Open Access Article
Open Access Article5-Selenocyanato and 5-trifluoromethanesulfonyl derivatives of 2′-deoxyuridine: synthesis, radiation and computational chemistry as well as cytotoxicity†
Samanta Makurat‡
a,
Magdalena Zdrowowicz‡a,
Lidia Chomicz-Mańka‡a,
Witold Kozak‡a,
Illia E. Serdiukb,
Paweł Wityka,
Alicja Kaweckaa,
Marta Sosnowskaa and
Janusz Rak *a
*a
aDepartment of Chemistry, University of Gdańsk, Wita Stwosza 63, 80-308 Gdańsk, Poland. E-mail: janusz.rak@ug.edu.pl
bFaculty of Mathematics, Physics and Informatics, University of Gdańsk, Wita Stwosza 57, 80-308 Gdańsk, Poland
First published on 12th June 2018
Abstract
5-Selenocyanato-2′-deoxyuridine (SeCNdU) and 5-trifluoromethanesulfonyl-2′-deoxyuridine (OTfdU) have been synthesized and their structures have been confirmed with NMR and MS methods. Both compounds undergo dissociative electron attachment (DEA) when irradiated with X-rays in an aqueous solution containing a hydroxyl radical scavenger. The DEA yield of SeCNdU significantly exceeds that of 5-bromo-2′-deoxyuridine (BrdU), remaining in good agreement with the computationally revealed profile of electron-induced degradation. The radiolysis products indicate, in line with theoretical predictions, Se–CN bond dissociation as the main reaction channel. On the other hand, the DEA yield for OTfdU is slightly lower than the degradation yield measured for BrdU, despite the fact that the calculated driving force for the electron-induced OTfdU dissociation substantially overpasses the thermodynamic stimulus for BrdU degradation. Moreover, the calculated DEA profile suggests that the electron attachment induced formation of 5-hydroxy-2′-deoxyuridine (OHdU) from OTfdU, while 2′-deoxyuridine (dU) is mainly observed experimentally. We explained this discrepancy in terms of the increased acidity of OTfdU resulting in efficient deprotonation of the N3 atom, which brings about the domination of the OTfdU(N3–H)− anion in the equilibrium mixture. As a consequence, electron addition chiefly leads to the radical dianion, OTfdU(N3–H)˙2−, which easily protonates at the C5 site. As a result, the C5–O rather than O–S bond undergoes dissociation, leading to dU, observed experimentally. A negligible cytotoxicity of the studied compounds toward the MCF-7 cell line at the concentrations used for cell labelling calls for further studies aiming at the clinical use of the proposed derivatives.
1. Introduction
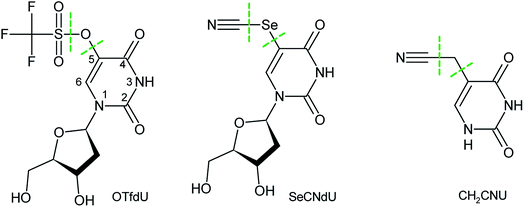
Cancer is the main cause of death in highly developed countries.1 Three essential methods of defeating it are surgery, chemotherapy and radiotherapy, with the latter being most commonly used.2 However, heavy hypoxia occurring in solid tumours, which accounts for 80% of the developed cases of cancer,3 decreases the efficacy of radiotherapy.4 Hence, in order to increase the efficiency of this modality and to minimize dangerous side effects of irradiation, including secondary cancer,5 radiosensitizers – molecules that are selectively transported to cancer cells and make them susceptible to ionizing radiation (IR) – have to be employed in radiotherapy.6Hall and Giaccia7,8 distinguished only two categories of radiosensitizers that might be employed in clinical treatment. According to their classification these two groups comprise the substances, which increase the sensitivity of hypoxic tumour cells toward ionizing radiation and the modified nucleosides that are incorporated into DNA during its biosynthesis.
In hypoxic cancer cells water radiolysis leads to the same amount of hydrated electrons and ˙OH radicals.9 To harness solvated electrons, which are inactive toward native DNA,10 2′-deoxyuridine derivatives substituted with an electrophilic residue in the C5-position might be used for DNA damage.11 Beside the necessity of undergoing enzymatic triphosphorylation and incorporation into DNA by enzymatic machinery of a cell, these derivatives should possess high electron affinity and should undergo dissociative electron attachment (DEA) that leads to reactive radicals inside the genome. The two most comprehensively studied examples of such type of 2′-deoxyuridines are 5-bromo-2′-deoxyuridine (BrdU) and 5-iodo-2′-deoxyuridine (IdU).2 In numerous works, it was demonstrated that the DEA process in the BrdU/IdU labelled oligonucleotides leads to the highly reactive uracil-5-yl radical that ultimately produces intra- and interstrand crosslinks,12–14 single strand breaks,15,16 alkali-labile lesions as well as induces base releasing in DNA.17
Even though a number of reports shows that BrdU and IdU are efficiently incorporated into the cellular DNA, they have not found any use for clinical practice so far.18 Therefore, it seems necessary to search for new modified nucleosides with radiosensitizing properties. There are two requirements that have to be considered to design an efficient radiosensitizer being a modification of dU: a presence of an electrophilic substituent in the 5-position of the nucleobase ring, which increases the electron affinity of the nucleoside, and the ability of the considered derivative to undergo a facile electron-induced dissociation. In our previous studies we have not only developed a method to propose a new potent radiosensitizer that consider the mentioned above requirements but also proposed several derivatives worth of further experimental studies.11,19
Prior to expensive and time-consuming DNA labelling20,21 and/or experiments on the labelled cells' response to IR, the chosen nucleoside has to be synthesized and its propensity to be damaged by ionizing radiation in an aqueous solution has to be confirmed. We have already synthesized 5-selenocyanatouracil (SeCNU), that was selected using our computational approach and proved that it undergoes efficient DEA, where mainly the Se–CN bond is broken due to the attachment of an electron. The calculated thermodynamic stimulus for the DEA degradation of SeCNU is significantly larger than that for BrU and, in accordance with this computational characteristic, the experimental yield of degradation turned out to be ca. 1.5 fold larger for SeCNU than for BrU.22
Since a nucleoside rather than a nucleobase is used for DNA labelling, we decided to verify whether the sugar moiety influences the radiosensitizing properties of SeCNU. Hence, in the current work we synthesized 5-selenocyanato-2′-deoxyuridine (SeCNdU) and determined its susceptibility to be damaged by IR in aqueous solution. We also carried out another radiolytic studies for a modified 2′-deoxyuridine derivative that was indicated by our computational model,11,19 namely for 5-trifluoromethanesulfonyl-2′-deoxyuridine (OTfdU). The latter compound has been chosen since it is characterized by an exceptionally large thermodynamic stimulus for the DEA process, leading from the OTfdU˙− radical anion to the 5-thio-2′-deoxyuridine radical. In order to confirm the accuracy of our computational model we also did the radiolysis of 5-cyanomethyluracil which according to the density functional calculations does not undergo DEA in an aqueous solution.
Both SeCNdU and OTfdU are characterized by the increased electron affinity and large thermodynamic stimuli for DEA. The radiolytic behaviour of SeCNdU is in line with the computational prediction. However, to our surprise, the decomposition yield of OTfdU was much lower than the one anticipated from the DEA driving force, while the products of radiolysis indicated the C5–O bond break rather than the dissociation of O–S bond indicated by the computational analysis. We interpreted those discrepancies with the increased pKa value of OTfdU due to the presence of the triflate residue with an extremely strong and dominant negative inductive effect23 and swift protonation of the OTfdU(N3–H)˙2− electron adduct, respectively. Finally, the cellular MTT test, carried out on the MCF-7 breast cancer cell line, proved no cytotoxicity of the studied derivatives at concentrations typical for cell labelling.
2. Methods
2.1. Experimental
1H NMR (Bruker AVANCE III, 500 MHz, CD3OD), δ: 8.72 (s, 1H), 6.25 (t, 1H), 4.40–4.46 (m, 1H), 3.99 (q, 1H), 3.87 (dd, 1H), 3.79 (dd, 1H), 2.33–2.41 (m, 1H), 2.23–2.32 (m, 1H); 13C NMR (125 MHz, DMSO), δ: 161.1, 150.4, 147.7, 104.5, 97.5, 88.2, 85.6, 70.4, 61.2, 40.8. HRMS (TripleTOF 5600+, SCIEX), m/z: [M − H]− calcd for C7H8F3N2O5 331.9863, found 331.9293; UV spectrum (water), λmax: 272 nm. For the synthesis scheme and NMR spectrum see Scheme S1, Fig. S7 and S8 in ESI.†
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (4
water (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The mixture was stirred for 3 h at room temperature. The solvents were evaporated in vacuo. The crude product was purified with semi-preparative HPLC using the same conditions described above. The resulting product was obtained as a white solid (40 mg) in a 24% yield.
1). The mixture was stirred for 3 h at room temperature. The solvents were evaporated in vacuo. The crude product was purified with semi-preparative HPLC using the same conditions described above. The resulting product was obtained as a white solid (40 mg) in a 24% yield.1H NMR (Bruker AVANCE III, 500 MHz, CD3OD), δ: 8.76 (s, 1H), 6.27 (t, 1H), 4.40–4.46 (m, 1H), 3.96–4.00 (m, 1H), 3.84 (dd, 1H), 3.77 (dd, 1H), 2.33–2.41 (m, 1H), 2.21–2.30 (m, 1H); 13C NMR (125 MHz, DMSO), δ: 158.0, 149.4, 135.2, 126.8, 118.6 (q), 88.1, 85.8, 70.0, 61.0, 40.8. HRMS (TripleTOF 5600+, SCIEX), m/z: [M − H]− calcd for C7H8F3N2O5 375.0188, found 375.0390; UV spectrum (water), λmax: 268 nm. For the synthesis scheme and NMR spectra see Scheme S2, Fig. S9 and S10 in ESI.†
1H NMR (Bruker AVANCE III, 500 MHz, CD3OD), δ: 8.21 (s, 1H), 3.43 (s, 3H), 3.34 (s, 3H); 13C NMR (125 MHz, CD3OD), δ: 158.2, 150.6, 138.5, 125.7, 119.9, 36.2, 27.4. HRMS (TripleTOF 5600+, SCIEX), m/z: [M + H]+ calcd for C7H8F3N2O5 289.0100, found 289.0009; UV spectrum (40% ACN in water), λmax: 271 nm. For the synthesis scheme and NMR spectra see Scheme S3, Fig. S11 and S12 in ESI.†
1H NMR (Bruker AVANCE III, 500 MHz, CD3OD), δ: 7.52 (s, 1H), 3.47 (s, 2H); 13C NMR (125 MHz, CD3OD), δ: 163.9, 151.9, 140.0, 117.0, 103.8, 14.3. HRMS (TripleTOF 5600+, SCIEX), m/z: [M − H]− calcd for C6H4N3O2 149.9833, found 150.0309); UV spectrum (water), λmax: 271 nm. For the synthesis scheme and NMR spectra see Scheme S4, Fig. S13 and S14 in ESI.†
In case of SeCNdU and OTfdU the linear gradient of 0–35% phase B in 30 min was used (mobile phase A: 0.1% formic acid and B: 80% ACN) and for BrdU and CH2CNU, separations were performed under isocratic conditions (mobile phase A: 0.1% formic acid, 2% ACN in water).
Chromatographic conditions were as follows: Kinetex column (Phenomenex, 1.7 μm, C18, 100 Å, 2.1 × 150 mm); flow rate of 0.2 mL min−1; the mobile phase A consisted of 0.1% HCOOH and deionized water, and the mobile phase B consisted of the same concentration of HCOOH and ACN in water (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, v/v); 10 μL injection of each sample was loaded onto the column and separated using the following gradient conditions (time [min], % mobile phase B): (0, 0) (2, 0) (30, 80); the column temperature was maintained at 20 °C. The effluent was diverted to waste for 2 min after injection during each analysis.
20, v/v); 10 μL injection of each sample was loaded onto the column and separated using the following gradient conditions (time [min], % mobile phase B): (0, 0) (2, 0) (30, 80); the column temperature was maintained at 20 °C. The effluent was diverted to waste for 2 min after injection during each analysis.
MS and MS/MS operation parameters were as follows: the spray voltage was −4.0 kV, the nebulizer gas (N2) pressure was 25 psi, the flow rate was 11 L min−1 and the source temperature was 300 °C. Each spectrum was obtained by averaging 3 scans and the time of each scan was 0.25 s.
Briefly, the MCF-7 cells were grown in RPMI medium supplemented with 10% FBS (fetal bovine serum) and 100 U mL−1 of antibiotics (penicillin and streptomycin). Cells were seeded at a density of 4 × 103 per well of 96-well plate and allowed to attach for 24 h. After this time, the medium was replaced with fresh medium and cells were treated with increasing concentration of SeCNdU and OTfdU (10−9 to 2 × 10−4 M). A concentrated stock solutions of compounds were prepared in sterile water. Cells were incubated with the studied analogs for a period of 24 and 48 h. Than the MTT salt (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) solution in concentration of 4 mg mL−1 was added to each well. After 4 h of incubation, medium was gently aspirated, and 200 μL of DMSO was added to dissolve the formazan crystals. The absorbance was measured at 570 nm (with reference wavelength 660 nm) using EnSpire (PerkinElmer) microplate reader. The results were analyzed with the use of GraphPad Prism software. The statistical evaluation of treated samples and untreated control was calculated using one-way analysis of variance (ANOVA) followed by Dunnett's multiple comparison test. The data were obtained from three independent experiments and each treatment condition assayed in triplicate. The differences were considered significant at P < 0.05.
2.2. Computational
The M06-2X26 functional with the 6-31++G(d,p) basis set27 have been employed to estimate the electron dissociative attachment profiles for the systems depicted in Fig. 1. Additionally, the B3LYP28–30 and ωB97XD31 functionals as well as the second-order Møller–Plesset method (MP2)32 were used for the estimation of DEA profile in OTfdU for which the M06-2X results did not agree with the outcome of radiolysis. Moreover, to simulate an aqueous environment, the polarized continuum model (PCM)33 was employed.The lowest energy structures for the DEA profile calculations resulted from the conformational scan on neutral geometries shown in Fig. 1. All stationary geometries on the DEA profiles have been fully optimized without geometrical constraints, and the analysis of harmonic frequencies proved that all of them are either geometrically stable (all force constants positive) or first-order saddle points (all but one force constants positive). In order to verify that the obtained transition state (TS) geometries connect the correct reactants, the intrinsic reaction coordinate (IRC)34 calculations have been performed for every saddle-point structure.
The energies of stationary geometries are expressed in terms of free energies. The latter were determined by correcting the values of electronic energies for zero-point vibration terms, thermal contributions to energy, the pV term and the entropy term. These terms were calculated in the rigid rotor-harmonic oscillator approximation35 for T = 298 K and p = 1 atm. Such methodology was successfully applied before.19,36–38
All calculations have been performed with the Gaussian 09 package39 while GaussView 5.0 was used for visualization.40
3. Results and discussion
3.1. Theoretical DEA profiles
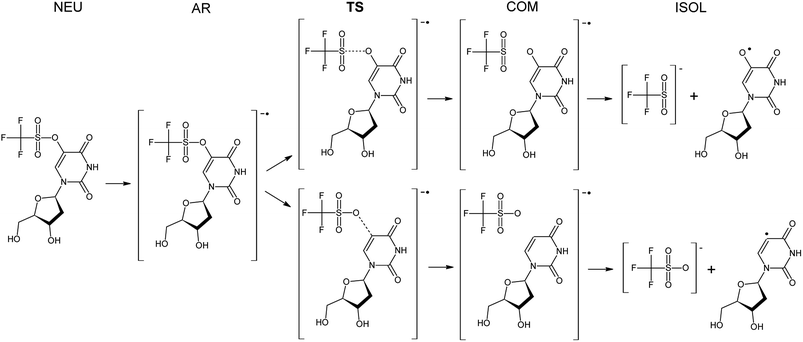
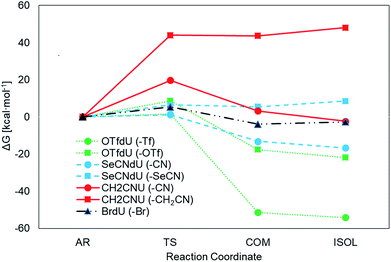
The Gibbs free energy changes along the pathways of DEA for the considered compounds (see Fig. 2 for the exemplar DEA for OTfdU) were calculated as the difference between particular stationary states (ΔG, shown in Fig. 3 and Table 1). Thus, the DEA profile comprises the following stationary geometries: neutral molecule (NEU), anion radical (AR) that forms as a result of electron attachment, transition state (TS) for DEA transformation and product complex (COM) with the respective bond broken. Additionally the isolated product (ISOL) was calculated for the fragments separated to infinity. In order to fully characterize the DEA process, also adiabatic electron affinities (AEA) are shown in Table 1. These values were calculated as the difference between Gibbs free enthalpy of the neutral molecules and their corresponding anion radicals in fully optimized geometries.| ΔG* | ΔGAR→COM | ΔGNEU→ISOL | AEA [eV] | ||
|---|---|---|---|---|---|
| OTfdU | Path A (–Tf) | 1.5 | −51.4 | −116.1 | 2.7 |
| Path B (–OTf) | 8.6 | −17.6 | −83.8 | ||
| SeCNdU | Path A (–CN) | 1.2 | −13.2 | −76.6 | 2.6 |
| Path B (–SeCN) | 6.4 | 5.5 | −51.5 | ||
| CH2CNU | Path A (–CN) | 19.7 | 3.2 | −55.23 | 2.3 |
| Path B (–CH2CN) | 44.0 | 43.7 | −4.9 | ||
| BrdU | –Br | 5.3 | −3.9 | −58.9 | 2.4 |
The results depicted in Fig. 3 and Table 1 clearly suggest that both OTfdU and SeCNdU are potent radiosensitizers, which easily undergo DEA process. It should result in bond-breakage in the substituent (–SeCN or –OTf) for the most favored pathways (path A for SeCNdU and OTfdU, see Table 1). DEA for CH2CNU, on the other hand, is connected with high kinetic barrier of 19.7 and 44.0 kcal mol−1 for path A and B, respectively and, in both cases, with unfavorable thermodynamic stimuli of 3.2 kcal mol−1 (path A) and 43.7 kcal mol−1 (path B). The OTfdU derivative most likely dissociates to ˙OdU and Tf− (ΔG* = 1.5 kcal mol−1), while dU˙ and OTf− products formation is connected with a higher kinetic barrier (8.6 kcal mol−1) and is less favored thermodynamically. Similar situation occurs for SeCNdU, where the transition state is connected to a small kinetic barrier for the CN− release (ΔG* = 1.2 kcal mol−1) and significantly larger for SeCN− production (ΔG* = 6.4 kcal mol−1); the latter reaction channel is also unfavored thermodynamically (ΔGAR→COM = 5.5 kcal mol−1). The results discussed for SeCNdU can be compared with those obtained by Rak et al.22 for SeCNU. They studied the DEA process at the G3B3 level and received activation barrier for the release of the CN− and SeCN− anions equal to 1.2 kcal mol−1 and 6.2 kcal mol−1, respectively, which strongly suggests that sugar moiety does not influence the DEA-induced degradation of the modified nucleosides.
3.2. Radiolysis
In order to check the efficiency of electron induced degradation of the studied analogs and to establish the end products of this process, we carried out radiolysis of the aqueous solutions of SeCNdU, OTfdU and CH2CNU and compared its outcome with the results obtained for the well-known radiosensitizer – BrdU. The solutions exposed to X-rays containing the ˙OH radical scavenger (to study only the effect of solvated electrons) were deoxygenated by bubbling with argon (to create hypoxia and prevent from electron scavenging by oxygen) and buffered (phosphate buffer, pH = 7.0). After irradiation with 140 Gy, the studied solutions were analyzed with HPLC and mass spectrometry.The chromatograms presented in Fig. 4 show that only CH2CNU is not degraded upon electron attachment. This finding remains in full agreement with our computational model. Indeed, only for CH2CNU the activation barriers and driving forces are unfavorable for both possible degradation paths (see Table 1). Such a result corroborates, thus, the accuracy of our quantum chemical model. The compounds predicted as DEA sensitive do undergo electron-induced degradation, while the compound forming the stable radical anion does not decay due to electron attachment (see Fig. 4).
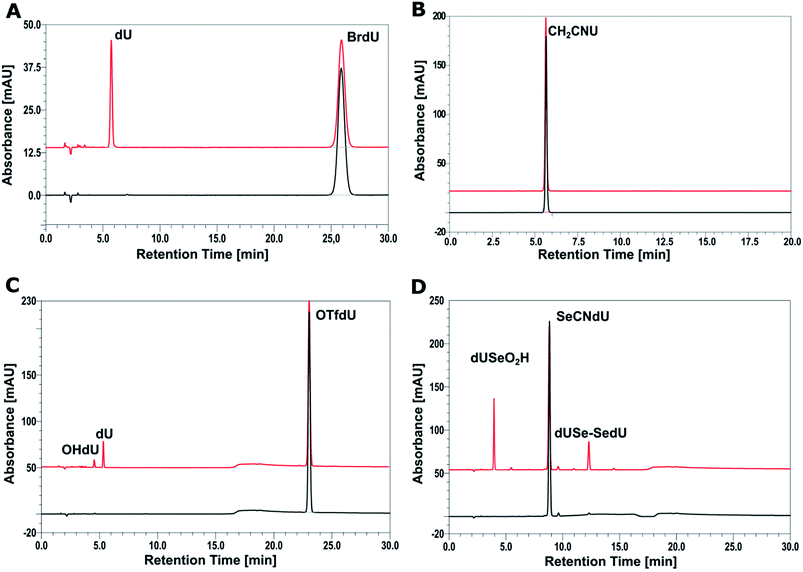 | ||
| Fig. 4 HPLC analysis of a solution of BrdU (A), CH2CNU (B), SeCNdU (C) and OTfdU (D) before (black chromatograms) and after irradiation with a dose of 140 Gy (red chromatograms). | ||
The chromatogram of the irradiated solution of SeCNdU demonstrates two main products, which elute at ca. 4 and 12 min, respectively (Fig. 4D). The data obtained from MS and tandem MS (for the MS/MS spectra and ion identities see Fig. S1, S2 and S3 in ESI†) allowed to determine that these radiolysis products are dUSe–SedU dimer (m/z = 613.0314) and SeO2HdU (m/z = 339.0155). The formation of such species indicates that the main product of dissociative electron attachment to SeCNdU is the dU–Se˙ radical, which is consistent with the theoretically predicted most favorable DEA pathway (see Table 1 and Fig. 3) and the previous observations for selenocyanatouracil.22 The dU–Se˙ radical can collide with the other one to give dUSe–SedU dimer or react with oxygen, produced during radiolysis, to give SeO2HdU (the formation of similar radiolysis product was observed during radiolysis of the SCNdU radiosensitizer).37
In case of the second proposed sensitizer – OTfdU – radiolysis leads to the formation of two stable products dU (m/z = 243.0653) and OHdU (m/z = 227.0705), eluting at ca. 4 and 5 min (Fig. 4C). The identities of these products were confirmed again by mass spectrometry (for the fragmentation spectra see Fig. S4– S6 in ESI†). These data demonstrate that the primary product of electron attachment to OTfdU – the OTfdU˙− radical anion – dissociates in two parallel ways, which produce eventually different stable products. The generation of dU is more favorable and this fact suggests that the main transient product triggered by dissociative electron attachment is dU˙. On the other hand, the presence of OHdU among the radiolysis products shows also the involvement of ˙OdU radical in the studied DEA process.
The steady-state radiolysis allowed to determine the extent of damage induced by X-rays for all the studied analogs (see Table 2). These quantities were calculated on the basis of comparison of the HPLC peak area for the substrate (BrdU, SeCNdU, OTfdU or OTfDMU) in the irradiated and non-irradiated solutions. The biggest decay was observed for SeCNdU (ca. 30%). The latter figure suggests that this derivative is more effective radiosensitizer than BrdU (ca. 17%). On the other hand, the radiolysis yield for OTfdU (ca. 16%) is lower than expected based on our theoretical predictions (see Table 1).
| Analog | Decay [%] |
|---|---|
| BrdU | 17.14 ± 0.27 |
| SeCNdU | 30.42 ± 0.72 |
| OTfdU | 16.25 ± 0.95 |
| OTfDMU | 26.60 ± 0.12 |
3.3. DEA driving forces and radiolysis yields
The measured yield of radiolysis do not reflect in the calculated DEA driving force for OTfdU as it is observed for SeCNdU and BrdU. It is worth noticing that the –OTf substituent in the pyrimidine base is located in the meta position against the N3 atom (see Fig. 1). At the same time, the Hammett σp and σm parameters for the –OTf substituent are 0.53 and 0.56, respectively.23 Taking into account the influence of resonance and inductive effect of –OTf moiety on the N3 atom, only the latter should be considered. Hence, the meta substituent constant, σm, for –OTf can be approximately equated to the inductive constant σI. From the σp and σm values it can be further deduced that the electron withdrawing inductive effect of this substituent highly dominates over its weak electron donating resonance properties. Eventually, it can be expected that the pKa value for OTfdU should drop significantly compared to the unsubstituted dU. This conclusion correlates with the pKa values calculated at the DFT/M06-2X/6-31++G(d,p) level according to the model published before41 and depicted by eqn (1) and (2) below. These calculations predicted the pKa value for OTfdU to be 5.9 (BrdU was taken as a reference compound, experimental pKa = 8.1).42
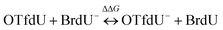 | (1) |
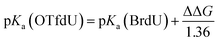 | (2) |
The pKa value for OTfdU suggests, that at the experimental conditions the N3 deprotonated structure should dominate in the equilibrium mixture. Indeed, at pH = 7.2 and for pKa = 5.6, the equilibrium concentration of the deprotonated OTfdU form is ca. 40 fold larger than that of the neutral one. The situation for BrdU is quite different. Its pKa = 8.1 leads to the ratio of the deprotonated to neutral forms equal to 0.13. Thus, only ca 10% of BrdU appears as the anionic N3-deprotonated structure while as much as 97.5% of OTfdU is in the form of the N3-deprotonated anion. This situation explains why despite much larger driving force for DEA in OTfdU, the yield of its radiolytic degradation is lower than that of BrdU. Namely, since in the OTfdU solution the anionic form prevails, electron attachment is hindered due to negative charge of the N3 deprotonated form anion. To verify this hypothesis, we synthesized and carried out radiolysis for the aqueous solution of 5-trifluoromethanesulfonyl-1,3-dimethyluracil (OTfDMU). In this compound, the N3 hydrogen is substituted with a methyl group, which blocks the proton equilibria possible for OTfdU. Thus, at pH = 7.2 OTfDMU occurs only in its neutral form, which should facilitate electron attachment compared to OTfdU. As indicated by data gathered in Table 1, the methylation in the N3 position in 5-trifluoromethanesulfonyluracil (OTfU) significantly increases the DEA yield with regard to that of OTfdU: from 16 to 26% (see Table 2), which confirms our hypothesis.
3.4. The C–O vs. O–S bond dissociation in the OTfdU˙− radical anion
The DEA mechanism resulted from our computational model seems to be inconsistent with the radiolytic experiment for OTfdU. While the main product of radiolysis is dU (see Fig. 4C), the preferred computational path indicates the O–S bond break followed by the formation of OHdU (see path A in Fig. 5). To make sure that this discrepancy is not due to methodological flaws, we did additional calculations using several other quantum chemical methods. Hence, we employed the hybrid B3LYP functional,28–30 ωB97XD31 functional, where dispersion is taken into account, and the conventional ab initio approach accounting for the correlation effects, i.e. the second-order Møller–Plesset method (MP2),32 (all the calculations were carried out with the 6-31++G(d,p) basis set and the PCM solvation model). While the reaction profiles differ slightly (see Table 3), the qualitative conclusions remain the same. According to all computational models employed, radiolysis of OTfdU should mainly lead to OHdU (see Table 3) rather than to dU observed experimentally (see Fig. 4C).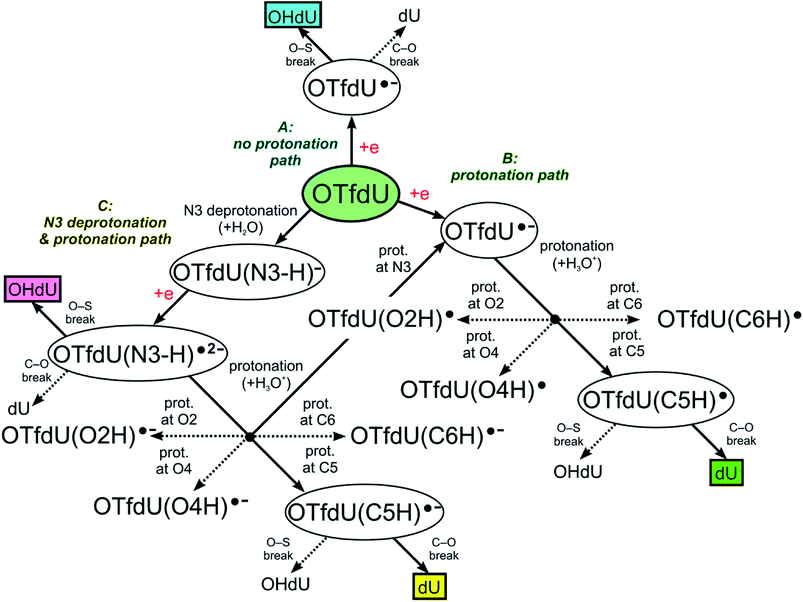 | ||
| Fig. 5 OTfdU electron induced degradation paths. Normal arrows lead to the main, while dotted arrows lead to the side products of each path. Final products of each path are framed. | ||
| M06-2X | B3LYP | ωB97XD | MP2 | ||
|---|---|---|---|---|---|
| a Due to the barier-free O–S bond break, the value calculated relative to OTfdU˙− anion radical with the freezed O–S bond.b Barrier-free O–S bond break. | |||||
| Path A (C–O bond break) | 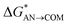 |
8.6 | 5.2 | 7.0 | 5.8 |
| ΔGAN→COM | −17.6 | −24.1a | −20.3 | −22.2 | |
| ΔGCOM→ISOL | −4.3 | −6.3 | −4.3 | −2.3 | |
| Path B (O–S bond break) | 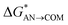 |
1.5 | BFb | 0.4 | 0.2 |
| ΔGAN→COM | −51.4 | −62.7a | −55.3 | −48.5 | |
| ΔGCOM→ISOL | −2.7 | −5.3 | −3.1 | 0.0 | |
The fact that different QM methodologies give the same qualitative results suggests that the assumed mechanism of electron induced degradation of OTfdU is too simple and something is lacking in our computational model.
Over 30 years ago von Sonntag and coworkers demonstrated within pulse radiolysis experiments on uracil and its derivatives, that the electron adduct formed due to transfer of solvated electron to the nucleobase undergoes a quick protonation at O2 or O4 and slower protonation at C6 leading finally to a radical protonated at C5.43 Therefore, we assumed that the anion radical (OTfdU˙−), formed after electron attachment to OTfdU, could be protonated in aqueous solution (see path B in Fig. 5). Hence, we analyzed OTfdU˙− protonation reactions at the carbonyl oxygens, O2 and O4, as well as at the C5 and C6 positions (for ring numbering see Fig. 1).
The most thermodynamically stable among the OTfdU˙ neutral protonated radicals is OTfdU(C5H)˙, protonated at C5. The radicals OTfdU(C6H)˙, OTfdU(O2H)˙ and OTfdU(O4H)˙ occurred to be 4.7, 22.8 and 10.7 kcal mol−1 less stable (in the free enthalpy scale). Taking into account the above-mentioned thermodynamics, only OTfdU(C5H)˙ should be present in the equilibrium mixture of radicals. Such radical is prone to further degradation, via breaking of the C–O bond (between the C5 atom and ester oxygen atom in the OTf substituent) or the O–S bond (between O and S atoms in the OTf substituent). The first pathway, leading to dU˙ radical and then to dU as a final product (green framed in Fig. 5), is both thermodynamically and kinetically favorable, with the thermodynamic stimulus ΔG = −11.76 kcal mol−1 and kinetic barrier as low as ΔG* = 0.12 kcal mol−1. The alternative degradation leading to OHdU as the final product is hindered due to high kinetic barrier for the O–S bond breakage (ΔG* = 21.79 kcal mol−1) and unfavorable thermodynamic stimulus (ΔG = 18.32 kcal mol−1). Hence, C5 protonation of the OTfdU˙− anion radical could explain why dU rather than OHdU is the major product observed in our radiolytic experiment.
As it was mentioned above, the –OTf moiety exerts strong inductive effect, which leads to significant lowering of pKa value related to the N3 deprotonation. As a result, the main OTfdU form under the neutral pH is the OTfdU(N3–H)− anion. Therefore, we also analyzed the possibility of protonation of the N3-deprotonated OTfdU(N3–H)− anion (see path C in Fig. 5). It occurred that OTfdU(N3–H)− is still able to attach an electron (AEA equal to 37.1 kcal mol−1, free enthalpy scale), producing the OTfdU(N3–H)˙2− dianion radical. This dianion radical can degrade directly via breaking the O–S bond (kinetic barrier almost equal to 0, comparing to kinetic barrier for C–O break equal to 7 kcal mol−1, free enthalpy scale), leading finally to OHdU product (see pink framed product, path C in Fig. 5). Moreover, such dianion radical species, due to its larger negative charge, is even more prone to protonation than OTfdU˙− (the thermodynamic stimuli for the protonation of dianion calculated for the most favored site is larger by as much as 25 kcal mol−1 than that for the protonation of monoanion in the free enthalpy scale). Also for OTfdU(N3–H)˙2− we analyzed four possible protonation sites: C5, C6, O2, O4 and additionally – intuitive N3 back protonation. As previously, protonation at positions C6, O2 and O4 occurred to be much less favored than protonation at C5 and N3. Thermodynamic stability of OTfdU(C6H)˙−, OTfdU(O2H)˙− and OTfdU(O4H)˙− occurred to be lower than back-N3-protonated OTfdU˙− anion radical by 7.3, 22.6 and 14.6 kcal mol−1, in the free enthalpy scale, respectively. Protonation at the N3 atom results in OTfdU˙− that may dissociate via the O–S bond breakage leading eventually to OHdU or may be protonated in path B giving dU as the final stable product. On the other hand, protonation of OTfdU(N3–H)˙2− at the C5 position leads to the barrier-free C5–O bond breakage and finally to the dU product too (see the yellow framed final product of path C, Fig. 5). Those two protonation routes are almost equally probable. As it is impossible to obtain stable OTfdU(C5H)˙− (due to its rapid degradation), we compared the stability of the OTfdU(C5H)˙− anion radical with the C5–O bond frozen (at the distance of 1.443 Å, characteristic for the optimum geometry of OTfdU(N3–H)˙2−) to the OTfdU(N3–H)˙− (“normal” OTfdU˙−) stability on the electronic energy scale. The difference in stability is as low as 1.7 kcal mol−1 (in favor of OTfdU˙−), but one should take into account that energy calculated for the OTfdU(C5H)˙− with the frozen C5–O bond was overestimated, so the actual difference is even lower. Moreover, the product of barrier-free C–O bond break reaction in OTfdU(C5H)˙− is by 24.1 kcal mol−1 more stable than that resulting from the OTfdU˙− dissociation (in free enthalpy scale).
Summing up, those OTfdU molecules, which are engaged in protonation are degrading to dU product (via path B or C, see Fig. 5), while the presence of OHdU product could be justified by the mechanism, discussed as path A.
3.5. Cytotoxicity
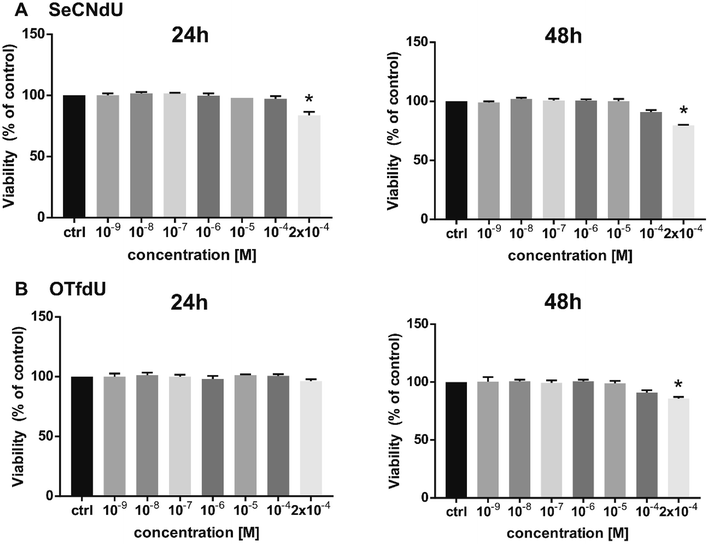
At the cellular level, the proposed radiosensitizers should act like a “Trojan Horse” – the lethal effects should be produced only as a result of interactions between radiation and nucleoside analogs incorporated into DNA. For this reason the studied analogs should exhibit minimal cytotoxicity until a damaging factor, i.e. ionizing radiation, appear and cytotoxic compounds cannot be used as useful radiosensitizers. Therefore, cytotoxic activity of SeCNdU and OTfdU was evaluated against human breast cancer cells (MCF-7 line). It was measured using the MTT test after 24 and 48 h incubation with test compounds in the concentration range of 10−9–2 × 10−4 M. Both SeCNdU and OTfdU showed toxicity to tumor cells in a dose and time dependent manner but their influence on the cells viability was rather slight.As shown in Fig. 6, statistically significant decrease in viability was observed only for concentration of tested analogs as high as 2 × 10−4 M. The treatment with SeCNdU, in the above-mentioned dose, results in cell viability decrease to 84% and 80% (compared to untreated culture) for 24 and 48 h incubation, respectively (Fig. 6A). In case of OTfdU, the number of viable cells was reduced to 86% for a contact time equal to 48 h (Fig. 6B). These results demonstrate that both SeCNdU and OTfdU have low cytotoxicity and can be considered as potential radiosensitizers.
4. Summary
Using a computational model based on the electron-triggered mechanism of DNA radiosensitization we proposed two 2′-deoxyuridine derivatives that could potentially be employed in anticancer treatment. The above-mentioned mechanism assumes an involvement of efficient DEA induced by electron addition to the 5-substituted-2′-deoxyuridine incorporated into DNA. In order to possess radiosensitizing properties, such a derivative should be modified with an electrophilic substituent (in order to increase its electron affinity) and the chemical bond between the pyrimidine ring and the substituent should be relatively weak. The –SeCN and –OTf residues seem to fulfill the above-mentioned requirements.We calculated the DEA profiles for SeCNdU and OTfdU at the M06/6-31++G(d,p) level and compared them to that of BrdU – a well-known radiosensitizer. Both new compounds possess thermodynamic characteristics that suggest their superiority over BrdU. In order to verify these computational results, we synthesized SeCNdU and OTfdU and carried out steady-state radiolytic studies on aqueous solutions containing the scrutinized derivatives. Additionally, to examine the accuracy of the computational model we synthesized CH2CNU – a compound for which the calculated DEA profile indicates no degradation induced by electron attachment. In full accordance with high activation barrier and unfavorable thermodynamic stimulus (see Table 1), we did not observe even traces of degradation triggered by the irradiation of CH2CNU solution with 140 Gy of IR (see Fig. 4D).
On the other hand, the steady state radiolysis of SeCNdU led, in agreement with the theoretical model, to efficient DEA with products indicating the electron-induced dissociation of the Se–CN bond. Its degradation yield surpassed that of BrdU, which remains in accordance with the larger thermodynamic stimulus of DEA calculated for the former compound. However, the experimental characteristics measured for OTfdU do not agree with our model. The radiolysis yield seems to be too low judging by the thermodynamic stimulus of DEA and degradation products show, unlike the theoretical model, that the C5–O bond breakage is favored over that of O–S. The DEA profile calculated with several other functionals and the MP2 method differed marginally, which proves that the observed discrepancy does not result from the theoretical model. Eventually, we explained the surprisingly low yield of damage by exceptionally high induction effect exerted by the –OTf substituent. As a result, most of the compound occurs in the N3-deprotonated anionic form with the reduced ability to attach an electron. This conclusion is confirmed by the significantly higher yield of DEA measured for OTfDMU that occurs exclusively in the neutral form. Moreover, the dianion formed due to solvated electron addition to OTfdU(N3–H)−, OTfdU(N3–H)˙2−, is easily protonated at the C5 position leading to the barrier-free dissociation of the C5–O bond. This explain why dU rather than OHdU is observed as the main degradation product in the irradiated solution of OTfdU.
Ultimately, both compounds demonstrate moderate cytotoxicity. Indeed, they are not harmful to the MCF-7 cells at the concentrations employed with labelling experiments, which suggests their suitability in further studies, aiming at clinical use of the proposed derivatives.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Polish National Science Center (NCN) under the Grant No. UMO-2014/14/A/ST4/00405 (J. R.). Calculations have been carried out in Wroclaw Center for Networking and Supercomputing (wcss.wroc.pl), grant No. 209 (L. C.-M.) and at a local computer cluster (Xeon E5-2623 v3 processors).References
- A. Jemal, F. Bray, M. M. Center, J. Ferlay, E. Ward and D. Forman, Ca-Cancer J. Clin., 2011, 61, 69–90 CrossRef PubMed.
- M. Joiner and A. van der Kogel, Basic Clinical Radiobiology, Hodder Arnold, London, UK, 4th edn, 2009 Search PubMed.
- J. E. Visvader and G. J. Lindeman, Nat. Rev. Cancer, 2008, 8, 755–768 CrossRef PubMed.
- S. Rockwell, I. T Dobrucki, E. Y. Kim, S. T. Marrison and V. T. Vu, Curr. Mol. Med., 2009, 9, 442–458 CrossRef PubMed.
- E. J. Hall and C. S. Wuu, Int. J. Radiat. Oncol., Biol., Phys., 2003, 56, 83–88 CrossRef.
- J. Rak, L. Chomicz, J. Wiczk, K. Westphal, M. Zdrowowicz, P. Wityk, M. Żyndul, S. Makurat and Ł. Golon, J. Phys. Chem. B, 2015, 119, 8227–8238 CrossRef PubMed.
- E. J. Hall and A. J. Giaccia, Radiobiology for the Radiologist, Lippincott Williams & Wilkins, Philadelphia, USA, 2006 Search PubMed.
- P. Wardman, Clin. Oncol., 2007, 19, 397–417 CrossRef PubMed.
- C. Von Sonntag, The Chemical Basis of Radiation Biology, Taylor & Francis London, 1987 Search PubMed.
- K. Westphal, J. Wiczk, J. Miloch, G. Kciuk, K. Bobrowski and J. Rak, Org. Biomol. Chem., 2015, 13, 10362–10369 Search PubMed.
- L. Chomicz, M. Zdrowowicz, F. Kasprzykowski, J. Rak, A. Buonaugurio, Y. Wang and K. H. Bowen, J. Phys. Chem. Lett., 2013, 4, 2853–2857 CrossRef.
- S. Cecchini, S. Girouard, M. A. Huels, L. Sanche and D. J. Hunting, Biochemistry, 2005, 44, 1932–1940 CrossRef PubMed.
- M. E. Dextraze, S. Cecchini, F. Bergeron, S. Girouard, K. Turcotte, J. R. Wagner and D. J. Hunting, Biochemistry, 2009, 48, 2005–2011 CrossRef PubMed.
- G. Lin, J. Zhang, Y. Zeng, H. Luo and Y. Wang, Biochemistry, 2010, 49, 2346–2350 CrossRef PubMed.
- S. Cecchini, S. Girouard, M. A. Huels, L. Sanche and D. J. Hunting, Radiat. Res., 2004, 162, 604–615 CrossRef PubMed.
- M. E. Dextraze, J. R. Wagner and D. J. Hunting, Biochemistry, 2007, 46, 9089–9097 CrossRef PubMed.
- Y. Park, K. Polska, J. Rak, J. R. Wagner and L. Sanche, J. Phys. Chem. B, 2012, 116, 9676–9682 CrossRef PubMed.
- M. D. Prados, C. Scott, H. Sandler, J. C. Buckner, T. Phillips, C. Schultz, R. Urtasun, R. Davis, P. Gutin and T. L. Cascino, Int. J. Radiat. Oncol., Biol., Phys., 1999, 45, 1109–1115 CrossRef.
- S. Makurat, L. Chomicz-Mańka and J. Rak, ChemPhysChem, 2016, 17, 2572–2578 CrossRef PubMed.
- M. Zdrowowicz, P. Wityk, B. Michalska and J. Rak, Org. Biomol. Chem., 2016, 14, 9312–9321 Search PubMed.
- K. Westphal, M. Zdrowowicz, A. Żylicz-Stachula and J. Rak, J. Photochem. Photobiol., B, 2017, 167, 228–235 CrossRef PubMed.
- M. Sosnowska, S. Makurat, M. Zdrowowicz and J. Rak, J. Phys. Chem. B, 2017, 121, 6139–6147 CrossRef PubMed.
- C. Hansch, A. Leo and R. W. Taft, Chem. Rev., 1991, 91, 165–195 CrossRef.
- L. B. Agenäs, Acta Chem. Scand., 1963, 17, 268–270 CrossRef.
- G. T. Crisp and B. L. Flynn, Tetrahedron, 1993, 49, 5873–5890 CrossRef.
- Y. Zhao and D. G. Truhlar, Theor. Chem. Acc., 2008, 120, 215–241 CrossRef.
- W. J. Hehre, R. Ditchfield and J. A. Pople, J. Chem. Phys., 1972, 56, 2257–2261 CrossRef.
- A. D. Becke, Phys. Rev. A, 1988, 38, 3098–3100 CrossRef.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B, 1988, 37, 785–789 CrossRef.
- J. D. Chai and M. Head-Gordon, Phys. Chem. Chem. Phys., 2008, 10, 6615–6620 RSC.
- M. J. Frisch, M. Head-Gordon and J. A. Pople, Chem. Phys. Lett., 1990, 166, 275–280 CrossRef.
- S. Miertuš, E. Scrocco and J. Tomasi, Chem. Phys., 1981, 55, 117–129 CrossRef.
- K. Fukui, Acc. Chem. Res., 1981, 14, 363–368 CrossRef.
- D. A. McQuarrie and J. D. Simon, Molecular Thermodynamics, University Science Books, Sausalito, CA, 1999 Search PubMed.
- L. Chomicz, J. Leszczynski and J. Rak, J. Phys. Chem. B, 2013, 117, 8681–8688 CrossRef PubMed.
- M. Zdrowowicz, L. Chomicz, M. Żyndul, P. Wityk, J. Rak, T. J. Wiegand, C. G. Hanson, A. Adhikary and M. D. Sevilla, Phys. Chem. Chem. Phys., 2015, 17, 16907–16916 RSC.
- K. Anil and M. D. Sevilla, J. Phys. Chem. A, 2017, 121, 4825–4829 CrossRef PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09 (Revision D.01), Gaussian, Inc., Wallingford, CT, 2013 Search PubMed.
- R. Dennington, T. Keith andJ. Millam, GaussView, version 5, Semichem, Inc., Shawnee Mission, KS, 2009 Search PubMed.
- N. L. Tran and M. E. Colvin, J. Mol. Struct.: THEOCHEM, 2000, 532, 127–137 CrossRef.
- R. M. C. Dawson, D. C. Elliott, W. H. Elliott and K. M. Jones, Data for Biochemical Research, Clarendon Press, Oxford, 3rd edn, 1986, pp. 262–263 Search PubMed.
- S. Das, D. J. Deeble, M. N. Schuchmann and C. von Sonntag, Int. J. Radiat. Biol., 1984, 46, 7–9 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra03172j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |