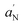Open Access Article
Open Access ArticleBioinspired hybrid eumelanin–TiO2 antimicrobial nanostructures: the key role of organo–inorganic frameworks in tuning eumelanin's biocide action mechanism through membrane interaction†
Giuseppe Vitiello ab,
Anna Zanfardinoc,
Olimpia Tammarobd,
Michela Di Napolic,
Maria Federica Casoe,
Alessandro Pezzella
ab,
Anna Zanfardinoc,
Olimpia Tammarobd,
Michela Di Napolic,
Maria Federica Casoe,
Alessandro Pezzella fgh,
Mario Varcamontic,
Brigida Silvestria,
Gerardino D'Erricobh,
Aniello Costantinia and
Giuseppina Luciani
fgh,
Mario Varcamontic,
Brigida Silvestria,
Gerardino D'Erricobh,
Aniello Costantinia and
Giuseppina Luciani *a
*a
aDepartment of Chemical, Materials and Production Engineering, University of Naples Federico II, p.le V. Tecchio 80, 80125 Naples, Italy. E-mail: luciani@unina.it; Fax: +39 081 76 82595; Tel: +39 081 76 82433
bCSGI, Consorzio Interuniversitario per lo Sviluppo dei Sistemi a Grande Interfase, Via della Lastruccia 3, 50019 Florence, Italy
cDepartment of Biology, University of Naples Federico II, Via Cintia 4, 80126 Naples, Italy
dCenter for Advanced Biomaterials for Health Care, Istituto Italiano di Tecnologia, Largo Barsanti e Matteucci, Naples, Italy
eNanofaber Spin-off at Italian National Agency for New Technologies, Energy and Sustainable Economic Development (ENEA), Casaccia Research Centre, Via Anguillarese 301, 00123 Rome, Italy
fInstitute for Polymers, Composites and Biomaterials (IPCB), CNR, Via Campi Flegrei 34, 80078 Pozzuoli (Na), Italy
gNational Interuniversity Consortium of Materials Science and Technology (INSTM), Piazza S. Marco, 4, 50121 Florence, Italy
hDepartment of Chemical Sciences, University of Naples Federico II, Via Cintia 4, 80126 Naples, Italy
First published on 7th August 2018
Abstract
Intrinsic biocide efficacy of eumelanins can be markedly enhanced through a templated formation in the presence of a TiO2-sol, leading to hybrid TiO2–melanin nanostructures. However, mechanisms and processes behind biocide activity still remain poorly understood. This paper discloses the fundamental mechanism of action of these systems providing mechanistic information on their peculiar interaction with Escherichia coli strains. To this purpose biocide characterization is combined with Electron Paramagnetic Resonance (EPR) spectroscopy to investigate radical species produced by the hybrids as well as their interactions with Gram(−) external bacterial membranes. Experimental results indicate that TiO2 mediated eumelanin polymerization leads to a peculiar mechanism of action of hybrid nanostructures, whose strong interactions with bacterial membranes enhance the action of reactive oxygen species (ROS) produced by eumelanin degradation itself, also concurring with the final biocide action. These findings provide strategic information for the development of eumelanin-based systems with enhanced activity against drug-resistant strains.
1. Introduction
Bacterial contamination is one of the major problems of human public health and food safety.1–4 Actually, fast development of drug resistant bacteria has been impairing efficacy of conventional antibiotics and high dose treatments are often required, rising toxicity concerns.5 This prompted the development of new efficient anti-bacterial systems, possibly acting through novel mechanisms that could impede the onset of bacterial resistance.6 To this purpose, bio-inspired approaches that look at nature's strategies against pathogen infections, can provide safe and high performance solutions.7,8 Accordingly, eumelanins, negatively charged and hydrophobic pigments found ubiquitously in many parts of living beings9 have been gaining great attention as bio-available compounds with huge technological potential.10 Eumelanins are truly multifunctional biopolymers,11 performing several biological functions such as photoprotection, photosensitization, free radical quenching, metal ion chelation and even intrinsic antimicrobial efficacy and biofilm inhibition without any photosensitization.1,12,13Notably, recent emerging evidence has disclosed and investigated the potential of eumelanin and eumelanin based materials as valuable antimicrobial agents,14–17 but mechanisms and processes behind biocide activity still remain poorly understood.1 Indeed some papers argue that antimicrobial action of eumelanins relies on ROS generation.18
Recent studies proved that eumelanins' biological functions can be markedly enhanced if the polymerization process of primarily monomers occurs in the presence of an inorganic nanostructured phase, acting as catalysts and structure directing agents in biopolymers building up.15–17,19 Following this approach eco-friendly, non-cytotoxic hybrid eumelanin–TiO2 nanostructures (NSs) were recently fabricated via a multistep process involving a sol–gel procedure (TiO2DHICA_ti nanosystems) and a subsequent exposition to ammonia vapors (TiO2DHICA_polym nanosystems), which showed up to 95% of antimicrobial activity against Escherichia coli DH5α.15–17 Actually, biocide efficacy of hybrid eumelanin–TiO2 nanostructures was even higher than bare eumelanin, suggesting a peculiar behavior of these systems and thus prompting investigation on the mechanism behind their antimicrobial action. Filling up this knowledge gap, will definitely provide significant contribution to shed light on biological action of these hybrid nanostructured pigments and boost progress towards a mature technology of eumelanin-based bioactive materials. This paper aims at elucidating processes and mechanisms underlying antimicrobial activity of eumelanin and hybrid eumelanin–TiO2 nanostructures. Actually, mechanisms of nanoparticles toxicity are very complicated, since several causes can be involved in the antimicrobial action including reactive oxygen species (ROS) production, bacterial membrane damage as well as cytotoxic moieties leaching.20,21 Each aspect was systematically addressed in this study through a joint chemical and biological investigation approach. Particularly, EPR spectroscopy provided key information to identify, by spin-trapping approach, the radical species produced by the nanostructures and to assess their interaction with lipid bilayers biomimicking the external membrane of Escherichia coli bacteria, by spin-labelling method. Results of EPR investigation were confirmed by biological assessment, also revealing the non-cytotoxic behavior of hybrid NSs. Filling this knowledge gap would allow to achieve the highest disinfection power of eumelanin–TiO2 based systems, providing the basis for their utilization as antimicrobial agents for biomedical and food-packaging fields.
2. Methods
2.1. Materials
Titanium isopropoxide (TTiP), isopropanol, acetic acid and triethylamine (TEA) were purchased from Sigma Aldrich (St. Louis, MO, USA) and used as received. 5,6-dihydroxyindole-2-carboxylic acid (DHICA) monomer was prepared as described elsewhere.21,22 Dichloromethane and methanol, HPLC-grade solvents, were obtained from Merck (Darmstadt, Germany).The phospholipids 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn glycero-3-[phospho-rac-(1-glycerol)] (DOPG) were obtained from Avanti Polar Lipids (Birmingham, AL, USA), while the lipopolysaccharides from Escherichia coli O111:B4 were obtained from Sigma-Aldrich. PBS buffer (10 mM phosphate buffer, 137 mM NaCl, 2.7 mM KCl, pH 7.4), 5,5-dimethyl-1-pyrroline-1-oxide (DMPO) spin-trap and spin-labelled phosphatidylcholine (5-PCSL) with the nitroxide group at 5 position in the sn-2 acyl chain were obtained from Sigma-Aldrich. The spin-label was stored at −20 °C in ethanol solutions at a concentration of 1 mg mL−1.
About the reagents for biological tests, Luria-Bertani (LB) agar, 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI), propidium iodide (PI), Dulbecco Modified Eagle Medium (DMEM) and penicillin–streptomycin were also obtained from Sigma-Aldrich, while fetal bovine serum was obtained from HyClone, GE Healthcare Lifescience (Chicago, IL).
Human immortalized non tumorigenic keratinocyte cell line HaCaT (Ethnicity, Caucasian; age, 62 years; gender, male and tissue, skin) was supplied by CLS Cell Lines Service, Germany.
The usage of human cells for the planned scientific analyses followed the ethical principles concerning human experimentation, approved in advance by the local ethic committee.
2.2. Synthesis of eumelanin–TiO2 nanostructures
Eumelanin–TiO2 nanostructures (NSs) were prepared by hydrothermal synthesis as previously described,15,17 by using TTiP and DHICA as precursors of the inorganic and organic phases, respectively. Suspensions of hybrid TiO2DHICA_polym nanostructures were obtained following a previously described procedure.15,17 Bare TiO2 and DHICA–eumelanin nanostructures were also prepared as ref. 15, 17 and 23.2.3. Antimicrobial assays
The antimicrobial activity of TiO2, TiO2DHICA_polym, TiO2DHICA_ti and DHICA–eumelanin samples was valuated against Escherichia coli DH5α. A single colony of this strain was resuspended in 5 mL of Luria-Bertani (LB) broth and incubated overnight at 37 °C.24 When the culture reached an OD600 of 1 unit, it was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in 20 mM phosphate buffer composed by NaCl 137 mM, KCl 2.7 mM, Na2HPO4 10 mM, KH2PO4 1.8 mM, CaCl2 1 mM, MgCl2·6H2O 0.5 mM at pH = 7.4. NSs suspensions were sonicated on ice with a tip-sonicator at 50% amplitude for 10 minutes (alternating 30 s on/off) before to prepare the mixture with bacteria. In dose–response curves, the samples were prepared by adding 1/25 of the volume of bacterial cells and NSs were used at different concentrations (from 50 to 400 μg mL−1), 500 μL final volume was reached with 20 mM phosphate buffer at pH = 7.4. Negative control was represented by cells with any treatment. Samples were incubated at 37 °C for 10 minutes or 4 hours, two dilutions (1
100 in 20 mM phosphate buffer composed by NaCl 137 mM, KCl 2.7 mM, Na2HPO4 10 mM, KH2PO4 1.8 mM, CaCl2 1 mM, MgCl2·6H2O 0.5 mM at pH = 7.4. NSs suspensions were sonicated on ice with a tip-sonicator at 50% amplitude for 10 minutes (alternating 30 s on/off) before to prepare the mixture with bacteria. In dose–response curves, the samples were prepared by adding 1/25 of the volume of bacterial cells and NSs were used at different concentrations (from 50 to 400 μg mL−1), 500 μL final volume was reached with 20 mM phosphate buffer at pH = 7.4. Negative control was represented by cells with any treatment. Samples were incubated at 37 °C for 10 minutes or 4 hours, two dilutions (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and 1
100 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) of all the samples were placed on solid medium LB agar and incubated overnight at 37 °C.25 The same assay was performed on Escherichia coli DH5α cells incubated with 200 μg mL−1 of TiO2DHICA_polym having different storage times (0, 2, 5, 15 and 21 days). In the end, the assay was carried out on Escherichia coli DH5α cells incubated with TiO2DHICA_polym and NSs supernatant (volume% corresponding to the NSs concentration of 200 μg mL−1), obtained by centrifugation at 13
1000) of all the samples were placed on solid medium LB agar and incubated overnight at 37 °C.25 The same assay was performed on Escherichia coli DH5α cells incubated with 200 μg mL−1 of TiO2DHICA_polym having different storage times (0, 2, 5, 15 and 21 days). In the end, the assay was carried out on Escherichia coli DH5α cells incubated with TiO2DHICA_polym and NSs supernatant (volume% corresponding to the NSs concentration of 200 μg mL−1), obtained by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and 4 °C for 2 hours, precipitation of the nanoparticles and filtration with filters of 0.22 μm. The following day the surviving cells were estimated by colony counting on each plate and compared with the controls. At the same way, the antimicrobial activity of H2O2 against Escherichia coli DH5α was tested. Samples were prepared by adding H2O2 at different concentrations (from 0.1 to 1000 μM). The following day, the surviving cells were estimated by colony counting on each plate and compared with the controls. Standard deviations were less than 5% for each experiment (which was performed at least in triplicate).
000 rpm and 4 °C for 2 hours, precipitation of the nanoparticles and filtration with filters of 0.22 μm. The following day the surviving cells were estimated by colony counting on each plate and compared with the controls. At the same way, the antimicrobial activity of H2O2 against Escherichia coli DH5α was tested. Samples were prepared by adding H2O2 at different concentrations (from 0.1 to 1000 μM). The following day, the surviving cells were estimated by colony counting on each plate and compared with the controls. Standard deviations were less than 5% for each experiment (which was performed at least in triplicate).
2.4. EPR investigation
Two sets of EPR measurements were performed, by using spin-trapping and spin-labelling methods. The first one was used to detect ROS production by eumelanin–TiO2 nanohybrids, while the second one allowed to investigate the interaction of antimicrobial nanostructures with lipid bilayers mimicking the external Gram-negative membranes.For the detection of ROS species in aqueous dispersions of hybrid NSs, samples were prepared by using the following procedure: a specific amount of a stock DMPO aqueous solution was added in 1 mL of hybrid NSs suspensions (at a concentration of 2 mg mL−1) in order to have a final spin-trap concentration of 20 mM. After 10 minutes from dispersion preparation, the samples were centrifuged and the supernatants were analyzed by EPR spectroscopy. For all samples, the time evolution of DMPO-adduct signals was monitored.
To study the interaction of antimicrobial nanostructures with mimicking biomembranes, liposomes of DOPE/DOPG/LPS 48/12/40 w/w/w (60/15/25 mol mol−1 mol−1) were prepared mixing appropriate amounts of DOPE, DOPG and LPS from Escherichia coli O111:B4, dissolved in CH2Cl2–CH3OH mixture (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v, 10 mg mL−1 lipid concentration), in a round-bottom test tube. In order to perform EPR measurements, a proper volume of the spin-labelled phosphatidylcholine (n-PCSL) stock solution in ethanol (1 mg mL−1) was also added to the lipid organic mixture in order to have a spin-label content equal to 1% by weight on total lipids. Then, a thin lipid film was produced by evaporating the solvents with dry nitrogen gas. Final traces of solvents were removed by subjecting the sample to vacuum desiccation for at least 3 h. The final dried film samples were then hydrated with 50 μL of 10 mM phosphate buffer (PBS, 137 mM NaCl, 2.7 mM KCl at pH 7.4) and repeatedly vortexed, obtaining a Multi-Lamellar Vesicles (MLVs) suspension, which was then extruded at room temperature (11 extrusions through a 100 nm polycarbonate membrane) to obtain a Large Unilamellar Vesicles (LUVs) suspension. Lipid samples also containing eumelanin–TiO2, bare TiO2 and DHICA–eumelanin nanostructures respectively were prepared by following the same procedure, adding a specific amount of the corresponding nanostructures suspension to the liposomes ones. The nanostructures–lipid ratio was 0.5
1 v/v, 10 mg mL−1 lipid concentration), in a round-bottom test tube. In order to perform EPR measurements, a proper volume of the spin-labelled phosphatidylcholine (n-PCSL) stock solution in ethanol (1 mg mL−1) was also added to the lipid organic mixture in order to have a spin-label content equal to 1% by weight on total lipids. Then, a thin lipid film was produced by evaporating the solvents with dry nitrogen gas. Final traces of solvents were removed by subjecting the sample to vacuum desiccation for at least 3 h. The final dried film samples were then hydrated with 50 μL of 10 mM phosphate buffer (PBS, 137 mM NaCl, 2.7 mM KCl at pH 7.4) and repeatedly vortexed, obtaining a Multi-Lamellar Vesicles (MLVs) suspension, which was then extruded at room temperature (11 extrusions through a 100 nm polycarbonate membrane) to obtain a Large Unilamellar Vesicles (LUVs) suspension. Lipid samples also containing eumelanin–TiO2, bare TiO2 and DHICA–eumelanin nanostructures respectively were prepared by following the same procedure, adding a specific amount of the corresponding nanostructures suspension to the liposomes ones. The nanostructures–lipid ratio was 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/w.
1 w/w.
EPR experiments were carried out by means of X-band (9 GHz) Bruker Elexys E-500 spectrometer (Bruker, Rheinstetten, Germany), equipped with a super-high sensitivity probe head. Liquid samples were transferred to flame-sealed glass capillaries which, in turn, were coaxially inserted in a standard 4 mm quartz sample tube. Measurements were performed at room temperature. The instrumental settings were as follows: sweep width, 100 G; resolution, 1024 points; modulation frequency, 100 kHz; modulation amplitude, 1.0 G. The spectra of surnatants and lipid samples were registered with an attenuation of 15 dB and several scans, respectively 512 and 32, were accumulated to improve the signal-to-noise ratio.
2.5. H2O2 detection by spectrophotometer method
Ferrous oxidation xylenol orange (FOX) assay was employed to detect H2O2 in nanostructures suspensions. More in detail, 0.2 mL of TiO2DHICA_polym nanoparticles suspension (1.5 mg mL−1) were incubated both in absence and with 0.2 mL of H2O2 0.25 μM (30 min). Each solution was then mixed with 4 mL of reagent mixture containing 0.10 μM xylenol orange, 0.25 μM ammonium iron(II) sulfate hexahydrate in 250 μM H2SO4 (30 mL) and 3.88 mM 1,1,3,3-tetramethoxypropane, butylated hydroxyanisole (BHA) in methanol (270 mL). After 30 min under stirring at room temperature, absorbance at 593 nm was measured. Control experiments were performed in absence of TiO2DHICA_polym or H2O2. A calibration curve was also prepared with H2O2.262.6. DAPI/PI dual staining and fluorescence microscopy image acquisition
For dual staining, 200 μL of bacterial culture (bacteria were grown to mid-logarithmic phase) was incubated in the dark for 10 minutes or 4 hours at 37 °C in agitation in the presence or absence of TiO2, and TiO2DHICA_polym at a concentration of 200 μg mL−1. After the incubation, 10 μL of bacterial culture was mixed with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) solution (1 μg mL−1 DAPI final concentration) and propidium iodide (PI) 20 μg mL−1. Samples were observed using an Olympus BX51 fluorescence microscope (Olympus, Tokyo, Japan) using a DAPI filter (excitation/emission: 358/461 nm). Standard acquisition times were 1000 ms for DAPI/PI dual staining. Images were captured using an Olympus DP70 digital camera.272.7. Eukaryotic cell cultures
HaCat (human keratinocytes) cells are a spontaneously transformed aneuploid immortal keratinocyte cell line from adult human skin, widely used in scientific research.28 These cells were maintained in Dulbecco Modified Eagle Medium (DMEM), supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Cells were cultured at 37 °C in humidified atmosphere of 5% CO2. The compound TiO2DHICA_polym were used for treatment at increasing concentrations in complete growth medium for the cytotoxicity assay.292.8. Cytotoxicity on mammalian cells
Cytotoxicity on HaCat cells was assessed by performing the 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide (MTT) reduction inhibition assay, the colorimetric assay for assessing cell metabolic activity. Cells were grown as previously described and plated on 96-well plates at a density of 5 × 103 cells per well, in 200 μL of medium containing TiO2DHICA_polym (100, 200, 400, 800 μg mL−1) for 24 hours. After 24 hours of treatment, the medium was aspirated and 10 μL of a stock MTT solution was added to the cells to a final concentration of 0.5 mg mL−1. After 4 h incubation the MTT solution was removed and the formazan salts were dissolved in 100 μL of 0.1 N HCl in anhydrous isopropanol. Cell survival was expressed as the absorbance of blue formazan measured at 570 nm with an automatic plate reader (Multi scan spectrum, Thermo Scientific, Waltham, MA, USA). Cytotoxicity test was performed at least 3 times. Standard deviations were always <5% for each experiment.302.9. Statistical analysis
Statistical analysis was performed using a Student's t-Test. Significant differences were indicated as (P < 0.05) and (P < 0.01).3. Results
3.1. Antimicrobial dose–response analysis
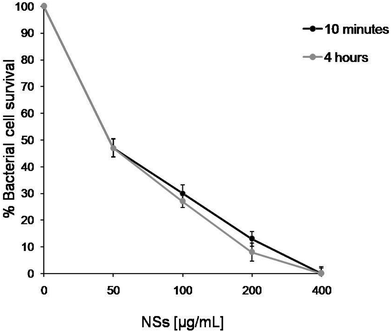
TiO2DHICA_polym freshly prepared have been used for dose–response curves, incubating E. coli cells with different nanostructures concentrations (from 0 to 400 μg mL−1) to test their activity. In previous studies15,17 hybrid NSs showed an excellent antimicrobial activity against the E. coli bacterial strain after 4 hours of treatment at 200 μg mL−1; in this study the NSs activity was confirmed (Fig. 1) and a very fast biocide action was observed since it occurred as early as 10 minutes following incubation of E. coli cells with TiO2DHICA_polym NSs. As shown in Fig. 1, the antimicrobial activity results are proportional to the concentration of the NSs, in addition the curves are almost completely overlapped, indicating that the antimicrobial activity was very similar for both used incubation time. A similar kinetics and dose dependent trends resulted after incubation of E. coli cells with DHICA–eumelanin and eumelanin–TiO2 nanostructures obtained without the exposition to ammonia vapors (TiO2DHICA_ti15,17), as reported in Fig. S1A and B.† However, our attention was focused on TiO2DHICA_polym NSs showing the highest antimicrobial activity and this formulation is considered the best candidate for future applications in the pharmacological field.3.2. Effect of storage on antimicrobial activity
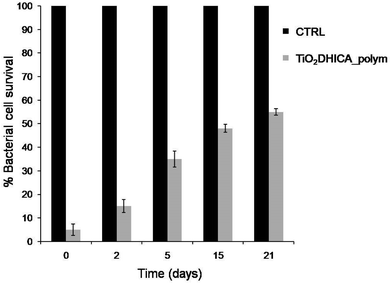
Antimicrobial activity was also checked using aged NSs (concentrated at 200 μg mL−1), after different storage time at room temperature before use. As reported in Fig. 2, the antimicrobial activity of freshly prepared NSs (TiO2DHICA_polym) was very high in the beginning, about 90% of the bacteria did not survive, but decreased slowly after several days' storage. However, after 21 days' storage the TiO2DHICA_polym NSs still caused a 50% of bacterial mortality.3.3. Antimicrobial test with NSs supernatants and NSs in dark conditions
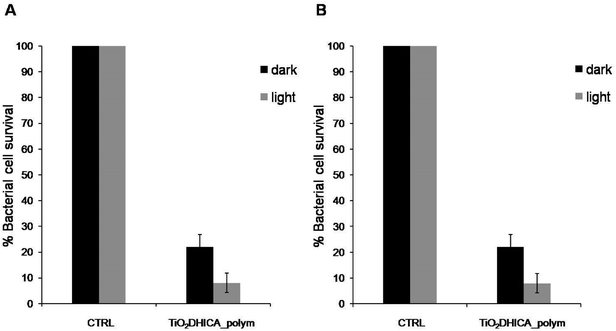
In order to evaluate if the nanostructures would release antimicrobial compounds in solution due to eumelanin degradation, we decided to separate TiO2DHICA_polym nanostructures from solvent, and to treat bacterial cells with the filtered supernatants. Our data showed that the supernatant did not have antimicrobial activity, which is therefore dependent on direct exposition of the bacteria to the nanoparticles (Fig. S2†).In addition, antimicrobial activity assay were performed at a fixed concentration of hybrid nanostructures (200 μg mL−1) used in dark conditions. As shown in Fig. 3, there was a slight decrease in the antimicrobial activity, compared to the same experiment carried out under light conditions. Same antimicrobial effects were observed after 4 hour or 10 minutes of NSs treatment (Fig. 3).
3.4. EPR spin-trapping experiments
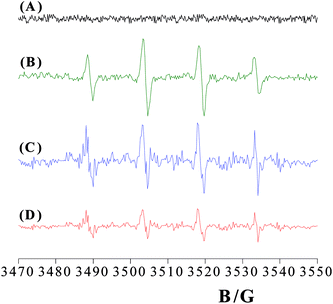
In order to assess if biocide action of eumelanin–TiO2 NSs was triggered by oxidative stress due to free radical production, generation of reactive oxygen species (ROS) in aqueous environment was investigated by EPR spin-trapping method. Indeed, the extremely high chemical reactivity of these species implies that they have too short a lifespan to be detected, so hampering direct measurements. EPR spin-trapping technique represented a useful method to overcome this problem.16,31,32 The reactive free radicals join with adequate reagents, the spin-traps, to yield spin-adducts, which are longer-living free radicals and can be monitored by EPR spectroscopy at room temperature (Fig. S3†). Preventively, we checked that DMPO in the NSs aqueous suspension does not produce any interfering signals (Fig. 4A). Both DHICA–eumelanin and TiO2DHICA_polym NSs in aqueous environment causes the appearance of a signal in the EPR spectrum showing a characteristic quartet with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 intensity ratio (Fig. 4B–D). It corresponds to the DMPO–OH adduct formed from the trapping of ·OH radical on DMPO.33 A quantitative analysis of this spectrum was realized determining the hyperfine coupling constants for the nitroxide nitrogen and for the β-proton, aN = aHβ = 14.8 ± 0.1 G. These values are consistent with those reported in the literature for the DMPO–OH adducts,34 confirming the formation of significant amounts of ·OH radicals in DHICA–eumelanin and TiO2DHICA_polym nanostructures suspensions (Fig. 4B–D). No signals were appreciated in the EPR spectrum of bare TiO2 nanostructures in aqueous suspension, thus confirming the absence of any radical species in the system.
1 intensity ratio (Fig. 4B–D). It corresponds to the DMPO–OH adduct formed from the trapping of ·OH radical on DMPO.33 A quantitative analysis of this spectrum was realized determining the hyperfine coupling constants for the nitroxide nitrogen and for the β-proton, aN = aHβ = 14.8 ± 0.1 G. These values are consistent with those reported in the literature for the DMPO–OH adducts,34 confirming the formation of significant amounts of ·OH radicals in DHICA–eumelanin and TiO2DHICA_polym nanostructures suspensions (Fig. 4B–D). No signals were appreciated in the EPR spectrum of bare TiO2 nanostructures in aqueous suspension, thus confirming the absence of any radical species in the system.
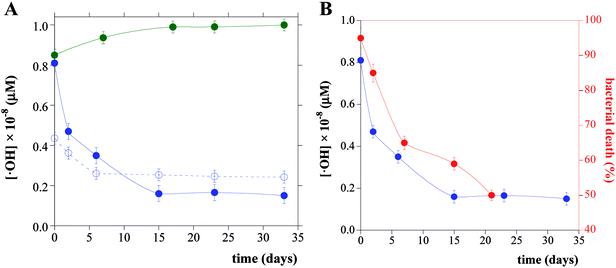
The concentration and the time evolution of ·OH produced by DHICA–eumelanin and TiO2DHICA_polym NSs, stored both under environment light and in the dark, was determined by evaluating the intensity of the adducts EPR spectra. Particularly, it was obtained by double integration of the first derivative signal using the standard methods reported in previous works.35 The experimental results, shown in Fig. 5A, indicated the formation of a low ·OH concentration (∼10−8 μM) for all considered systems. Specifically, in the case of DHICA–eumelanin and TiO2DHICA_polym nanostructures stored in the dark, no significant changes have been observed in time, while a slight decrease was observed in the first 15 days in the case of TiO2DHICA_polym stored under environment light. In Fig. 5B, the time evolution of ·OH concentration produced by TiO2DHICA_polym nanostructures stored under environment light was related to the corresponding biocide activity, showing a similar decreasing trend.
3.5. H2O2 generation and its effect on NSs antimicrobial activity
In order to assess oxidative potential of NSs, possible generation of H2O2 was also checked because of the potential toxicity of hydrogen peroxide. The assay based on the FOX method revealed 0.10 ± 0.02 μM H2O2 formation at 30 min incubation under oxygen streaming of a 1.5 mg mL−1 particle suspension. Although the H2O2 formation was observed, the detected level appeared too little for possible biocide action, because of the highly oxidizing conditions required (i.e. the oxygen stream). In order to understand the hydrogen peroxide contribution to the antimicrobial activity of NSs, a dose response curves was carried out by adding to the bacterial cells H2O2 at different concentrations (from 0 to 1000 μM). The cellular mortality was proportional to the increase in H2O2 concentration as shown in ESI† (Fig. S4†). At the concentration found in the NSs solution about 0.10 ± 0.02 μM, the antimicrobial activity of the hydrogen peroxide was absent. This experiment has shown that H2O2 alone, at measured concentration levels in solution was not sufficient to achieve high levels of mortality in solution.3.6. EPR spin-labelling experiments
In order to assess interactions of NSs with bacterial cell membranes, spin-labelling experiments were carried out investigating the effect of the eumelanin-based nanostructures on the microstructure of bio-mimicking external bacterial membrane. DOPE and DOPG were chosen to reproduce the zwitterionic and cation phospholipids usually present into external bacterial membranes while Escherichia coli O111:B4 lipopolysaccharide represents the characteristic LPS molecule consisting of a glycolipid moiety and a saccharide portion covalently linked playing the key role in the pathogenesis of Gram(−) infections.36,37 The phosphatidylcholine spin-labelled on the 5-position of the sn-2 chain (5-PCSL) was incorporated in lipid bilayers of DOPE/DOPG/LPS 48/12/40 w/w/w. The presence of spin-labelled lipid did not causes changes in the liposomes mesoscopic properties, as demonstrated by DLS analysis (Fig. S5†). 5-PCSL monitors the bilayer region just underneath the hydrophilic interface and is the most indicative to evaluate changes in the bilayer microstructure. 5-PCSL spectra in DOPE/DOPG/LPS membranes in the absence and presence of TiO2DHICA_polym nanostructures are shown in Fig. 6. The effect of bare TiO2 and DHICA–eumelanin was also investigated and the corresponding EPR spectra are shown in Fig. 6.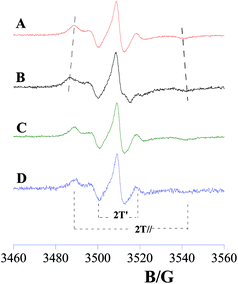 | ||
| Fig. 6 EPR spectra of 5-PCSL in DOPE/DOPG/LPS membranes before nanostructures addition (A) and after addition of TiO2DHICA_polym (B), bare TiO2 (C) and DHICA–eumelanin nanostructures (D). | ||
A well-resolved anisotropic lineshape is observed in all cases, indicating that the rotational motion of the labelled molecules along one axis is different from that in the other two directions. This lineshape is typical of molecules associated to layered structures such as lipid bilayers, in which the rotation along the normal to the bilayer interface is different with respect to those along the axes lying on it. Thus, the EPR spectra clearly demonstrate that bare TiO2 (Fig. 6C) and DHICA–eumelanin (Fig. 6D) do not cause significant changes in the spectrum of 5-PCSL in DOPE/DOPG/LPS bilayers. In contrast, a strong increase in the anisotropic character was observed in the spectrum after TiO2DHICA_polym addition (Fig. 6B), suggesting a significant perturbation of the lipid bilayers caused by the hybrid nanostructures. This effect was also confirmed by the decrease in the DOPE/DOPG/LPS liposomes, after the addition of TiO2DHICA_polym nanostructures (Fig. S5†), probably inducing a partial rearrangement of the lipid aggregates. A quantitative analysis of EPR spectra was performed by determining, from the splittings between the minimum and maximum points, the two parameters 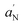 and S.38–40 S is the order parameter and is related to the angular amplitudes of motion of the label, which in turn reflects the motion of the acyl chain segment to which the label is bound, while
and S.38–40 S is the order parameter and is related to the angular amplitudes of motion of the label, which in turn reflects the motion of the acyl chain segment to which the label is bound, while 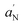 is an index of the micropolarity experienced by the nitroxide.
is an index of the micropolarity experienced by the nitroxide. 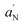 and S were calculated according to the following relations:
and S were calculated according to the following relations:
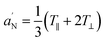 | (1) |
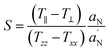 | (2) |
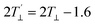 ). Txx and Tzz are the principal elements of the real hyperfine splitting tensor in the spin Hamiltonian of the spin-label, which can be measured from the corresponding single-crystal EPR spectrum and are reported in the literature (Txx = 6.1 G and Tzz = 32.4 G). aN is the isotropic hyperfine coupling constant for the spin-label in crystal state, given by:
). Txx and Tzz are the principal elements of the real hyperfine splitting tensor in the spin Hamiltonian of the spin-label, which can be measured from the corresponding single-crystal EPR spectrum and are reported in the literature (Txx = 6.1 G and Tzz = 32.4 G). aN is the isotropic hyperfine coupling constant for the spin-label in crystal state, given by:
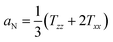 | (3) |
The 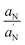 ratio in eqn (2) corrects the order parameter for polarity differences between the crystal state and the membrane. To obtain reliable values of T‖ and T⊥ splittings, a home-made MATLAB-based software routine was used. The values of the spectral parameters are collected in Table 1.
ratio in eqn (2) corrects the order parameter for polarity differences between the crystal state and the membrane. To obtain reliable values of T‖ and T⊥ splittings, a home-made MATLAB-based software routine was used. The values of the spectral parameters are collected in Table 1.
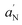 , of DOPE/DOPG/LPS 60/5/25 bilayers before and after nanostructures addition. The evident increase in both S and
, of DOPE/DOPG/LPS 60/5/25 bilayers before and after nanostructures addition. The evident increase in both S and 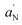 values after the addition of TiO2DHICA_polym NSs to lipid bilayers indicates an increase in the ordering and micropolarity of biomembranes due to the presence of hybrid nanostructures
values after the addition of TiO2DHICA_polym NSs to lipid bilayers indicates an increase in the ordering and micropolarity of biomembranes due to the presence of hybrid nanostructures
3.7. Fluorescence microscopy with NSs
From EPR spin-labelling experiments, a clear interaction between the bacterial membrane and the nanostructures was proved. To support these data, fluorescence microscopy experiments were performed to verify the effect of NSs on the integrity of bacterial membranes. Escherichia coli cells were used and stained with DAPI, fluorescent stain for DNA, and propidium iodide, able to enter only cells with damaged membranes and therefore it is considered an indicator of cell membrane disruption. Results of these experiments are reported in Fig. 7. When membranes are intact, bacterial cells appear blue because of DAPI fluorescence (Fig. 7C). After TiO2DHICA_polym (200 μg mL−1) treatment for 10 minutes (Fig. 7D) a significant amount of cells developed a red fluorescence suggesting the disruption of membrane integrity. Fig. 7B shows adhesion of bacterial cells on nanostructured aggregate.3.8. Cytotoxicity assays
Cytotoxicity of NSs against eukaryotic cells was evaluated by MTT assay according to the procedure described in the method section. The cytotoxic effect of TiO2DHICA_polym on human cell lines HaCat (keratinocytes) has been verified. Cells were treated with NPs at various concentrations (from 0 to 800 μg mL−1) for 24 hours, and no toxicity was found (Fig. S5†).4. Discussion
EPR technique allowed to detect the presence of ·OH radicals in aqueous suspensions of DHICA–eumelanin and TiO2DHICA_polym NSs. Furthermore, UV spectroscopic assay revealed a small amount of H2O2. Since no radical species could be appreciated in TiO2 suspensions, melanin must be involved in ROS formation. Several studies report that melanin photo-degradation in aerated environment involves ROS production through the interaction of the excited state of melanin molecules with molecular oxygen.41 In aqueous solutions these pigments undergo to reversible red-ox reactions, with hydroquinone (HQ) moieties coexisting in equilibrium with quinone (Q) species and semiquinone (Q.) radicals.42 Semiquinones would further induce the generation of ROS according to the pathway previously proposed.43–45More in detail, SQ would transfer electron to O2 forming superoxide anion (·O2−), that can react with SQ themselves or dismutate to produce H2O2, that was revealed by spectroscopic assay. Hydrogen peroxide can be consequently involved in Fenton-like reactions to produce ·OH radicals, as confirmed by EPR analysis.44 Furthermore, it must be considered that DHICA can act as a ligand for Ti4+ ions through its catechol group, thus forming a ligand to metal charge transfer complex (LMCTC).16,17 TiO2DHICA-LMCTC is incorporated into the hybrid structure; thus it could induce ROS production even upon visible light irradiation.16,17 Therefore, a possible reason to explain the antimicrobial NSs mechanism of action is the effect of light on melanin which could give rise to the formation of reactive oxygen species such as hydrogen peroxide, a toxic compound for bacterial cells if present over determinate concentrations.46 Particularly, among detected ROS, ·OH species are expected to play a major role in biological damage, since measured H2O2 concentration (only 0.10 ± 0.02 μM) is too low to determine any cytotoxicity effect, as confirmed by biological tests carried out in the presence of H2O2 in similar concentration.
To verify this hypothesis, an antimicrobial activity assay has been performed at a fixed concentration of nanoparticles (200 μg mL−1) in dark conditions where production of ·OH species is strongly depressed, as shown by EPR data reported in Fig. 5. As shown in Fig. 3, NSs showed significant yet lower antimicrobial action even in these conditions, compared to the same experiment carried out under light conditions. Moreover, bare DHICA–eumelanin nanostructures displayed a lower antimicrobial action despite a comparable ROS production, as appreciated by EPR spectra (Fig. 5B). Finally, hybrid NSs kept a residual biocide efficacy even after 5 days' soaking in aqueous solutions, when ROS production was markedly depressed.
On these bases, ROS formation could only partly account for antibacterial activity of TiO2DHICA nanostructures. Thus, some other causes must concur to overall antimicrobial performance of TiO2DHICA_polym NSs. In order to outline the exact antimicrobial mechanism of TiO2DHICA, elucidating all factors for toxicity against Gram(−) bacteria, results of spin-labelling experiments were compared and integrated with fluorescence microscopy observations.
Optical microscopy images in Fig. 7 show bacterial adhesion upon TiO2DHICA_polym aggregates formed within the culture batch (Fig. 7D). As reported in literature, melanogenic precursors such as dopamine and L-dopa display high stable binding to membrane lipids, mediated through H-bonds.46 The same behavior is expected for DHICA molecules in TiO2DHICA hybrids. Indeed, previous studies reveal additional oxidation of eumelanin pigment in TiO2DHICA_polym nanostructures, with a relative increase of carboxyl forms when the eumelanin–TiO2 nanohybrids produced by hydrothermal synthesis were incubated in the oxygen/ammonia atmosphere.17 The presence of a higher number of carboxylic group should increase the number of hydrogen bonds with lipid head groups, thus improving affinity and binding to bacterial membranes.47 This higher interaction to cytoplasmic membrane should account for changes in its structure, as revealed by spin-labelling experiments (Fig. 5), thus increasing permeability to ROS cytotoxic species and enhancing their action, ultimately leading to membrane damage and cells death, as confirmed by optical microscope images (Fig. 7).
5. Conclusions
TiO2 nanoparticle templated eumelanin polymerization confers to the final hybrid nanostructures a peculiar mechanism of activity against Gram(−) bacteria. Antibacterial activity of these nanostructures results from a combined effect of cell wall destabilization and oxidative stress due to ROS produced during melanin degradation. Particularly, the enhanced interaction of TiO2DHICA_polym nanostructures with bacterial biomembranes, also related to the high oxidation degree of eumelanin component with relative increase of carboxyl forms in the considered hybrids, results in membrane destructuring and increased permeability, even enhancing ROS action and ultimately producing membrane disruption. This combined mechanism is expected to prohibit microbes from developing antibiotic resistance to these nanostructures, thus addressing a serious limitation of other more conventional antimicrobial agents. High biocide efficacy of these systems joined with the absence of any cytotoxic effect observed on eukaryotic cells set their huge potential as topical antimicrobial agents for wound care. The picture here disclosed concerning the biocide activity of eumelanin–TiO2 hybrid nanostructures clearly outlines the valence of ceramic templated melanin formation as well as melanin oxidation degree in modulating interactions with bacterial membrane and overall biological action. This will provide strategic guidelines for future design of melanin based antimicrobial surfaces with relevant antimicrobial activity for biomedical and food-packaging applications.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This work was supported by University of Naples “Federico II” Research Grant (Finanziamento per la Ricerca di Ateneo, BioDressMel, DR/2017/408-07/02/2017).References
- C. Xu, J. Li, L. Yang, F. Shi, L. Yang and M. Ye, Food Control, 2017, 73, 1445–1451 CrossRef.
- C. Liu, J. Guo, X. Yan, A. Tang, S. Mazumder, Y. Wu and Y. Yan Liang, Environ. Rev., 2017, 25, 225–244 CrossRef.
- I. Finore, P. Orlando, P. Di Donato, L. Leone, B. Nicolaus and A. Poli, Int. J. Syst. Evol. Microbiol., 2016, 66, 1554–1560 CrossRef PubMed.
- S. Zaidi, L. Misba and A. U. Khan, Nanomedicine: Nanotechnology, Biology and Medicine, 2017, 13, 2281–2301 CrossRef PubMed.
- A. S. Abd-El-Aziz, C. Agatemor and N. Etkin, Biomaterials, 2017, 118, 27–50 CrossRef PubMed.
- F. Rabanal, A. Grau-Campistany, X. Vila-Farrés, J. Gonzalez-Linares, M. Borràs, J. Vila, A. Maresa and Y. Cajal, Sci. Rep., 2015, 5, 10558 CrossRef PubMed.
- I. Malagurski, S. Levic, M. Pantic, D. Matijasevic, M. Mitric, V. Pavlovic and S. Dimitrijevic-Brankovic, Carbohydr. Polym., 2017, 165, 313–321 CrossRef PubMed.
- G. Vitiello, B. Silvestri and G. Luciani, Curr. Top. Med. Chem., 2018, 18, 22–41 CrossRef PubMed.
- G. Prota, Melanins and Melanogenesis, Melanoma Res., 1992, 3, 1–290 Search PubMed.
- M. d'Ischia, K. Wakamatsu, F. Cicoira, E. Di Mauro, J. C. Garcia-Borron, S. Commo, I. Galván, G. Ghanem, K. Kenzo, P. Meredith, A. Pezzella, C. Santato, T. Sarna, J. D. Simon, L. Zecca, F. A. Zucca, A. Napolitano and S. Ito, Pigm. Cell Melanoma Res., 2015, 28, 520–544 CrossRef PubMed.
- M. d'Ischia, A. Napolitano, A. Pezzella, P. Meredith and T. Sarna, Angew. Chem., Int. Ed. Engl., 2009, 48, 3914–3921 CrossRef PubMed.
- G. S. Kiran, S. A. Jackson, S. Priyadharsini, A. D. W. Dobson and J. Selvin, Sci. Rep., 2017, 7, 9167 CrossRef PubMed.
- S. Yasar Yildiz, A. Anzelmo, T. Ozer, N. Radchenkova, S. Genc, P. Di Donato, B. Nicolaus, E. Toksoy Oner and M. Kambourova, J. Appl. Microbiol., 2014, 116, 314–324 CrossRef PubMed.
- A. Pezzella, L. Capelli, A. Costantini, G. Luciani, F. Tescione, B. Silvestri, G. Vitiello and F. Branda, Mater. Sci. Eng., C, 2013, 33, 347–355 CrossRef PubMed.
- G. Vitiello, A. Pezzella, A. Zanfardino, M. Varcamonti, B. Silvestri, A. Costantini, F. Branda and G. Luciani, J. Mater. Chem. B, 2015, 3, 2808–2815 RSC.
- G. Vitiello, A. Pezzella, V. Calcagno, B. Silvestri, L. Raiola, G. D'Errico, A. Costantini, F. Branda and G. Luciani, J. Phys. Chem. C, 2016, 120, 6262–6268 CrossRef.
- G. Vitiello, A. Pezzella, A. Zanfardino, B. Silvestri, P. Giudicianni, A. Costantini, F. Branda and G. Luciani, Mater. Sci. Eng., C, 2017, 75, 454–462 CrossRef PubMed.
- H. Liu, X. Qu, E. Kim, M. Lei, K. Dai, X. Tan, M. Xu, J. Li, Y. Liu, X. Shi, P. Li, G. F. Payn and C. Liu, Biomaterials, 2018, 162, 109–122 CrossRef PubMed.
- B. Silvestri, G. Vitiello, G. Luciani, G. Calcagno, V. Costantini, M. Gallo, S. Parisi, S. Paladino, M. Iacomino, G. D'Errico, M. F. Caso, A. D. Pezzella and M. D'Ischia, ACS Appl. Mater. Interfaces, 2017, 9, 37615–37622 CrossRef PubMed.
- H. L. Su, C. C. Chou, D. J. Hung, S. H. Lin, I. C. Pao, J. H. Lin, F. L. Huang, R. X. Dong and J. J. Lin, Biomaterials, 2009, 30, 5979–5987 CrossRef PubMed.
- H. Van Acker and T. Coenye, Trends Microbiol., 2017, 25, 456–466 CrossRef PubMed.
- R. Edge, Pigm. Cell Res., 2006, 19, 443–450 CrossRef PubMed.
- A. Pezzella, D. Vogna and G. Prota, Tetrahedron: Asymmetry, 2003, 14, 1133–1140 CrossRef.
- E. Pizzo, M. Varcamonti, A. Di Maro, A. Zanfardino, C. Giancola and G. D'Alessio, FEBS J., 2008, 275, 1283–1295 CrossRef PubMed.
- A. Zanfardino, E. Pizzo, A. Di Maro, M. Varcamonti and G. D'Alessio, FEBS J., 2010, 277, 921–1928 CrossRef PubMed.
- J. Nourooz-Zadeh, J. Tajaddini-Sarmadi and S. P. Wolff, J. Agric. Food Chem., 1995, 43, 17–21 CrossRef.
- E. Pizzo, A. Zanfardino, A. M. A. Di Giuseppe, A. Bosso, N. Landi, S. Ragucci, M. Varcamonti, E. Notomista and A. Di Maro, FEBS Lett., 2015, 589, 2812–2818 CrossRef PubMed.
- M. D. Seo, T. J. Kang, C. H. Lee, A. Y. Lee and M. Noh, Biomol. Ther., 2012, 20, 171–176 CrossRef PubMed.
- K. Pane, V. Sgambati, A. Zanfardino, G. Smaldone, V. Cafaro, A. Angrisano, E. Pedone, S. Di Gaetano, D. Capasso, E. F. Haney, V. Izzo, M. Varcamonti, E. Notomista, R. E. W. Hancock, A. Di Donato and E. Pizzo, FEBS J., 2016, 283, 2115–2131 CrossRef PubMed.
- A. Zanfardino, G. Criscuolo, B. DiLuccia, E. Pizzo, M. L. Ciavatta, E. Notomista, A. Carpentieri, A. Pezzella and M. Varcamonti, Benefic. Microbes, 2017, 8, 133–141 CrossRef PubMed.
- Z. Wang, W. Ma, C. Chen, H. Ji and J. Zhao, Chem. Eng. J., 2011, 170, 353–362 CrossRef.
- D. Spasiano, R. Marotta, I. Gargano, I. Di Somma, G. Vitiello, G. D'Errico and R. Andreozzi, Chem. Eng. J., 2014, 249, 130–142 CrossRef.
- S. Satyro, R. Marotta, L. Clarizia, I. Di Somma, G. Vitiello, M. Dezotti, G. Pinto, R. F. Dantas and R. Andreozzi, Chem. Eng. J., 2014, 251, 257–268 CrossRef.
- R. Robert, S. Barbati, N. Ricq and M. Ambrosio, Water Res., 2002, 36, 4821–4829 CrossRef PubMed.
- A. B. Munoz-Garcia, F. Sannino, G. Vitiello, D. Pirozzi, L. Minieri, A. Aronne, P. Pernice, M. Pavone and G. D'Errico, ACS Appl. Mater. Interfaces, 2015, 7, 21662–21667 CrossRef PubMed.
- G. D'Errico, A. Silipo, G. Mangiapia, A. Molinaro, L. Paduano and R. Lanzetta, Phys. Chem. Chem. Phys., 2009, 11, 2314–2322 RSC.
- G. D'Errico, A. Silipo, G. Mangiapia, G. Vitiello, A. Radulescu, A. Molinaro, R. Lanzetta and L. Paduano, Phys. Chem. Chem. Phys., 2010, 12, 13574–13585 RSC.
- A. Luchini, G. D'Errico, S. Leone, Z. Vaezi, A. Bortolotti, L. Stella, G. Vitiello and L. Paduano, Colloids Surf., B, 2018, 168, 2–9 CrossRef PubMed.
- G. Vitiello, D. Musumeci, A. Koutsioubas, L. Paduano, D. Montesarchio and G. D'Errico, Biochim. Biophys. Acta, Biomembr., 2017, 1859, 2392–2401 CrossRef PubMed.
- G. Vitiello, A. Falanga, A. A. Petruk, A. Merlino, G. Fragneto, L. Paduano, S. Galdiero and G. D'Errico, Soft Matter, 2015, 11, 3003–3016 RSC.
- S. Ito, M. Kikuta, S. Koike, G. Szewczyk, T. Sarna, A. Zadlo, T. Sarna and K. Wakamatsu, Pigm. Cell Melanoma Res., 2016, 29, 340–351 CrossRef PubMed.
- A. Pezzella, L. Panzella, O. Crescenzi, A. Napolitano, S. Navaratnam, R. Edge, E. J. Land, V. Barone and M. d'Ischia, J. Org. Chem., 2009, 74, 3727–3734 CrossRef PubMed.
- A. J. Nappi and E. Vass, Melanoma Res., 1996, 6, 341–349 CrossRef PubMed.
- W. Qin, Y. Wang, G. Fang, C. Liu, Y. Sui and D. Zhou, Chem. Eng. J., 2016, 285, 69–76 CrossRef.
- H. P. Misra and I. Fridovich, J. Biol. Chem., 1972, 247, 188–192 Search PubMed.
- J. A. Imlay, S. M. Chin and S. Linn, Science, 1988, 240, 640–642 CrossRef PubMed.
- A. Orłowski, M. Grzybek, A. Bunker, M. Pasenkiewicz-Gierula, I. Vattulainen, P. T. Männistö and T. Róg, J. Neurochem., 2012, 122, 681–690 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra04315a |
| This journal is © The Royal Society of Chemistry 2018 |