Open Access Article
Open Access ArticleA multifunctional oxidosqualene cyclase from Tripterygium regelii that produces both α- and β-amyrin†
Yun Lu a,
Jiawei Zhoua,
Tianyuan Hua,
Yifeng Zhangab,
Ping Sub,
Jiadian Wangab,
Wei Gao*acd and
Luqi Huangb
a,
Jiawei Zhoua,
Tianyuan Hua,
Yifeng Zhangab,
Ping Sub,
Jiadian Wangab,
Wei Gao*acd and
Luqi Huangb
aSchool of Traditional Chinese Medicine, Capital Medical University, Beijing 100069, China. E-mail: weigao@ccmu.edu.cn; Fax: +86 10 83911627; Tel: +86 10 83916572
bState Key Laboratory of Dao-di Herbs, National Resource Center for Chinese MateriaMedica, China Academy of ChineseMedical Sciences, Beijing, China
cBeijing Key Lab of TCM Collateral Disease Theory Research, Capital Medical University, Beijing, China
dAdvanced Innovation Center for Human Brain Protection, Capital Medical University, Beijing, China
First published on 27th June 2018
Abstract
Tripterygium regelii is a rich source of triterpenoids, containing many types of triterpenes with high chemical diversity and interesting pharmacological properties. The cDNA of the multifunctional oxidosqualene cyclase (TrOSC, GenBank accession number: MH161182), consisting of a 2289 bp open reading frame and coding for 762 amino acids, was cloned from the stems and roots of Tripterygium regelii. Phylogenetic analysis using OSC genes from other plants suggested that TrOSC might be a mixed-amyrin synthase. The coding sequence was cloned into the expression vector pYES2 and transformed into the yeast Saccharomyces cerevisiae. The resulting products were analysed by GC-MS. Surprisingly, although it showed 76% sequence identity to lupeol synthase from Ricinus communis, TrOSC was found to be a multifunctional triterpene synthase producing both α- and β-amyrin, the precursors of ursane and oleanane type triterpenes, respectively. qRT-PCR analysis revealed that the transcript of TrOSC accumulated mainly in roots and stems. Taken together, our findings contribute to the knowledge of key genes in the pentacyclic triterpene biosynthesis pathway.
Introduction
Tripterygium regelii is one of the most commonly used traditional chinese medicinal herbs. It is native to Korea and Japan1 and is also distributed throughout northeast China. The plants in the Tripterygium genus of the family Celastraceae, such as Tripterygium regelii and Tripterygium wilfordii, are well known as a rich source of triterpenoids.2 An analysis of the phylogenetic relationships within the Tripterygium genus showed that Tripterygium wilfordii and Tripterygium hypoglaucum cluster together, while Tripterygium regelii represents a separate cluster,3 indicating that there are some unique aspects to Tripterygium regelii, with potential values meriting further study.Many triterpenes isolated from Tripterygium regelii showed various pharmacological activities, such as anti-cancer activity4 and potential SARS-CoV 3CLpro inhibitory effects.5 In addition, celastrol, a special pentacyclic triterpene, is a promising agent for the pharmacological treatment of obesity.6 Pentacyclic triterpenes are C30 terpenes consisting of six isoprene units7 and the therapeutic utility of pentacyclic triterpenoids as multifunctional adjuvants in cancer chemotherapy is highlighted.8 Alpha- and β-amyrin, as well as lupeol are the most common pentacyclic triterpenoid precursors in plants, which can be further converted into ursane, oleanane and lupane-type pentacyclic triterpenoids, respectively.9 For example, an oxidosqualene cyclase gene encoding a β-amyrin synthase from P. tenuifolia catalyses the first committed step in the biosynthesis of oleanane-type triterpene saponins, such as onjisaponins and polygalasaponins.10 Numerous studies indicated that oleanane- and ursane-triterpenoids exert anti-inflammatory and anti-tumour activities.11,12 Moreover, ursane-type triterpenoids are potential candidates for the design of multi-target bioactive compounds, with focus on their anti-cancer effects.13 Furthermore, 3-O-β-chacotriosyl oleanane-type triterpenes have been evaluated as H5N1 entry inhibitors.14 However, new functions of triterpenes need further exploration. A study on triterpene synthesis in oats revealed a role for β-amyrin in determining root epidermal cell patterning.15 Nine triterpene synthase genes (VvTTPSs) from Vitis vinifera cultivars were found to play differential roles in the plant's responses to environmental stress factors.16
In higher plants, isoprenoids are derived from the cytoplasmic mevalonate (MVA) and the plastidial methylerythritol phosphate (MEP) pathways.17 The two biosynthetic pathways have evolved to generate DMAPP and isopentenyl pyrophosphate (IPP), the universal terpenoid precursors, and synergy between the two pathways was demonstrated for isoprene production in Escherichia coli.18 However, the relative contribution of each pathway to the biosynthesis of isoprenoids is different, so that C5 units from the MEP pathway are used to form monoterpenes (C10), phytol side chains (C20) and carotenoids (C40), while C5 units from the MVA pathway are used to form sesquiterpenes (C15), terpenoid aldehydes (C15 and C25) and steroids/triterpenoids (C30).19 The first step in the biosynthesis of all triterpenes is the cyclization of 2,3-oxidosqualene20 (Fig. 1). Subsequently, the chair–boat–chair conformation leads to a protosteryl cation intermediate, the precursor of the sterols, which have important functions in membranes and signaling,14 via the formation of cycloartenol or lanosterol in plants.21 The chair-chair-chair conformation directs the cyclization to yield the dammarenyl cation, which subsequently forms diverse triterpene skeletons such as ursane, oleanane, and lupane.22 Because of their potential ability to modify the chemical structures of terpenoids, OSCs (oxidosqualene cyclases) have attracted the attention of many investigators.23 Many types of OSCs have been cloned and identified, such as β-amyrin synthase,24,25 which catalyses the formation of the most popular pentacyclic triterpene among higher plants,26 cycloartenol synthase,27 as well as multifunctional triterpene synthase.28 In addition, pathway engineering has been used to produce β-amyrin and cycloartenol in Escherichia coli.29 Here, we describe the identification and functional characterization of TrOSC, a mixed-amyrin synthase from Tripterygium regelii responsible for the production of α-amyrin and β-amyrin, which will lay the foundation for future research on the biosynthesis pathway of triterpenoids.
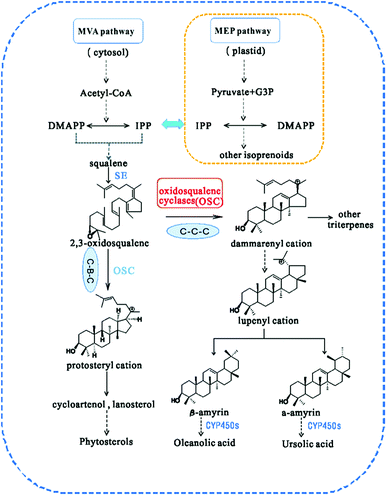 | ||
| Fig. 1 Biosynthesis pathway of triterpenes catalysed by different types of oxidosqualene cyclases (OSCs). CYP450s, cytochrome P450s. | ||
Results and discussion
Sequence analysis of TrOSC
The cDNA contained an ORF of 2289 bp,which was deduced to code for a 762 amino acid protein (Mw: 87.842 kDa, pI: 6.09). The encoded protein was soluble and was predicted to most probably be located in peroxisome or the nucleus. The amino acid sequence of TrOSC had 76, 74 and 71% identity to RcLUS (lupeol synthase; NP_001310684), KcMS (multifunctional triterpene synthase; BAF35580) and EtAS (β-amyrin synthase; BAE43642), respectively. In addition, five motifs were identified in the amino acid sequences of TrOSC (Fig. 2), including a QXXXGXXXW motif, a DCTAE motif and three QXXXGXW motifs. The QXXXGXW motifs were probable to stabilize carbocation intermediates in the process of cyclization of OSCs.30 By contrast, the DCTAE motif might be related to substrate binding,31 since it is a putative initiation site for the polycyclization reaction. In fact, it has been demonstrated that the acidic carboxyl residue Asp485 of β-amyrin synthase from Euphorbia tirucalli is responsible for initiating the polycyclization reaction, while Cys564 plays a role in hydrogen bond formation.32 The secondary structure of the protein was mainly composed of alpha helices (40.81%) and random coils (31.50%), along with extended strands (17.06%) and beta turns (10.63%). Protein secondary structure prediction can provide vital information about the functions and 3D structure prediction of a given protein.33 Studies have shown that secondary structure of proteins can be changed by pulsed electric field (PEF) treatment, which affects the antioxidant activity of pentapeptides.34Phylogenetic analysis
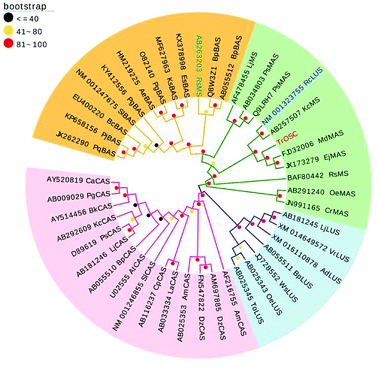
The phylogenetic tree was generated on the basis of the deduced amino acid sequences of other plant OSCs downloaded from NCBI (www.ncbi.nlm.nih.Gov/). These OSCs were grouped into four main branches (Fig. 3): β-amyrin synthase, multifunctional triterpene synthase/mixed-amyrin synthase, lupeol synthase and cycloartenol synthase. TrOSC was found to be phylogenetically related to mixed-amyrin synthase from Malus domestica, which has been reported to produce both α-amyrin and β-amyrin when expressed in yeast.28 The phylogenetic trees can encompass a wide variety of triterpene skeletal types, allowing us to predict the likely functions of uncharacterized OSCs.22 Accordingly, TrOSC was predicted to most likely be a mixed-amyrin synthase, even if this assertion needed to be verified experimentally. However, it was also phylogenetically related to lupeol synthase from Ricinus communis, with which it shares the highest sequence identity (76%), indicating that the observed differences of catalytic functions between multifunctional triterpene synthase and lupeol synthase might be caused by small differences of their respective sequence.Functional expression of TrOSC in yeast
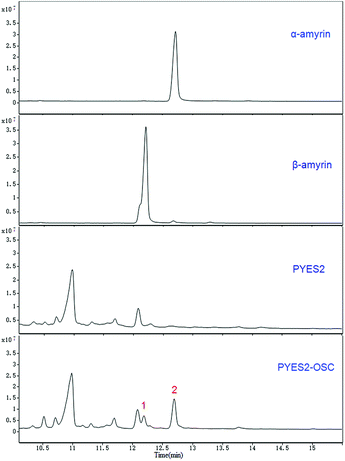
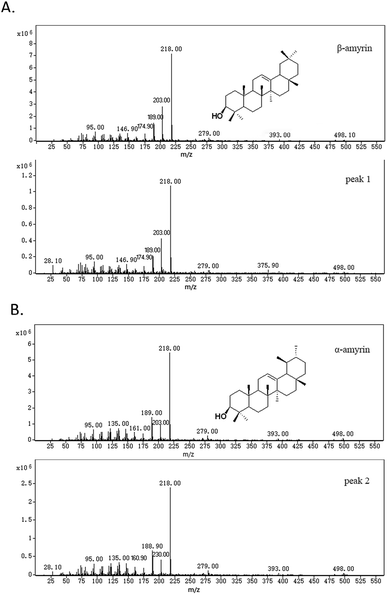
Two compounds (Fig. 4; peaks 1 and 2) were found in extracts of cells expressing TrOSC compared with the control group. They were identified as β-amyrin and α-amyrin, respectively, by comparing their retention times and mass fragmentation patterns with those of authentic standards (Fig. 5), which confirmed that TrOSC is indeed a mixed-amyrin synthase. The enzyme's main product was α-amyrin, with a ratio to β-amyrin of about 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
The product specificity of OSCs is controlled by the key amino acid residue, which will result in a major change in triterpene cyclization by mutation.35 For example, it has been shown that mutations of the trp259 residue in the MWCYCR motif of the β-amyrin synthase (PNY) lead to the production of lupeol as a major product, whereas mutations of the Leu256 residue in the MLCYCR motif of lupeol synthase (OEW) lead to almost exclusive β-amyrin production with only minor amount of lupeol, demonstrating that certain amino acid residues are responsible for product specificity.36 Recently, a study on the triterpene synthases of Oryza species also revealed that three key amino acid residues determine the specificity of the chair–semi(chair)–chair and chair–boat–chair interconversions.37 These experiments shed light on the mechanism of OSC cyclization and provide a useful strategy to identify key residues that determine the specificity for α-amyrin and β-amyrin or other products.
Since the diversity of triterpene skeletons produced by different oxidosqualene cyclases (OSCs), the biosynthetic pathway of the large varieties of triterpenes found in plants need further investigation. Cytochrome P450s (CYP450s) play critical roles in the production of highly functionalized terpenoids.38 After cyclization, these enzymes introduce functional groups, such as hydroxyl, carbonyl and carboxyl groups, into the carbon skeletons by a series of oxidation reactions.39 For example, β-amyrin can be converted to oleanolic acid catalysed by CYP450s.37 Thus, further research on cytochrome P450 genes involved in the triterpene biosynthetic pathway is necessary for us to understand the mechanisms of triterpenoid biosynthesis, which in turn will provide useful molecular tools for synthetic biology to produce desirable triterpenoids from specific precursors.
Relative expression of TrOSC in different tissues of Tripterygium regelii
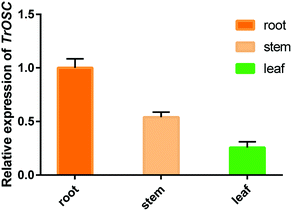
The relative expression levels of TrOSC in roots, stems and leaves of Tripterygium regelii were determined to investigate whether there is tissue-specific expression. As shown in Fig. 6, the expression was highest in roots, followed by stems and lowest in leaves, showing that the transcript of TrOSC, which encodes the mixed-amyrin synthase, accumulated mainly in the roots and stems. Consequently, we chose the roots and stems as the starting materials to obtain cDNA for cloning. What's more, a number of studies have reported on the isolation of triterpenoids from the roots and stems of Tripterygium regelii but rarely from leaves. For instance, regelin and regelinol, as well as new ursane-type triterpenoids with anticancer activity, were isolated from the roots of Tripterygium regelii,40 and new triterpenoids were also isolated from the stems of this plant.2,41 Thus, it can be speculated that roots and stems are important organs of Tripterygium regelii to obtain triterpenoids.Experimental introduction
RNA isolation
Plant material was collected from Baijiyao Forest Park in Tonghua, JiLin, P. R. China (longitude 126.07 degrees E, latitude 41.571 degrees N) and stored at −80 °C. Total RNA was extracted from the roots and stems of Tripterygium regelii using the cetyltrimethylammonium bromide (CTAB) method.42Cloning of cDNA from Tripterygium regelii
Total RNA was used as template to be reverse transcribed into cDNA using the SMARTer™ RACE cDNA Amplification Kit (Clontech Laboratories Inc., USA) according to the manufacturer's instructions. Specific primers (Table S1†) were designed by Primer Premier 5.0 software according to the selected sequence. The cDNA served as a template for amplifying the open reading frame using 2X Phusion HF Master Mix (NEB, USA) with a procedure of 98 °C for 30 s; followed by 35 cycles of 98 °C for 10 s, 56 °C for 15 s and 72 °C for 1 min; a final elongation at 72 °C for 7 min and 4 °C hold. PCR products were purified and ligated into the pEASY-T3 vector (TransGen Biotech, China) and transformed into E. coli DH5α competent cells (TransGen Biotech, China). Positive bacterial colonies were selected on ampicillin plates and confirmed by sequencing.Bioinformatic analysis of the TrOSC gene sequence
The open reading frame was searched using ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). The nucleotide and amino acid sequence was blasted against the NCBI database (http://www.ncbi.NLM.NIH<?NIH?>.gov). The theoretical molecular weight (Mw) and isoelectric point (pI) were determined using the Compute pI/Mw tool (http://Web.ExPASy.org/compute_pi/). Prediction of transmembrane helices was conducted using TRMHMM server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Subcellular localization was predicted by Wolf psort (http://wolfpsort.org/). Secondary structures were predicted by PRABI-GERLAND (https://npsa-prabi.ibcp.fr/) and the 3-dimensional structural model was constructed by Swiss-Model (http://swissmodel.expasy.org/). Multiple sequence alignments were carried out using DNAMAN software. Phylogenetic analysis was performed using MEGA 6.0 software.43Expression of TrOSC in yeast
In order to investigate the function of TrOSC, the yeast expression system was used. TrOSC was amplified using the 2X Phusion HF Master Mix (NEB, USA), and ligated directly into the pYES2 expression vector (Invitrogen, USA) using pEASY®-Uni Seamless Cloning and Assembly Kit (TransGen Biotech, China) with the designed primers (Table S1†) to obtain the recombinant plasmid pYES2-OSC. The resulting plasmid was introduced into the lanosterol synthase-deficient Saccharomyces cerevisiae (-erg7,-ura; ATCC, USA) using Frozen-EZ Yeast Transformation II™ kit (ZYMO RESEARCH, USA), along with the same transformation of empty vector pYES2 as a control. Positive yeast transformants were incubated in 20 mL SC-U medium (Ura minus medium 8 g L−1; FunGenome, China) and glucose (20 g L−1) at 30 °C with shaking (220 rpm). Two days later, the cells were collected by centrifugation at low speed (2000g, 4 min) and resuspended in SC-U without glucose, supplemented with Ura minus medium (8 g L−1) and galactose (20 g L−1) at 30 °C with shaking (220 rpm) for 16 h. After that, the cells were collected at low speed (2000g, 4 min) and resuspended in 0.1 M potassium phosphate (pH 7.0) with 3% glucose and cultured under the same conditions for 24 h.GC-MS analysis
Cells were harvested by centrifugation at 5000g for 4 min and extracted by ultrasonic extraction in 20% KOH/50% EtOH (10 mL) for 2 h, then extracted 3 times with equal volumes of hexane. The organic phase was collected, evaporated to dryness, and re-dissolved in 1.5 mL hexane. After drying under nitrogen gas, the sample was derivatized with N,O-bis (trimethylsilyl)trifluoroacetamide and pyridine at 70 °C for 2 h. Finally, the extracts were concentrated under a stream of nitrogen gas and dissolved in 1 mL CHCl3. The analysis was performed on an Agilent 7890B gas chromatograph with a DB-5ms column (15 m × 250 μm × 0.1 μm). One microliter of the concentrated organic phase was injected under an He flow rate of 1 mL min−1 with a temperature program of 1 min at 50 °C, followed by a gradient from 50 to 260 °C at 50 °C min−1, rising to 272 °C at 1 °C min−1, with a 4 min hold at 272 °C.44 The ion trap heating temperature was 250 °C. The electron energy was 70 eV. Spectra were recorded in the range of 10–550 m/z.qRT-PCR analysis
Total RNA from roots, stems and leaves was isolated and reverse transcribed using the Fast King RT Kit (Tiangen, China). qRT-PCR was carried out using the QuantStudio™ 5 Real-Time PCR System (Applied Biosystems, USA), in conjunction with the KAPA SYBR FAST qPCR Master Mix Kit (KAPA Biosystems, USA). The conditions of real-time PCR were 95 °C for 3 min, followed by 40 cycles at 95 °C for 3 s and 60 °C for 30 s. The melting curve conditions were 95 °C for 1 s, 60 °C for 20 s and 95 °C for 1 s. The relative expression value of TrOSC was estimated using the 2−ΔΔCt method45 with three biological replicates and three technical replicates. The EF-α gene was used as an endogenous control. The primers are listed in Table S1.†Conclusions
The multifunctional oxidosqualene cyclase from Tripterygium regelii (TrOSC) was cloned and characterized by heterologous expression in yeast. TrOSC was found to be a mixed-amyrin synthase, whose products were identified as α-amyrin and β-amyrin, the precursors of ursane and oleanane type triterpenes, respectively, by GC-MS analysis. And the biochemical results were in agreement with the phylogenetic analysis. The relative expression level of TrOSC was higher in roots and stems than in leaves. This is the first study on oxidosqualene cyclase from Tripterygium regelii, which will lay a solid foundation for studying the triterpenoid biosynthesis pathway of this important medicinal plant.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (81773830 to W. G.), Beijing Natural Science Foundation Program and Scientific Research Key Program of Beijing Municipal Commission for Education (KZ201710025022), High-level Teachers in Beijing Municipal Universities in the Period of the 13th Five-year Plan program (CIT&TCD20170324 to W. G.), and National Program for Special Support of Eminent Professionals.References
- B. S. Choi, K. Sapkota, S. Kim, H. J. Lee, H. S. Choi and S. J. Kim, Neurochem. Res., 2010, 35, 1269–1280 CrossRef PubMed.
- D. Fan, S. Parhira, G. Y. Zhu, Z. H. Jiang and L. P. Bai, Fitoterapia, 2016, 113, 69–73 CrossRef PubMed.
- B. Ma, T. Hu, P. Li, Q. Yuan, Z. Lin, Y. Tu, J. Li, X. Zhang, X. Wu and X. Wang, Ecol. Evol., 2017, 7, 8612–8623 CrossRef PubMed.
- H. S. Yang, J. Y. Kim, J. H. Lee, B. W. Lee, K. H. Park, K. H. Shim, M. K. Lee and K. I. Seo, Food Chem. Toxicol., 2011, 49, 527–532 CrossRef PubMed.
- Y. B. Ryu, S. J. Park, Y. M. Kim, J. Y. Lee, W. D. Seo, J. S. Chang, K. H. Park, M. C. Rho and W. S. Lee, ChemInform, 2010, 20, 1873–1876 Search PubMed.
- J. Liu, J. Lee, M. A. Salazar Hernandez, R. Mazitschek and U. Ozcan, Cell, 2015, 161, 999–1011 CrossRef PubMed.
- C. M. Andre, S. Legay, A. Deleruelle, N. Nieuwenhuizen, M. Punter, C. Brendolise, J. M. Cooney, M. Lateur, J. F. Hausman and Y. Larondelle, New Phytol., 2016, 211, 1279–1294 CrossRef PubMed.
- S. M. Kamble, S. N. Goyal and C. R. Patil, RSC Adv., 2015, 45, 33370–33382 Search PubMed.
- S. Jäger, H. Trojan, T. Kopp, M. N. Laszczyk and A. Scheffler, Molecules, 2009, 14, 2016–2031 CrossRef PubMed.
- M. L. Jin, D. Y. Lee, Y. Um, J. H. Lee, C. G. Park, R. Jetter and O. T. Kim, Plant Cell Rep., 2014, 33, 511–519 CrossRef PubMed.
- N. R. Parikh, A. Mandal, D. Bhatia, K. S. Siveen, G. Sethi and A. Bishayee, Phytochem. Rev., 2014, 13, 793–810 CrossRef PubMed.
- S. Jar, A. S. Leal, A. S. Valdeira, G. Bmf, A. Dps, F. Sac, S. M. Silvestre and M. Vis, Eur. J. Med. Chem., 2017, 142, 95–130 CrossRef PubMed.
- J. A. Salvador, V. M. Moreira, B. M. Gonçalves, A. S. Leal and Y. Jing, Nat. Prod. Rep., 2012, 29, 1463–1479 RSC.
- G. Song, X. Shen, S. Li, H. Si, Y. Li, H. Luan, J. Fan, Q. Liang and S. Liu, RSC Adv., 2015, 5, 39145–39154 RSC.
- A. C. Kemen, S. Honkanen, R. E. Melton, K. C. Findlay, S. T. Mugford, K. Hayashi, K. Haralampidis, S. J. Rosser and A. Osbourn, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 8679–8684 CrossRef PubMed.
- F. Pensec, A. Szakiel, C. Pączkowski, A. Woźniak, M. Grabarczyk, C. Bertsch, M. J. Fischer and J. Chong, J. Plant Res., 2016, 129, 499–512 CrossRef PubMed.
- K. Skorupinska-Tudek, J. Poznanski, J. Wojcik, T. Bienkowski, I. Szostkiewicz, M. Zelman-Femiak, A. Bajda, T. Chojnacki, O. Olszowska and J. Grunler, Chem. Phys. Lipids, 2008, 154, 21024–21035 CrossRef.
- C. Yang, X. Gao, Y. Jiang, B. Sun, F. Gao and S. Yang, Metab. Eng., 2016, 37, 79–91 CrossRef PubMed.
- S. Opitz, W. D. Nes and J. Gershenzon, Phytochemistry, 2014, 98, 110–119 CrossRef PubMed.
- D. R. Phillips, J. M. Rasbery, B. Bartel and S. P. Matsuda, Curr. Opin. Plant Biol., 2006, 9, 305–314 CrossRef PubMed.
- M. Suzuki, T. Xiang, K. Ohyama, H. Seki, K. Saito, T. Muranaka, H. Hayashi, Y. Katsube, T. Kushiro and M. Shibuya, Plant Cell Physiol., 2006, 47, 565–571 CrossRef PubMed.
- Z. Xue, L. Duan, D. Liu, J. Guo, S. Ge, J. Dicks, P. ÓMáille, A. Osbourn and X. Qi, New Phytol., 2012, 193, 1022–1038 CrossRef PubMed.
- M. Basyuni, H. Oku, E. Tsujimoto, K. Kinjo, S. Baba and K. Takara, FEBS J., 2010, 274, 5028–5042 CrossRef PubMed.
- R. Sun, S. Liu, Z. Z. Tang, T. R. Zheng, T. Wang, H. Chen, C. L. Li and Q. Wu, FEBS Open Bio, 2017, 7, 1575–1585 CrossRef PubMed.
- Y. Liu, Z. Zhao, Z. Xue, L. Wang, Y. Cai, P. Wang, T. Wei, J. Gong, Z. Liu and J. Li, Sci. Rep., 2016, 6, 33364 CrossRef PubMed.
- T. Kushiro, M. Shibuya and Y. Ebizuka, Eur. J. Biochem., 1998, 256, 238–244 CrossRef PubMed.
- G. Calegario, J. Pollier, P. Arendt, L. S. D. Oliveira, C. Thompson, A. R. Soares, R. C. Pereira, A. Goossens and F. L. Thompson, PLoS One, 2016, 11, e0165954 CrossRef PubMed.
- C. Brendolise, Y. K. Yauk, E. D. Eberhard, M. Wang, D. Chagne, C. Andre, D. R. Greenwood and L. L. Beuning, FEBS J., 2011, 278, 2485–2499 CrossRef PubMed.
- M. Takemura, R. Tanaka and N. Misawa, Appl. Microbiol. Biotechnol., 2017, 101, 6615–6625 CrossRef PubMed.
- K. Poralla, A. Hewelt, G. D. Prestwich, I. Abe, I. Reipen and G. Sprenger, Trends Biochem. Sci., 1994, 19, 157–158 CrossRef PubMed.
- I. Abe and G. D. Prestwich, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 9274–9278 CrossRef.
- R. Ito, Y. Masukawa and T. Hoshino, FEBS J., 2013, 280, 1267–1280 CrossRef PubMed.
- C. Fang, Y. Shang and D. Xu, Proteins: Struct., Funct., Bioinf., 2018, 86, 592–598 CrossRef PubMed.
- R. Liang, S. Cheng and X. Wang, Food Chem., 2018, 254, 170–184 CrossRef PubMed.
- M. Salmon, R. B. Thimmappa, R. E. Minto, R. E. Melton, R. K. Hughes, P. E. O'Maille, A. M. Hemmings and A. Osbourn, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E4407–E4414 CrossRef PubMed.
- T. Kushiro, M. Shibuya, A. Kazuo Masuda and Y. Ebizuka, J. Am. Chem. Soc., 2000, 122, 6816–6824 CrossRef.
- Z. Xue, Z. Tan, A. Huang, Y. Zhou, J. Sun, X. Wang, R. B. Thimmappa, M. J. Stephenson, A. Osbourn and X. Qi, New Phytol., 2018, 218, 1076–1088 CrossRef PubMed.
- C. Zhan, S. Ahmed, S. Hu, S. Dong, Q. Cai, T. Yang, X. Wang, X. Li and X. Hu, Biochem. Biophys. Res. Commun., 2017, 495, 1271–1277 CrossRef PubMed.
- K. Tamura, Y. Teranishi, S. Ueda, H. Suzuki, N. Kawano, K. Yoshimatsu, K. Saito, N. Kawahara, T. Muranaka and H. Seki, Plant Cell Physiol., 2017, 58, 874–884 CrossRef PubMed.
- H. Hori, G.-M. Pang, K. Harimaya, Y. Iitaka and S. Inayama, Chem. Pharm. Bull., 1987, 35, 2125–2128 CrossRef PubMed.
- G. M. Pang, C. J. Zhao, H. Hori and S. Inayama, Acta Pharm. Sin., 1989, 24, 75–79 Search PubMed.
- S. G. Del, G. Manfioletti and C. Schneider, BioTechniques, 1989, 7, 514–520 Search PubMed.
- Y. J. Zhao, X. Chen, M. Zhang, P. Su, Y. J. Liu, Y. R. Tong, X. J. Wang, L. Q. Huang and W. Gao, PLoS One, 2015, 10, e0125415 CrossRef PubMed.
- T. Xiang, M. Shibuya, Y. Katsube, T. Tsutsumi, M. Otsuka, H. Zhang, K. Masuda and Y. Ebizuka, Org. Lett., 2006, 8, 2835–2838 CrossRef PubMed.
- X. Rao, X. Huang, Z. Zhou and L. Xin, Biostatistics, Bioinformatics and Biomathematics, 2013, 3, 71–85 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra03468k |
| This journal is © The Royal Society of Chemistry 2018 |