2), 2.67–2.69 (m, 1H, CH2), 2.53–2.60 (m, 1H, CH2), 2.47–2.51 (m, 1H, CH), 2.33–2.40 (m, 1H, CH2), 1.19–1.20 (d, 3H, J = 9.6 Hz, CH3). 13C NMR (150 MHz, CDCl3): 197.5, 166.0, 163.6, 157.4, 131.0, 130.5, 129.7 (2C), 129.6 (2C), 128.1 (2C), 128.0, 122.0, 114.4 (2C), 64.8, 45.7, 36.9, 28.3, 20.8; IR (KBr, cm−1): 3035–2850 (C–H), 1770, 1665 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1586(C
O), 1586(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1132 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 359.1362, found 359.1254.
C), 1132 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 359.1362, found 359.1254.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH2–O), 2.63 (s, 2H, CH2), 2.52 (s, 2H, CH2), 1.22 (s, 6H, –(CH3)2). 13C NMR (150 MHz, CDCl3): 197.4, 166.1, 162.6, 157.3, 130.9, 129.9, 129.6 (4C), 128.2 (2C), 128.0, 122.0, 114.4 (2C), 64.7, 51.4, 42.7, 32.6, 28.2 (2C); IR (KBr, cm−1) ν: 3033–2843 (C–H), 1758, 1666 (C
C–CH2–O), 2.63 (s, 2H, CH2), 2.52 (s, 2H, CH2), 1.22 (s, 6H, –(CH3)2). 13C NMR (150 MHz, CDCl3): 197.4, 166.1, 162.6, 157.3, 130.9, 129.9, 129.6 (4C), 128.2 (2C), 128.0, 122.0, 114.4 (2C), 64.7, 51.4, 42.7, 32.6, 28.2 (2C); IR (KBr, cm−1) ν: 3033–2843 (C–H), 1758, 1666 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1585 (C
O), 1585 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1137 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 351.1518, found 351.1519.
C), 1137 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 351.1518, found 351.1519.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH2–O), 2.74–2.78 (t, 2H, J = 8.2 Hz, CH2), 2.60–2.65 (t, 2H, J = 8.9 Hz, CH2), 2.14–2.23 (m, 2H, CH2), 2.12 (s, 3H, O
C–CH2–O), 2.74–2.78 (t, 2H, J = 8.2 Hz, CH2), 2.60–2.65 (t, 2H, J = 8.9 Hz, CH2), 2.14–2.23 (m, 2H, CH2), 2.12 (s, 3H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH3); 13C NMR (100 MHz, CDCl3): 197.5, 170.0, 164.9, 164.1, 130.9, 130.6, 129.5 (2C), 128.0 (2C), 127.9, 60.2, 37.5, 28.8, 20.6, 20.2; IR (KBr, cm−1): 3058–2893 (C–H), 1755, 1680 (C
C–CH3); 13C NMR (100 MHz, CDCl3): 197.5, 170.0, 164.9, 164.1, 130.9, 130.6, 129.5 (2C), 128.0 (2C), 127.9, 60.2, 37.5, 28.8, 20.6, 20.2; IR (KBr, cm−1): 3058–2893 (C–H), 1755, 1680 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1599 (C
O), 1599 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1150 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 311.0998, found 311.0890. 3(S14).
C), 1150 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 311.0998, found 311.0890. 3(S14).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH2–O), 2.71–2.73 (m, 1H, CH2), 2.67–2.69 (m, 1H, CH2), 2.55–2.62 (m, 1H, CH2), 2.46–2.51 (m, 1H, CH), 2.31–2.38 (m, 1H, CH2), 2.13 (s, 3H, O
C–CH2–O), 2.71–2.73 (m, 1H, CH2), 2.67–2.69 (m, 1H, CH2), 2.55–2.62 (m, 1H, CH2), 2.46–2.51 (m, 1H, CH), 2.31–2.38 (m, 1H, CH2), 2.13 (s, 3H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH3), 1.18–1.20 (d, 3H, J = 9.7 Hz, CH3); 13C NMR (150 MHz, CDCl3): 197.6, 170.1, 165.0, 163.6, 130.8, 130.2, 129.5 (2C), 128.1 (2C), 128.0, 60.2, 45.7, 36.8, 28.4, 20.8, 20.3; IR (KBr, cm−1): 3034–2851 (C–H), 1742, 1667 (C
C–CH3), 1.18–1.20 (d, 3H, J = 9.7 Hz, CH3); 13C NMR (150 MHz, CDCl3): 197.6, 170.1, 165.0, 163.6, 130.8, 130.2, 129.5 (2C), 128.1 (2C), 128.0, 60.2, 45.7, 36.8, 28.4, 20.8, 20.3; IR (KBr, cm−1): 3034–2851 (C–H), 1742, 1667 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1143 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 325.1154, found 325.1046.
O), 1143 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 325.1154, found 325.1046.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH2–O), 2.62 (s, 2H, CH2), 2.49 (s, 2H, CH2), 2.11 (s, 3H, O
C–CH2–O), 2.62 (s, 2H, CH2), 2.49 (s, 2H, CH2), 2.11 (s, 3H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH3), 1.20 (s, 6H, –(CH3)2); 13C NMR (100 MHz, CDCl3): 197.5, 170.1, 165.1, 162.5, 130.8, 129.6, 129.5 (2C), 128.1 (2C), 128.0, 60.2, 51.4, 42.5, 32.6, 28.1 (2C), 20.3; IR (KBr, cm−1): 3058–2872 (C–H), 1756, 1680 (C
C–CH3), 1.20 (s, 6H, –(CH3)2); 13C NMR (100 MHz, CDCl3): 197.5, 170.1, 165.1, 162.5, 130.8, 129.6, 129.5 (2C), 128.1 (2C), 128.0, 60.2, 51.4, 42.5, 32.6, 28.1 (2C), 20.3; IR (KBr, cm−1): 3058–2872 (C–H), 1756, 1680 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1587 (C
O), 1587 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1145 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 339.1311, found 339.1203.
C), 1145 (C–O); HRMS (ESI): m/z [M + Na+] calcd for monoisotopic mass 339.1311, found 339.1203.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1647 (C
O), 1647 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1173 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 543.9478 found, 543.9551.
C), 1173 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 543.9478 found, 543.9551.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1542 (C
O), 1542 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1171 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 557.9634, found 557.9707.
C), 1171 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 557.9634, found 557.9707.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1639 (C
O), 1639 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1185 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 571.9791, found 571.9864.
C), 1185 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 571.9791, found 571.9864.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH3), 2.06–2.22 (m, 2H, CH2); 13C NMR (100 MHz, CDCl3): 197.5, 177.3, 164.3, 162.5, 158.6, 131.5, 130.9, 129.9, 129.5 (2C), 129.2 (2C), 128.1 (2C), 128.1 (2C), 128.0, 127.7, 107.0, 37.4, 29.0, 20.7, 13.5; IR (KBr, cm−1): 3058–2871 (C–H), 1739, 1679 (C
C–CH3), 2.06–2.22 (m, 2H, CH2); 13C NMR (100 MHz, CDCl3): 197.5, 177.3, 164.3, 162.5, 158.6, 131.5, 130.9, 129.9, 129.5 (2C), 129.2 (2C), 128.1 (2C), 128.1 (2C), 128.0, 127.7, 107.0, 37.4, 29.0, 20.7, 13.5; IR (KBr, cm−1): 3058–2871 (C–H), 1739, 1679 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1595 (C
O), 1595 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1142 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 374.1314, found 374.1387.
C), 1142 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 374.1314, found 374.1387.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH3), 2.30–2.37 (m, 1H, CH2), 1.67 (s, 1H, CH), 1.17–1.19 (d, 3H, J = 9.6 Hz, CH3). 13C NMR (150 MHz, CDCl3): 197.6, 177.4, 163.8, 162,5, 158.7, 131.4, 130.4, 130.0, 129.5 (2C), 129.2 (2C), 128.2 (2C), 128.1 (2C), 128.0, 127.7, 107.0, 45.7, 37.2, 28.5, 20.9, 13.5; IR (KBr, cm−1): 3058–2874 (C–H), 1739, 1681 (C
C–CH3), 2.30–2.37 (m, 1H, CH2), 1.67 (s, 1H, CH), 1.17–1.19 (d, 3H, J = 9.6 Hz, CH3). 13C NMR (150 MHz, CDCl3): 197.6, 177.4, 163.8, 162,5, 158.7, 131.4, 130.4, 130.0, 129.5 (2C), 129.2 (2C), 128.2 (2C), 128.1 (2C), 128.0, 127.7, 107.0, 45.7, 37.2, 28.5, 20.9, 13.5; IR (KBr, cm−1): 3058–2874 (C–H), 1739, 1681 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1595 (C
O), 1595 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1065 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 388.1471, found 388.1543.
C), 1065 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 388.1471, found 388.1543.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH3), 1.21 (s, 6H, –(CH3)2); 13C NMR (150 MHz, CDCl3): 197.5, 177.4, 162.6, 162.5, 158.7, 131.3, 130.0, 129.7, 129.5 (2C), 129.2 (2C), 128.2 (2C), 128.1 (2C), 128.0, 127.7, 107.1, 51.4, 42.8, 32.7, 28.2 (2C), 13.5; IR (KBr, cm−1): 3065–2870 (C–H), 1723, 1657 (C
C–CH3), 1.21 (s, 6H, –(CH3)2); 13C NMR (150 MHz, CDCl3): 197.5, 177.4, 162.6, 162.5, 158.7, 131.3, 130.0, 129.7, 129.5 (2C), 129.2 (2C), 128.2 (2C), 128.1 (2C), 128.0, 127.7, 107.1, 51.4, 42.8, 32.7, 28.2 (2C), 13.5; IR (KBr, cm−1): 3065–2870 (C–H), 1723, 1657 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1597 (C
O), 1597 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1064 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 402.1627, found 402.1700.
C), 1064 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 402.1627, found 402.1700.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH3), 2.12–2.19 (m, 2H, CH2); 13C NMR (150 MHz, CDCl3): 197.6, 177.1 (2C), 161.9, 159.4, 157.7, 155.3, 135.1, 131.9, 131.0, 129.3 (2C), 128.0 (2C), 127.9, 125.3, 116.9, 114.3, 108.5, 37.5, 28.8, 20.7, 13.2; IR (KBr, cm−1): 3059–2850 (C–H), 1726, 1666 (C
C–CH3), 2.12–2.19 (m, 2H, CH2); 13C NMR (150 MHz, CDCl3): 197.6, 177.1 (2C), 161.9, 159.4, 157.7, 155.3, 135.1, 131.9, 131.0, 129.3 (2C), 128.0 (2C), 127.9, 125.3, 116.9, 114.3, 108.5, 37.5, 28.8, 20.7, 13.2; IR (KBr, cm−1): 3059–2850 (C–H), 1726, 1666 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1587 (C
O), 1587 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1071 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 426.0830 found 426.0903.
C), 1071 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 426.0830 found 426.0903.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH3), 2.42–2.46 (m, 1H, CH), 2.27–2.34 (m, 1H, CH2), 1.16–1.17 (d, 3H, J = 4.8 Hz, –CH3); 13C NMR (150 MHz, CDCl3): 197.5, 177.2, 163.4, 159.4, 157.7, 155.3, 135.1, 131.8, 130.9, 130.2, 129.3 (2C), 128.0 (2C), 127.9, 125.3, 116.8, 114.2, 108.5, 45.6, 36.7, 28.6, 20.6, 13.1; IR (KBr, cm−1): 3054–2864 (C–H), 1738, 1679 (C
C–CH3), 2.42–2.46 (m, 1H, CH), 2.27–2.34 (m, 1H, CH2), 1.16–1.17 (d, 3H, J = 4.8 Hz, –CH3); 13C NMR (150 MHz, CDCl3): 197.5, 177.2, 163.4, 159.4, 157.7, 155.3, 135.1, 131.8, 130.9, 130.2, 129.3 (2C), 128.0 (2C), 127.9, 125.3, 116.8, 114.2, 108.5, 45.6, 36.7, 28.6, 20.6, 13.1; IR (KBr, cm−1): 3054–2864 (C–H), 1738, 1679 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1599 (C
O), 1599 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1071 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 440.0987, found 440.1059.
C), 1071 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 440.0987, found 440.1059.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH3), 2.46 (s, 2H, CH2), 1.18 (s, 6H, –(CH3)2); 13C NMR (150 MHz, CDCl3): 196.9, 177.1, 162.7, 159.3 (2C), 155.5, 135.1, 131.8, 130.8, 129.4, 129.3 (2C), 128.0 (2C), 127.7, 125.2, 116.9, 114.2, 108.5, 51.6, 42.2, 32.8, 27.8 (2C), 13.1; IR (KBr, cm−1): 3084–2872 (C–H), 1738, 1680 (C
C–CH3), 2.46 (s, 2H, CH2), 1.18 (s, 6H, –(CH3)2); 13C NMR (150 MHz, CDCl3): 196.9, 177.1, 162.7, 159.3 (2C), 155.5, 135.1, 131.8, 130.8, 129.4, 129.3 (2C), 128.0 (2C), 127.7, 125.2, 116.9, 114.2, 108.5, 51.6, 42.2, 32.8, 27.8 (2C), 13.1; IR (KBr, cm−1): 3084–2872 (C–H), 1738, 1680 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1600 (C
O), 1600 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1071 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 454.1143, found 454.1216.
C), 1071 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 454.1143, found 454.1216.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1540 (C
O), 1540 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1168 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 365.1035, found 365.1108.
C), 1168 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 365.1035, found 365.1108.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1542 (C
O), 1542 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1171 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 379.1191, found 379.1264.
C), 1171 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 379.1191, found 379.1264.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–N), 7.14–7.35 (m, 5H, Ar–H), 3.92 (s, 3H, N–CH3), 2.72 (s, 2H, CH2), 2.52 (s, 2H, CH2), 1.24 (s, 6H, –(CH3)2); 13C NMR (150 MHz, CDCl3): 197.6, 162.7, 157.1, 142.5, 136.8, 131.2, 129.7, 129.6 (2C), 127.9 (2C), 127.7, 111.4, 100.0, 51.5, 42.8, 39.9, 32.7 (2C), 28.2; IR (KBr, cm−1): 3133–2873 (C–H), 1749, 1677 (C
CH–N), 7.14–7.35 (m, 5H, Ar–H), 3.92 (s, 3H, N–CH3), 2.72 (s, 2H, CH2), 2.52 (s, 2H, CH2), 1.24 (s, 6H, –(CH3)2); 13C NMR (150 MHz, CDCl3): 197.6, 162.7, 157.1, 142.5, 136.8, 131.2, 129.7, 129.6 (2C), 127.9 (2C), 127.7, 111.4, 100.0, 51.5, 42.8, 39.9, 32.7 (2C), 28.2; IR (KBr, cm−1): 3133–2873 (C–H), 1749, 1677 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1543 (C
O), 1543 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1146 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 393.1348, found 393.1421.
C), 1146 (C–O); HRMS (ESI): m/z [M + H+] calcd for monoisotopic mass 393.1348, found 393.1421.X-ray diffraction
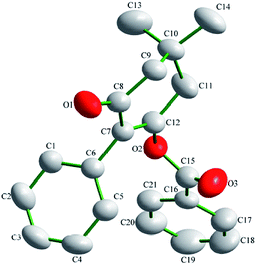
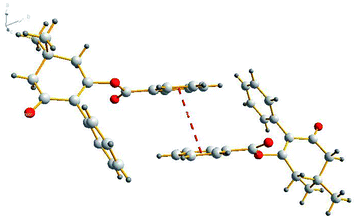
Suitable single-crystals of compound 3(S3) were recrystallized from ethanol. The molecular structure of compound 3(S3) is shown in Fig. 1. The π–π stacking interaction is shown in Fig. 2. The X-ray data were collected on a Rigaku R-AXIS RAPID diffractometer with graphite-monochromator Mo Kα radiation (λ = 0.071073 nm) at 293(2) K. A total of 6888 reflections were measured, of which 3054 independent reflections (Rint = 0.0233) were obtained in the range of 3.14° < θ < 25.00° (h, 9 to 9; k, −10 to 10; l, −15 to 15), and 2357 observed reflections with I > 2σ(I) were used in the refinement on F2. The structure was solved by direct methods using SHELXS-97 and refined by the least-squares procedures on F2 (SHELXL-97) in the full matrix anisotropic approximation for all non-hydrogen atoms.28,29 Symmetry equivalent reflections were used to optimize the crystal shape and size. The crystallographic data has been deposited at the Cambridge Crystallographic Data Centre as supplementary publication number CCDC 1588796.Biological assays
The concentration of the safener and compounds applied in the bioassay was determined after a preliminary screening. Maize seeds were soaked in solutions of synthesized compounds at a concentration of 1 μmol L−1 for 12 h, and the control was soaked in water. Then, the seeds were germinated in incubators for 24 h. The seeds were sown in paper cups (10 cm × 15 cm), with seven seeds per cup, and incubated in an incubator with a 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 h photoperiod, 26.5 ± 1 °C, and 75% relative humidity. The clethodim concentration was set to 10 mL hm−2 to cause injury to the maize. The spraying treatment was conducted when the maize had reached the two-leaf stage. After 6 days, the chlorophyll content of the maize was determined. Each treatment was replicated three times.
12 h photoperiod, 26.5 ± 1 °C, and 75% relative humidity. The clethodim concentration was set to 10 mL hm−2 to cause injury to the maize. The spraying treatment was conducted when the maize had reached the two-leaf stage. After 6 days, the chlorophyll content of the maize was determined. Each treatment was replicated three times.
Computational methods
The three-dimensional structures of compound 3(S24), compound 3(S12), clethodim and cloquitocet-mexyl were constructed using the sketch module of SYBYL-X 2.0.30 Subsequently, the molecules were optimized, and the Gasteiger–Huckel charges were calculated. The crystal structure of ACCase was taken from the Protein Data Bank (PDB ID 3K8X). Docking modeling was achieved using the CDOCKER method in Accelrys Discovery Studio 2.5.31 Before docking, the protein structure was given the CHARMM force field, and water and other co-crystallized small molecules were removed. After the protein preparation, the active site was defined, with a subset region of 13.0 Å from the centre of the known ligand. During the docking process, the top 10 conformations were saved for each ligand based on the −CDOCKER_ENERGY after the energy minimization using the smart minimize method in DS 2.5, and the default values were used for the remaining parameters.Results and discussion
Chemistry
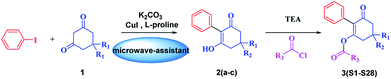
The synthetic route is depicted in Scheme 2. A series of 3-hydroxy-2-phenylcyclohex-2-en-1-one derivatives (2) were synthesized from substituted 1,3-cyclohexanedione (1) and iodobenzene in DMSO under microwave irradiation, the conditions of which were optimized. Initially, the intermediates 2a–c were synthesized by a coupling reaction in CH2Cl2 using L-proline and CuI as the catalyst with 25.6–43.8% yields. This promising result indicated that it was feasible to synthesize 3-hydroxy-2-phenylcyclohex-2-en-1-one through a coupling reaction. Subsequently, we investigated the effects of microwave power, time, base and solvents on the coupling reaction. It was found that microwave power was crucial for this reaction. The reaction cannot proceed with a power less than 200 W. Moreover, the reaction yields constantly improved upon increasing the power, and the yields were highest at 800 W. In addition, the optimum yields were achieved by adjusting the reaction time from 5 to 40 min. Furthermore, the bases Et3N, NaOH and anhydrous K2CO3 were employed as the acid-binding agents, and anhydrous K2CO3 was found to be the best acid-binding agent for this coupling reaction. To further improve the reaction yields, we evaluated the effects of different solvents on the yields. The experimental results indicated that DMSO provided higher yields than CH2Cl2, THF, and 1,4-dioxane due to polar aprotic solvents being the best at promoting this coupling reaction. The optimum conditions were as follows: 800 W as the power, 40 min as the reaction time, and DMSO as the solvent. The yields were higher, and the reaction time was much shorter than that of the reported conventional method32 (Table 1).| Compound | Conventional method | Microwave irradiation method | ||
|---|---|---|---|---|
| Time (h) | Yield (%) | Time (min) | Yield (%) | |
| 2a | 48 h | 64 | 40 min | 79 |
| 2c | 48 h | 35 | 40 min | 88 |
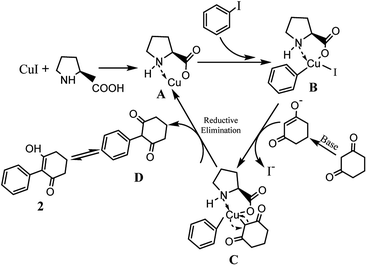
The possible reaction mechanism outlined in Scheme 3 is similar to that reported.33,34 L-proline and Cu(I) formed complex A, and the chelation of L-proline with Cu(I) makes the Cu(I) more reactive towards an oxidative addition and stabilizes the formed intermediate B, facilitating the coupling reaction. Subsequently, the iodine coordinated to the copper was exchanged for the carbanion of the activated methylene group of 1,3-cyclohexanedione, forming the intermediate C via a reductive elimination, releasing the coupling product D and regenerating the active Cu(I) catalyst A.
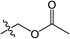
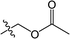
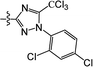
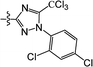
The substituted 2-phenyl-2-cyclohexanedione enol ester derivatives 3 were prepared from the intermediates 2 and different acyl chlorides, with yields of 33–91% (Table 2). Et3N was employed to promote the acylation in the positive direction as an acid-attaching agent.
In general, there were significant effects on the yields of the target products caused by the presence of electron-donating groups. Notably, the presence of a methyl group at the 5-position of the cyclohexanedione increased the yields significantly. Two methyl groups in the 5-position of cyclohexanedione provided better yields than one methyl group, and the yields were lowest when there was no substitution. For example, the yield of 3(S28) was significantly higher than that of 3(S26).
All of the structures were characterized by IR, 1H NMR, 13C NMR and HRMS spectroscopies. The peaks at 1732 and 1677 cm−1 in the IR for compound 3(S3) confirmed the presence of the carbonyl groups. The 1H NMR spectrum also confirmed the proposed structure. The signals at δ 6.88–7.45 ppm were related to the benzene rings. The single signal observed at δ 1.26 ppm was characteristic of the two methyl groups linked to the cyclohexanedione.
Structure analysis
The molecular structure of compound 3(S3) is shown in Fig. 1. Compound 3(S3) contains a cyclohexanedione ring and two benzene rings. Cyclohexanedione adopts a half-chair conformation structure. The cyclohexanedione and benzene ring II [C16, C17, C18, C19, C20 and C21] are not coplanar, with a dihedral angle of 63.803 (62)°. Benzene ring II [C16, C17, C18, C19, C20 and C21] is also not coplanar with benzene ring I [C1, C2, C3, C4, C5 and C6] with a dihedral angle of 62.933 (65)°. The π–π stacking interaction is shown in Fig. 2. Within the double chain, weak π–π packing interactions exist between the two benzene rings. These interactions further reinforce the supramolecular structural stability. No significant hydrogen bonding was found in the crystal structure.Biological activity
The in vivo safener activities of the title compound (1 μmol L−1) against clethodim were evaluated (Table 3). The chlorophyll content was measured after treatment with clethodim for 6 d.| Compound | Chlorophyll content (mg g−1) | Compound | Chlorophyll content (mg g−1) | Compound | Chlorophyll content (mg g−1) |
|---|---|---|---|---|---|
| a Data are the means of three replicates.b Water treated was used as a contrast and the herbicide is clethodim. | |||||
| Clethodim | 14.9 ± 0.3 | 3(S9) | 15.5 ± 0.5 | 3(S19) | 16.3 ± 1.0 |
| Cloquitocet-mexyl | 17.6 ± 0.8 | 3(S10) | 16.3 ± 0.4 | 3(S20) | 16.8 ± 0.6 |
| 3(S1) | 16.7 ± 1.2 | 3(S11) | 15.8 ± 0.9 | 3(S21) | 17.3 ± 0.4 |
| 3(S2) | 15.6 ± 0.9 | 3(S12) | 13.8 ± 1.4 | 3(S22) | 18.2 ± 1.2 |
| 3(S3) | 17.6 ± 1.3 | 3(S13) | 16.3 ± 1.2 | 3(S23) | 19.9 ± 0.7 |
| 3(S4) | 15.1 ± 1.1 | 3(S14) | 14.3 ± 0.6 | 3(S24) | 20.5 ± 0.5 |
| 3(S5) | 15.8 ± 0.3 | 3(S15) | 16.1 ± 0.8 | 3(S25) | 15.3 ± 1.1 |
| 3(S6) | 16.5 ± 0.6 | 3(S16) | 15.7 ± 0.5 | 3(S26) | 16.1 ± 0.6 |
| 3(S7) | 15.5 ± 0.9 | 3(S17) | 16.1 ± 1.1 | 3(S27) | 15.9 ± 0.9 |
| 3(S8) | 16.1 ± 1.2 | 3(S18) | 16.6 ± 1.2 | 3(S28) | 15.3 ± 1.0 |
Photosynthesis is an indispensable physiological activity in the process of plant growth. The chlorophyll content in leaves directly affects photosynthesis sufficiently to promote plant growth and plant nutrient accumulation. Clethodim can provoke an obvious decrease in the chlorophyll content of maize, but significant differences were observed after the initial introduction of the compounds. Compounds 3(S1–S28) showed some recovery of chlorophyll content. Among the compounds tested, compound 3(S24) showed the best activity against the injury of clethodim, even better than that of the commercial safener cloquitocet-mexyl.
From a structural perspective, the introduction of the subunit of 5-methyl-3-phenylisoxazole exhibited excellent safener activity in protecting maize. The 5-position substituents on the cyclohexanedione were further studied. It was noticeable that the presence of a methyl group at the 5-position gave rise to a positive improvement in the safener activity. Two methyl groups at the 5-position provided higher safener activity than one methyl group, and the safener activity was the lowest when there was no substitution. The results indicate that the 5-position substituents were greatly associated with safener activity.
The greenhouse experiments showed that compound 3(S24) exhibited the best safener activity, and compound 3(S12) showed the worst. To prove the hypothesis that safeners may compete for the target site with herbicides, the chemical properties of the compounds 3(S12) and 3(S24), clethodim and cloquitocet-mexyl were compared, such as the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p, aromatic rings, surface area and electronegativity (Table 4). It was observed that the log
p, aromatic rings, surface area and electronegativity (Table 4). It was observed that the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p, aromatic rings, surface area and electronegativity of compound 3(S24) were all similar to those of the safener cloquitocet-mexyl. The properties of compound 3(S12) regarding the log
p, aromatic rings, surface area and electronegativity of compound 3(S24) were all similar to those of the safener cloquitocet-mexyl. The properties of compound 3(S12) regarding the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p, surface area and electronegativity shared a similarity with the herbicide clethodim. This indicated that, in terms of the investigated features and SAR theory, compound 3(S24) may be a lead candidate compound for use as a novel safener, and compound 3(S12) might be a candidate for a potential herbicide.
p, surface area and electronegativity shared a similarity with the herbicide clethodim. This indicated that, in terms of the investigated features and SAR theory, compound 3(S24) may be a lead candidate compound for use as a novel safener, and compound 3(S12) might be a candidate for a potential herbicide.
| Compounds | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pa pa |
Aromatic ringsa | Surface areab | Rotatable bondsb | Molecular weighta | Electronegativityc |
|---|---|---|---|---|---|---|
a The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p, numbers of aromatic rings and molecular weights were predicted by ChemBioOffice 2014.b The surface areas and number of rotatable bonds were predicted by Discovery Studio 2.5.c The electronegativity was predicted by Sybyl-X 2.0 (Tripos Inc., St. Louis, MO). p, numbers of aromatic rings and molecular weights were predicted by ChemBioOffice 2014.b The surface areas and number of rotatable bonds were predicted by Discovery Studio 2.5.c The electronegativity was predicted by Sybyl-X 2.0 (Tripos Inc., St. Louis, MO). |
||||||
| Clethodim | 2.23 | 0 | 363.73 | 9 | 359.90 |  |
| Cloquitocet-mexyl | 4.47 | 2 | 332.46 | 9 | 335.12 |  |
| 3(S24) | 4.59 | 3 | 322.51 | 5 | 424.83 |  |
| 3(S12) | 2.85 | 2 | 400.15 | 6 | 336.38 |  |
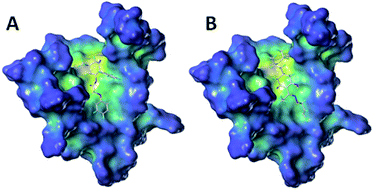
To further study the effect of the safener on the target enzyme of the herbicides, a molecular docking experiment was conducted. It showed that both compound 3(S24) and cloquitocet-mexyl had no interactions with the surrounding residues. Interestingly, the docking score of both compound 3(S24) and cloquitocet-mexyl were negative in value, representing a bad docking result. However, compound 3(S12) and clethodim were well matched to the active site of ACCase (Fig. 3).
Compound 3(S12) and clethodim have a similar binding position at the active site, and this indicated the excellent similarity between the activities of compound 3(S12) and clethodim. As shown in Fig. 4, the ester carbonyl formed two H-bond interactions with Ala1627 and ILE1735 in the protein hinge region. Similarly, the cyclohexanedione group of clethodim generated two H-bond interactions with Ala1627 and ILE1735. Based on the facts mentioned above, the theory of competing with the herbicide at the active site might be ruled out; in connection with the results of greenhouse experiments, we speculated that antagonism potentially existed between compound 3(S24) and clethodim in maize, which protected the maize from injury by clethodim.
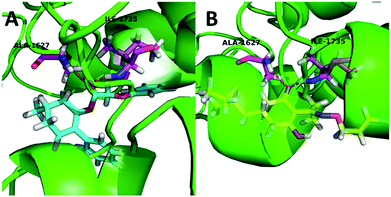 | ||
| Fig. 4 The receptor–ligand interaction of 3(S12) and clethodim with the active site of ACCase, and 3(S12) and clethodim are shown in cyan and yellow, respectively. | ||
Conclusion
In conclusion, a novel class of substituted 2-phenyl-2-cyclohexanedione enol ester derivatives were designed and synthesized based on the SAR and active substructure combination. The intermediate 3-hydroxy-2-phenylcyclohex-2-en-1-one derivatives were synthesized using microwave irradiation, and the reaction conditions were optimized. The bioactivity results demonstrated that most of the compounds showed excellent safener activity; compound 3(S24) displayed inspired safener activity in protecting maize from clethodim injury comparable to that of the commercial safener cloquitocet-mexyl. Molecular docking experiments revealed that the promising safener potency may be ascribed to the antagonism between the compounds 3(S24) and clethodim. The present results provide a powerful complement to the SAR in designing bioactive molecules. Thus, substituted 2-phenyl-2-cyclohexanedione enol ester derivatives have emerged as novel lead compounds for developing new safener agents.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
We are very grateful to the National Nature Science Foundation of China (31572042), the Natural Science Foundation of Heilongjiang Province (ZD2017002), and the Research Science Foundation in Technology Innovation of Harbin (2015RAXXJ032). The authors are grateful to Prof. Jia-Zhong Li (Lanzhou University) for assistance with the molecular docking analyses.References
- Q. Zhou, X. He, F. Liu, Y. Li, C. Chen and Q. Peng, J. Mol. Liq., 2017, 244, 521–527 CrossRef.
- L. S. Shergill, J. Malone and P. Boutsalis, Pest Manage. Sci., 2017, 73, 1638–1647 CrossRef PubMed.
- M. Laforest, B. Soufiane, M. J. Simard, K. Obeid, E. Page and R. E. Nurse, Pest Manage. Sci., 2017, 73, 2227–2235 CrossRef PubMed.
- Y. Sasaki and Y. Nagano, Biosci., Biotechnol., Biochem., 2004, 68, 1175–1184 CrossRef PubMed.
- M. T. Elmore, J. T. Brosnan, G. R. Armel, J. J. Vargas and G. K. Breeden, Weed Technol., 2015, 29, 550–560 CrossRef.
- A. B. Mcquarters and M. W. Wolf, Angew. Chem., 2014, 126, 4846–4848 CrossRef.
- D. E. Riechers, K. Kreuz and Q. Zhang, Plant Physiol., 2010, 153, 3–13 CrossRef PubMed.
- P. Jeschke, Pest Manage. Sci., 2016, 72, 433–455 CrossRef PubMed.
- R. Busi, N. K. Nguyen, B. S. Chauhan, F. Vidotto, M. Tabacchi and S. B. Powles, Pest Manage. Sci., 2016, 73, 71–77 CrossRef PubMed.
- M. M. Williams II and J. K. Pataky, Weed Sci., 2010, 58, 289–294 CrossRef.
- J. A. Bunting, C. L. Sprague and D. E. Riechers, Weed Sci., 2004, 52, 711–717 CrossRef.
- A. Paporisch and B. Rubin, Pestic. Biochem. Physiol., 2017, 138, 22–28 CrossRef PubMed.
- I. Cummins, D. P. Dixon, S. Freitag-Pohl, M. Skipsey and R. Edwards, Drug Metab. Rev., 2011, 43, 266–280 CrossRef PubMed.
- A. C. Cataneo, L. C. Ferreira, M. M. Mischan, E. D. Velini, N. Corniani and A. L. Cerdeira, Planta Daninha, 2013, 31, 387–393 CrossRef.
- I. Jablonkai, Herbicide Safeners: Effective Tools to Improve Herbicide Selectivity, ed. A. J. Price and J. A. Kelton, InTech, Croatia, 2013, ch. 23, pp. 589–620 Search PubMed.
- H. Kraehmer, B. Laber, C. Rosinger and A. Schulz, Plant Physiol., 2014, 166, 1119–1131 CrossRef PubMed.
- B. D. Del, L. Scarponi and L. Espen, Phytochemistry, 2007, 68, 2614–2624 CrossRef PubMed.
- G. R. Stephenson and F. Y. Chang, Comparative activity and selectivity of herbicide antidotes, ed. F. Pallos, Elsevier Inc., Canada, 1978, pp. 35–61 Search PubMed.
- Y. Zheng, B. Liu, Z. Gou, Y. Liu, X. Zhang and Y. Wang, Bioorg. Med. Chem., 2015, 25, 791–794 CrossRef PubMed.
- H. Yu, Y. Cheng, M. Xu, Y. Q. Song, Y. M. Luo and B. Li, J. Agric. Food Chem., 2016, 64, 9586–9591 CrossRef PubMed.
- H. Ahrens, G. Lange, T. Muller, C. Rosinger, L. Willms and A. van Almsick, Angew. Chem., Int. Ed., 2013, 52, 9388–9398 CrossRef PubMed.
- Z. Hou, L. F. Zhu, X. C. Yu, M. Q. Sun, F. Miao and L. Zhou, J. Agric. Food Chem., 2016, 64, 2847–2854 CrossRef PubMed.
- S. K. Li, D. D. Li, T. F. Xiao, S. S. Zhang, Z. H. Song and H. Y. Ma, J. Agric. Food Chem., 2016, 64, 8927–8934 CrossRef PubMed.
- Y. Fu, J. Y. Wang, D. Zhang, Y. F. Chen, S. Gao, L. X. Zhao and F. Ye, Molecules, 2017, 22, 1601 CrossRef PubMed.
- F. Ye, C. Wang, P. Ma, L. X. Zhao, S. Gao and Y. Fu, J. Heterocycl. Chem., 2018, 55, 335–341 CrossRef.
- Y. Fu, M. X. Wang, D. Zhang, Y. W. Hou, S. Gao and L. X. Zhao, RSC Adv., 2017, 7, 46858–46865 RSC.
- A. Goldblum and R. Mechoulam, J. Chem. Soc., Perkin Trans. 1, 1977, 8, 1889–1898 RSC.
- G. M. Sheldrick, SHELXS-97, Program for X-ray Crystal Structure Solution, University of Göttingen, Göttingen, Germany Search PubMed.
- G. M. Sheldrick, SHELXS-97, Program for X-ray Crystal Structure Refinement, University of Göttingen, Göttingen, Germany Search PubMed.
- Sybyl, Version 6.9, Tripos Inc., St. Louis, MO Search PubMed.
- Catalyst, Version 4.10, Accelrys Inc., San Diego, CA, USA, 2005 Search PubMed.
- C. S. Reddy, I. Khan and H. W. Lam, Angew. Chem., Int. Ed., 2012, 51, 12115–12119 CrossRef PubMed.
- E. Sperotto, G. P. van Klink, K. G. Van and J. G. de Vries, Dalton Trans., 2010, 39, 10338–10351 RSC.
- F. Monnier and M. Taillefer, Angew. Chem., 2009, 48, 6954–6971 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: CCDC 1588796. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8ra02647e |
| This journal is © The Royal Society of Chemistry 2018 |