Open Access Article
Open Access ArticleThe CRISPR-Cas9 system: a promising tool for discovering potential approaches to overcome drug resistance in cancer
Jiayu Zhang,
Wenlong Zhou,
Xiaoxuan Wang and
Lihui Wang *
*
Department of Pharmacology, Shenyang Pharmaceutical University, 110016, Shenyang, PR China. E-mail: lhwang@syphu.edu.cn
First published on 28th September 2018
Abstract
The CRISPR-Cas system was identified in bacteria as an immune defense mechanism against threats from the external environment. A common form of this system, called CRISPR-Cas9, is now widely used in gene editing, especially in mammalian cells. Through CRISPR-Cas9, gene knock-ins or knock-outs have become more feasible, thus deepening our understanding of the mechanisms of human diseases, including cancers, and suggesting possible treatment strategies. In this review, we discuss how CRISPR-Cas9 can be used as a tool to discover more about drug-resistance in cancers, including both the underlying mechanisms and ways to overcome them.
1. The discovery of the CRISPR-Cas system
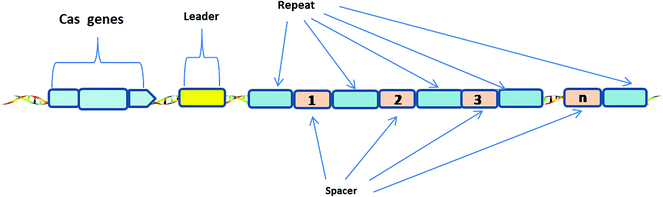
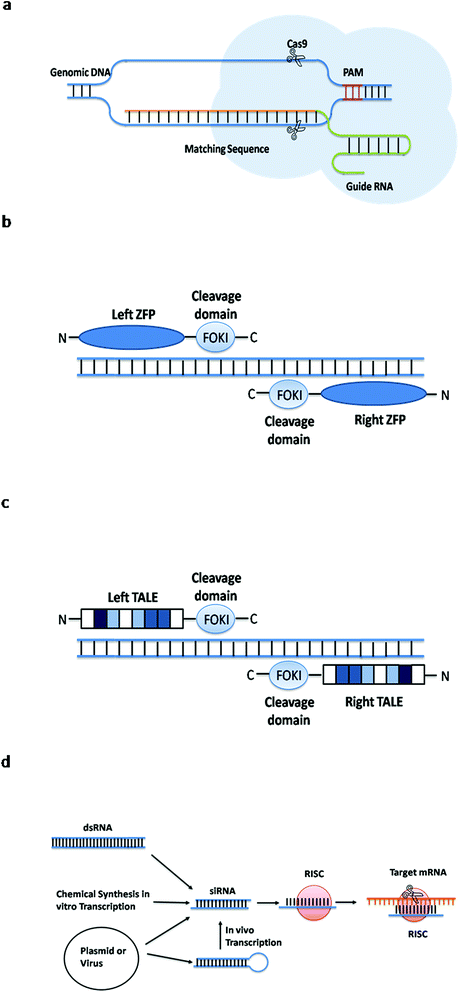
The CRISPR-Cas system consists of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and associated Cas proteins (Figure 1).1 It exists in prokaryotes, approximately half of bacteria and most archaea.2 CRISPR arrays are composed of continuous 24–47 base pair (bp) repeats, which are separated by unique sequences (spacers). The length of these spacers sequences is similar.3 These peculiar loci of the CRISPR system were first discovered in 1987,4 then Bolotin et al. proposed that the formation of the CRISPR structures involves DNA fragmentation by enzymes encoded by the associated Cas genes, and the function of CRISPR is to protect prokaryotes from invasion by foreign DNA in 2005.5 Two years later, CRISPR was confirmed as an adaptive immune system6 and the CRISPR-based defense mechanism was also discovered.7 Subsequently, scientists put forward the idea that the CRISPR-Cas system is a potential method for RNA-programmable genome editing.8 The successful applications of the CRISPR-Cas system in mammalian cells demonstrate the easy programmability and wide applicability of this technology.9 In 2015, it was shown that CRISPR-Cas9 controls the cleavage of target DNA.10 Knight et al. found that Cas9 protein can successfully navigate the chromatin structure of mammalian genomes.11 In mammalian cells, the CRISPR-Cas9 system is largely divided into two parts: the chimeric single guide RNA (gRNA) and the Cas9 nuclease protein. The gRNA targets the 20 bp nucleotide region next to a short DNA sequence called the protospacer-adjacent motif (PAM). The gRNA then directs the Cas9 protein to the position of the target sequence. Cas9 protein has two domains with endonuclease activity, HNH and RuvC, which cut the two strands of the target gene.12 After the DNA is cleaved, cells attempt to repair the break using the non-homologous end joining (NHEJ) or homologous recombination (HR) methods, and this eventually achieves the goal of gene editing.13 Consequently, the CRISPR-Cas9 system has become a current research hotspot.A CRISPR locus consists of Clustered Regularly Interspaced Short Palindromic Repeats (continuous 24–47 bp repeats), which are separated by unique spacer segments (integrated foreign DNA sequences, 30–40 bp in length), and CRISPR-associated (Cas) genes, which surround the CRISPR locus and encode Cas enzymes. The leader sequence (a conserved sequence of several hundred base pairs) acts as a promoter for transcription of the CRISPR array.1
1.1 Mechanism of CRISPR interference
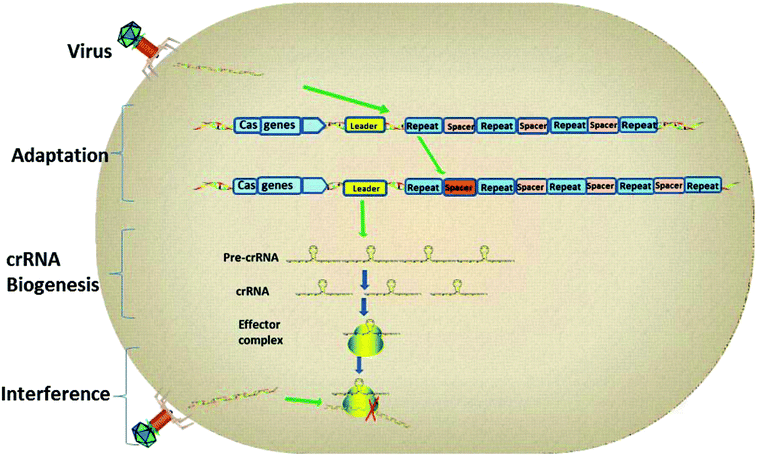
As the research moved along, CRISPR immunity was divided into three phases (Fig. 2).14 Firstly, during the adaptation stage, the CRISPR system will gain new spacers by integrating a portion of the foreign genome into the CRISPR sequence. Secondly, the CRISPR array will be transcribed to produce small guide RNAs, also known as CRISPR RNAs (crRNAs). Finally, the crRNA guides the interference and cuts the foreign genes by way of homology-dependent cleavage to achieve silencing of the target DNA.15CRISPR loci contain clusters of repeats and spacers that are flanked by a leader sequence and Cas genes. The three stages of CRISPR immunity are as follows: (1) the CRISPR system gains new spacers by integrating a portion of the foreign plasmid or virus genome into the CRISPR sequence; (2) transcription of the CRISPR array generates pre-crRNA, which is processed to give mature crRNAs; (3) the effector complex combines with foreign genetic elements to interfere with them and degrade them.
1.2 Biological function of the CRISPR-Cas system
1.3 The differences between CRISPR and other gene editing techniques
2. Revealing the underlying mechanisms in drug resistance with CRISPR-Cas9
Drug resistance, especially acquired resistance, is the most common problem in long-term drug treatment. Usually, the emergence of drug resistance is a slow and gradual process, suggesting the gene manipulation tool to explore the underlying mechanisms of drug resistance should be a more stable. Compared to RNA interference, CRISPR-Cas9 can permanently remove the interesting genes. Moreover, gene mutation is considered as one of the important reason to induce drug resistance in cancer. In contrast to ZFNs and TALENs technologies, CRISPR-Cas9 is easy to operate and can knock in multiple gene mutations simultaneously, which greatly improves the experimental efficiency. Therefore, accompany with the coming of CRISPR-Cas9 era, the investigation of drug resistance in cancers also made a great progression.2.1 ARID1A in HER2-targeting antibody resistance
HER2-targeting antibody, such as trastuzumab, is an efficacy drug for treatment with breast cancers with HER2 positive expression. However, resistance to trastuzumab reduces the long-term efficacy of this treatment, and it is therefore important to reveal the underlying resistance mechanism. Berns et al. found that directly targeting the PI3K/mTOR pathway seems to be the key to reversing the drug resistance. Recurrent inactivating mutations in ARID1A have been identified at high frequencies in human cancers.31,32 Loss of ARID1A activates Annexin A1 (ANXA1), a member of the annexin calcium/phospholipid-binding protein family which is involved in cell proliferation and tumor progression,33 then activates AKT in the HER2/PI3K/mTOR pathway, which finally produces drug resistance. It was verified that completely knocking out ARID1A in BT474 cells by the CRISPR-Cas9-based genome editing technique activated the expression of ANXA1, and high expression of ANXA1 increased the resistance to trastuzumab.34 The above results indicate that ANXA1 might be a novel target for treatment with HER2-targeting antibody resistance.2.2 Rpn13 and CXCR in proteasome inhibitors resistance
Proteasome inhibitors, such as bortezomib, is used clinically for multiple myeloma (MM) therapy,35 but the development of drug resistance limits its long-term efficacy. Song et al. demonstrated that the expression of the proteasome-related ubiquitination receptor Rpn13 in MM cells was higher than that in normal blood cells. The function of Rpn13 in MM was confirmed by knockout Rpn13 using CRISPR-Cas9 technology. They found Rpn13 would meditated cell proliferation and contributed to further study showed that knockout of Rpn13 by CRISPR-Cas9 would contributed to bortezomib resistance, suggesting targeting Rpn13 might be a novel approach to overcome proteasome inhibitor resistance.36 Recently, another group also demonstrated the role of CXCR/SDF-1 signaling pathway in bortezomib resistance. Chen et al. used the CRISPR-Cas9 system to knockout the CXCR gene, and found CXCR4 silencing in MCL cells led to a significant reduction in proliferation, cell adhesion, and colony formation. Furthermore, they elucidated a novel bortezomib resistance mechanism which is triggered by ROS release, following by CXCR/SDF-1 pathway activation, next resulted in autophagy increasing, and last induced drug resistance.37 This example shows that the CRISPR-Cas9 system can improve our understanding of the pathways which are involved in cancer, and then provide new means for the treatment of cancers by targeting the molecules involved in these pathways.2.3 p57 in HDAC inhibitors resistance
HDAC (histone deacetylase) is considered as the crucial epigenetic enzyme, which regulates histone acetylation and affects the target genes expression. Recently, HDAC was reported to associated with development and progression of various tumors.38 In addition, several HDAC inhibitors have been approved by FDA to treat cancers. In a recent study on pancreatic ductal adenocarcinoma of mice, Mazur et al. found that HDAC inhibitor (SAHA) induced tumor cell death by de-repression p57 gene, also known as CDKN1C.39 Further, using CRISPR-Cas9 system to delete the p57 gene, they found that p57-knockout pancreatic tumors were less responsive to the HDAC inhibitor, with reduced apoptosis. Thus, this study shows how the CRISPR-Cas9 approach is used to clarify the mechanism underlying a novel epigenetic-related therapeutic strategy.2.4 ASXL1 in imatinib resistance
As a tyrosine kinase inhibitor (TKI), imatinib is widely used for treatment with chronic myelocytic leukemia (CML) patients. Similar to other TKIs, resistance is inevitable in the long-term treatment. Recently study showed ASXL1, a tumor suppressor gene which frequently mutates in CML, might be involved into imatinib resistance. Re-express ASXL1 protein using CRISPR-Cas9 technology in the CML cell line KBM5, which originally lacked ASXL1 expression, could result in the inhibition of cell growth, the enhancement of cell differentiation, and the increasing of drug sensitivity,40 suggesting ASXL1 would be a potential target for reversing imatinib resistance.2.5 Kinesin-5 A133P mutation in ispinesib resistance
Kinesin-5 is a molecular motor protein that is essential in mitosis. Accumulation of studies have showed that kinesin-5 is involved into tumor progression and is considered as a promising target for cancer treatment. Ispinesib, an inhibitor of kinesin-5, has entered clinical trials as an anticancer agent. Recently, a CRISPR-Cas9 based screening study indicated that gene mutation of kinesin-5 A133P, but not overexpression of kinesin-12 or EGFR activation that was regarded as resistance mechanism in previous study,41 contributed to ispinesib resistance.42 This work demonstrated that drug resistance related gene mutation might be deepened by CRISPR-Cas9 system.2.6 p53 and CTR in cisplatin resistance
Cisplatin is a commonly used cytotoxic drug for the treatment of various tumor types, such as oesophageal adenocarcinoma (OAC). The p53 is a critical tumor suppressor and is mutated frequently in many cancers, often leading to chemoresistance and poor survival. About 70% of OACs have p53 mutations and develop resistance, so that many patients are not sensitive to long-term treatment with cisplatin, and may relapse after treatment.43 APR-246 is a first-in-class reactivator of mutant p53, which can upregulate the p53 gene, induce cell cycle arrest, inhibit cancer cell growth, and trigger apoptosis of OAC cells carrying a p53 mutation.44 Recent study showed ectopic expression of mutant p53 sensitized p53-null cells to APR-246, while p53 gene knockout diminished drug activity. Furthermore, they demonstrated that reactivation of p53 would restore chemosensitivity to a cisplatin resistant xenograft model.44 Another investigation on the mechanisms of cisplatin resistance is related to copper transporter genes (CTR1 and CTR2) or copper chaperone genes (ATOX1 and CCS). Bompiani et al. knocked out these genes with CRISPR-Cas9 and found that the sensitivity to cisplatin did not change much compared with the untreated cells. This indicates that these genes are not necessary for the process by which cisplatin enters cells and carries out its function.45 The above studies show that CRISPR-Cas9 can be used to explore the role of specific genes in drug resistance in tumor cells, and can also be used to engineer drug-resistant cells so that they regain their sensitivity to drugs.2.7 MLL in epirubicin resistance
Epirubicin is a cytotoxic agent for the treatment of bladder cancer (BC). BC is characterized by a high rate of postoperative recurrence. Studies have found that changes in the MLL gene are involved in BC recurrence. MLL encodes a transcription coactivation factor which regulates gene expression during early embryonic development and hematopoiesis. The MLL protein contains a SET (suppressor of variegation, enhancer of zeste, trithorax) domain at the C-terminus, and is a member of the MLL/trx family, which specifically methylates lysine 4 on histone H3 (H3K4).46 H3K4 methylation is typically associated with transcriptionally active chromatin regions. Recent study indicated that MLL-mutant bladder cancer cells obtained using CRISPR-Cas9 technology are more resistant to epirubicin than wild-type cells.47 Thus, targeting MLL may improve the treatment of recurrent bladder cancer.2.8 Rsf-1 in paclitaxel resistance
Paclitaxel is a classic drug for the treatment of lung cancer, but it readily induces resistance.48 Therefore, new ways to overcome drug resistance are urgently needed. The Rsf-1 (remodeling and spacing factor 1) gene is located on chromosome 11 at position q13.5 and encodes a histone-binding protein. It forms an ISWI chromatin remodeling complex with hSNF2H (human sucrose nonfermenting protein 2 homologue), and this complex can participate in transcriptional regulation and chromatin remodeling.49 Rsf-1 expression is high in lung cancer and increases paclitaxel tolerance through the NF-κB pathway. Many other genes are also involved in drug resistance, such as genes that affect apoptosis and the cell cycle.50 Chen and colleagues knocked out Rsf-1 in lung cancer cells and xenograft mice using CRISPR-Cas9 technology, and tested whether Rsf-1 affects the sensitivity of lung cancer to paclitaxel by regulating the activation of the NF-κB pathway and the expression of its downstream genes. The data indicated that paclitaxel reduced the ability of Rsf-1-knockout NCI-H1299 and NCI-H460 cells to migrate and proliferate, and increased apoptosis. In mice with xenograft tumors derived from Rsf-1-knockout cells, the anti-tumor effect of paclitaxel was enhanced. Therefore, targeting Rsf-1 by CRISPR-Cas9 technology has also become one of the approaches for treating lung cancer.512.9 P-glycoprotein in doxorubicin resistance
P-glycoprotein (P-gp) is the product of the multi-drug resistance gene (MDR1), and is considered as a classic drug resistant protein. Recently, the role of P-gp in drug resistance is also confirmed by CRISPR-Cas9 technology. Ha et al. demonstrated that knockout of MDR1 gene by transfecting the Cas9-sgRNA complex into doxorubicin-resistant breast cancer cells could enhance the sensitivity of transfected cells to doxorubicin.28 Thus, the CRISPR-Cas9 system might be a valuable and feasible method to confirm previous identified targets involved to drug resistance.2.10 PBAF in immunotherapy resistance
Immunotherapy has been widely applied in the treatment of leukemia and solid tumors, and obtain a good efficacy. This therapy is usually targeted to the PD-1 receptor, which is an inhibitory receptor on T cells. Cytotoxic T cells also express T cell receptors which recognize peptide antigens bound to MHC class I proteins.52 In the normal immune response, this interaction leads to activation of the cytotoxic T cells, which find and clear abnormal cells expressing the antigen, including cancer cells. However, most cancer patients do not respond to immunotherapy, and the mechanism underlying this lack of response is poorly understood.53 To help clarify the mechanism, Pan et al. used CRISPR-Cas9 to inactivate more than 100 genes in mouse B16F10 melanoma cells, and then they tested the sensitivity of the cells to T cell-mediated killing. Cells lacking the genes Pbrm1, Arid2 and Brd7 were found to be more sensitive to cytotoxic T cells. These three genes encode proteins in the PBAF form of the SWI/SNF tumor suppressor complex, which regulates transcription factor recognition of chromatin. When PBAF was inactivated by CRISPR-Cas9, the B16F10 cells increased their sensitivity to interferon-gamma and their secretion of chemokines. Therefore, inactivation of the above three genes in human tumors may increase the effectiveness of PD-1 blockade, adoptive T cell therapies or cancer vaccines. In addition, T cell-mediated cytotoxicity is more lethal to PBAF-deficient tumor cells. Thus, targeting the intrinsic resistance mechanisms of tumor cells may represent a new method for improving the response to immunotherapy (Table 1).54| Therapeutic agents | Molecules involved in resistance | Cancer | Result | |
|---|---|---|---|---|
| Molecular targeted agents | Trastuzumab | HER2 | Breast cancer | Reverses drug resistance34 |
| Bortezomib | Rpn13 | Multiple myeloma | Reduces the survival of MM cells and inhibits proliferation36 | |
| SAHA | p57 | Pancreatic ductal adenocarcinoma | Reverses drug resistance39 | |
| Imatinib | ASXL1 | Chronic myelocytic leukemia | Slows down cell growth and enhances in differentiation ability40 | |
| Ispinesib | Kinesin-5 A133P | Cervical cancer | Overthrows the previous resistance mechanism42 | |
| Cytotoxic agents | Cisplatin | p53,CTR | Oesophageal adenocarcinoma | Induces cell cycle arrest and inhibits cancer cells growth44 |
| Epirubicin | MLL | Bladder cancer | Reverses drug resistance47 | |
| Paclitaxel | Rsf-1 | Lung cancer | Reverses drug resistance51 | |
| Doxorubicin | P-glycoprotein | Breast cancer | Increases sensitivity to doxorubicin28 | |
| Immunotherapy | Cancer vaccines and adoptive T cell therapies | PBAF | Melanoma | Increases sensitivity to immunotherapy54 |
3. The problems and challenges of the CRISPR-Cas9 system
The CRISPR-Cas9 system can edit genomes accurately, it has been rapidly and widely applied to molecular biology and cell research. However, due to the fact that the specificity of CRISPR-Cas9 is determined by base pairing, a low targeting specificity can result in uncertain research results and the waste of manpower and material resources. Therefore, the biggest problem with the system is off-target effect. It affects the efficiency of the system, and can lead to a series of adverse consequences such as oncogene activation in the genome or other important mutations. For clinical applications and gene therapy in particular, off-target effects can cause great harm.55 In addition, the plasmids needed for CRISPR-Cas9 editing are large, and there are difficulties with the transfection process. Furthermore, the gene deletion rate is low, and even if the vectors are successfully imported into the cells, a lack of effective screening methods is also a problem. In spite of the successful generation of gene-knockout cell lines, the biological functions of cell lines formed by different monoclonal cells may not be exactly the same. This may lead to poor reproducibility in subsequent experiments with such cell lines. Moreover, the gene editing of human embryonic and germ cells has aroused strong opposition from many scientists. The rational application of this technology requires extensive discussions in the scientific, medical, legal and ethical communities.56Despite these problems, CRISPR-Cas9 technology has many advantages compared with other gene editing tools, such as simple operation, high targeting efficacy and robust sequence specificity. It has a broad range of applications in site-directed genome modification. In drug-resistance research, its most important application is in making tumor cells regain their sensitivity to drugs. It is believed that with the rapid development of the technology, researchers can rapidly identify and select drug-resistance-related genes in different tumor types. The CRISPR-Cas9 system provides new ideas and a theoretical basis for figuring out drug-resistance mechanisms and for developing new treatments to overcome drug resistance. It has the potential to change all aspects of tumor biology and biomedicine. Recently CRISPR-Cpf1, as a novel class II CRISPR-Cas systems, is designed by an RNA-directed endonuclease, which is a smaller and simpler endonuclease than Cas9. It can overcome the limitations of some CRISPR-Cas9 systems and is expected to become a new generation of gene editing technology. Moreover, recent study showed another more sensitivity system, called specific high-sensitivity enzymatic reporter unLOCKing (SHERLOCK),57 could be used to detect tumor circulating DNA mutations, which provide a possibility to find biomarkers for diagnosis and drug resistance using circulating tumor cells or cell-free nucleic acids.58 Therefore, with the gradual application of CRISPR-Cas9 technology in clinical practice, it will be possible to achieve cancer gene therapy by carrying out genome repairs in tumor cells to correct mutations, chromosomal defects, copy number variations, and abnormal gene regulation and expression.59 In summary, the CRISPR-Cas9 system will provide many possibilities for overcoming drug resistance in tumors.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by National Natural Science Foundation of China (No. 81572947, 81773216), Natural Science Foundation of Liaoning Province (No. 20170540841) and Scientific Research Fund of Liaoning Provincial Education Department (No. 2017LFW01).References
- F. V. Karginov and G. J. Hannon, The CRISPR system: small RNA-guided defense in bacteria and archaea, Mol. Cell, 2010, 37(1), 7–19 CrossRef CAS PubMed.
- V. Kunin, R. Sorek and P. Hugenholtz, Evolutionary conservation of sequence and secondary structures in CRISPR repeats, Genome Biol., 2007, 8(4), R61 CrossRef PubMed.
- I. Grissa, G. Vergnaud and C. Pourcel, The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats, BMC Bioinf., 2007, 8, 172 CrossRef PubMed.
- Y. Ishino, H. Shinagawa, K. Makino, M. Amemura and A. Nakata, Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product, J. Bacteriol., 1987, 169(12), 5429–5433 CrossRef CAS PubMed.
- A. Bolotin, B. Quinquis, A. Sorokin and S. D. Ehrlich, Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin, Microbiology, 2005, 151(8), 2551–2561 CrossRef CAS PubMed.
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero and P. Horvath, CRISPR provides acquired resistance against viruses in prokaryotes, Science, 2007, 315(5819), 1709–1712 CrossRef CAS PubMed.
- J. Van der Oost, M. M. Jore, E. R. Westra, M. Lundgren and S. J. Brouns, CRISPR-based adaptive and heritable immunity in prokaryotes, Trends Biochem. Sci., 2009, 34(8), 401–407 CrossRef CAS PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity, Science, 2012, 337(6096), 816–821 CrossRef CAS PubMed.
- L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang, L. A. Marraffini and F. Zhang, Multiplex genome engineering using CRISPR/Cas systems, Science, 2013, 339(6121), 819–823 CrossRef CAS PubMed.
- S. H. Sternberg, B. LaFrance, M. Kaplan and J. A. Doudna, Conformational control of DNA target cleavage by CRISPR-Cas9, Nature, 2015, 527(7576), 110–113 CrossRef CAS PubMed.
- S. C. Knight, L. Xie, W. Deng, B. Guglielmi, L. B. Witkowsky, L. Bosanac, E. T. Zhang, M. El Beheiry, J. B. Masson, M. Dahan, Z. Liu, J. A. Doudna and R. Tjian, Dynamics of CRISPR-Cas9 genome interrogation in living cells, Science, 2015, 350(6262), 823–826 CrossRef CAS PubMed.
- I. M. Slaymaker, L. Gao, B. Zetsche, D. A. Scott, W. X. Yan and F. Zhang, Rationally engineered Cas9 nucleases with improved specificity, Science, 2016, 351(6268), 84–88 CrossRef CAS PubMed.
- K. E. Savell and J. J. Day, Applications of CRISPR/Cas9 in the Mammalian Central Nervous System, Yale J. Biol. Med., 2017, 90(4), 567–581 Search PubMed.
- W. Jiang and L. A. Marraffini, CRISPR-Cas: New Tools for Genetic Manipulations from Bacterial Immunity Systems, Annu. Rev. Microbiol., 2015, 69, 209–228 CrossRef CAS PubMed.
- C. R. Hale, P. Zhao, S. Olson, M. O. Duff, B. R. Graveley, L. Wells, R. M. Terns and M. P. Terns, RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex, Cell, 2009, 139(5), 945–956 CrossRef CAS PubMed.
- M. Babu, N. Beloglazova, R. Flick, C. Graham, T. Skarina, B. Nocek, A. Gagarinova, O. Pogoutse, G. Brown, A. Binkowski, S. Phanse, A. Joachimiak, E. V. Koonin, A. Savchenko, A. Emili, J. Greenblatt, A. M. Edwards and A. F. Yakunin, A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair, Mol. Microbiol., 2011, 79(2), 484–502 CrossRef CAS PubMed.
- K. S. Makarova, Y. I. Wolf, O. S. Alkhnbashi, F. Costa, S. A. Shah, S. J. Saunders, R. Barrangou, S. J. Brouns, E. Charpentier, D. H. Haft, P. Horvath, S. Moineau, F. J. Mojica, R. M. Terns, M. P. Terns, M. F. White, A. F. Yakunin, R. A. Garrett, J. van der Oost, R. Backofen and E. V. Koonin, An updated evolutionary classification of CRISPR-Cassystems, Nat. Rev. Microbiol., 2015, 13(11), 722–736 CrossRef CAS PubMed.
- M. E. Zegans, J. C. Wagner, K. C. Cady, D. M. Murphy, J. H. Hammond and G. A. O'Toole, Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa, J. Bacteriol., 2009, 191(1), 210–219 CrossRef CAS PubMed.
- P. Viswanathan, K. Murphy, B. Julien, A. G. Garza and L. Kroos, Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats, J. Bacteriol., 2007, 189(10), 3738–3750 CrossRef CAS PubMed.
- J. E. DiCarlo, J. E. Norville, P. Mali, X. Rios, J. Aach and G. M. Church, Genome engineering in Saccharomyces cerevisiae using CRISPR-Cassystems, Nucleic Acids Res., 2013, 41(7), 4336–4343 CrossRef CAS PubMed.
- G. Gasiunas, R. Barrangou, P. Horvath and V. Siksnys, Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(39), E2579–E2586 CrossRef CAS PubMed.
- Y. G. Kim, J. Cha and S. Chandrasegaran, Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain, Proc. Natl. Acad. Sci. U. S. A., 1996, 93(3), 1156–1160 CrossRef CAS.
- T. Gaj, C. A. Gersbach and R. C. Barbas, ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering, Trends Biotechnol., 2013, 31(7), 397–405 CrossRef CAS PubMed.
- D. Hockemeyer, H. Y. Wang, S. Kiani, C. S. Lai, Q. Gao, J. P. Cassady, G. J. Cost, L. Zhang, Y. Santiago, J. C. Miller, B. Zeitler, J. M. Cherone, X. D. Meng, S. J. Hinkley, E. J. Rebar, P. D. Gregory, F. D. Urnov and R. Jaenisch, Genetic engineering of human pluripotent cells using TALE nucleases, Nat. Biotechnol., 2011, 29(8), 731–734 CrossRef CAS PubMed.
- Q. W. Shan, Y. P. Wang, K. L. Chen, Z. Liang, J. Li, Y. Zhang, K. Zhang, J. X. Liu, D. F. Voytas, X. L. Zheng, Y. Zhang and C. X. Gao, Rapid and efficient gene modification in rice and Brachypodium using TALENs, Mol. Plant, 2013, 6(4), 1365–1368 CrossRef CAS PubMed.
- S. Guo and K. J. Kemphues, par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed, Cell, 1995, 81(4), 611–620 CrossRef CAS PubMed.
- C. Lipardi, Q. Wei and B. M. Paterson, RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs, Cell, 2001, 107(3), 297–307 CrossRef CAS PubMed.
- J. S. Ha, J. Byun and D. R. Ahn, Overcoming doxorubicin resistance of cancer cells by Cas9-mediated gene disruption, Sci. Rep., 2016, 6, 22847 CrossRef CAS PubMed.
- S. J. Kim, J. Kyung Hee, S. Mi Kwon, L. Joo Han and H. Soon-Sun, Synergistic anticancer effects through tumor vessel normalization by PI3K inhibitors, Eur. J. Cancer, 2016, 69, S129 CrossRef.
- F. Wang, T. Guo and H. Jiang, et al., A comparison of CRISPR/Cas9 and siRNA-mediated ALDH2 gene silencing in human cell lines, Mol. Genet. Genomics, 2018, 293(3), 1–15 Search PubMed.
- K. C. Wiegand, A. F. Lee and O. M. Al-Agha, et al., Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas, J. Pathol., 2011, 224(3), 328 CrossRef CAS PubMed.
- S. Jones, T. L. Wang and S. Iem, et al., Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma, Science, 2010, 330(6001), 228–231 CrossRef CAS PubMed.
- R. Belvedere, V. Bizzarro and A. Popolo, et al., Role of intracellular and extracellular annexin A1 in migration and invasion of human pancreatic carcinoma cells, BMC Cancer, 2014, 14(1), 961 CrossRef PubMed.
- K. Berns, A. Sonnenblick and A. Gennissen, et al., Loss of ARID1A activates ANXA1, which serves as a predictive biomarker for trastuzumab resistance, Clin. Cancer Res., 2016, 22(21), 5238 CrossRef CAS PubMed.
- P. Richardson, S. Jagannath and N. Raje, et al., A phase 2 trial of lenalidomide, bortezomib, and dexamethasone in patients with relapsed and relapsed/refractory myeloma, Blood, 2014, 123(10), 1461–1469 CrossRef CAS PubMed.
- Y. Song, A. Ray and S. Li, et al., Targeting proteasome ubiquitin receptor Rpn13 in multiple myeloma, Leukemia, 2016, 30(9), 1877 CrossRef CAS PubMed.
- Z. Chen, A. E. Teo and N. McCarty, ROS-Induced CXCR4 Signaling Regulates Mantle Cell Lymphoma (MCL) Cell Survival and Drug Resistance in the Bone Marrow Microenvironment via Autophagy, Clin. Cancer Res., 2016, 22(1), 187–199 CrossRef CAS PubMed.
- A. C. West and R. W. Johnstone, New and emerging HDAC inhibitors for cancer treatment, J. Clin. Invest., 2014, 124(1), 30–39 CrossRef CAS PubMed.
- P. K. Mazur, A. Herner and S. S. Mello, et al., Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma, Nat. Med., 2015, 21(10), 1163–1171 CrossRef CAS PubMed.
- V. Simona, D. Hamid and B. Matthias, et al., ASXL1mutation correction by CRISPR/Cas9 restores gene function in leukemia cells and increases survival in mouse xenografts, OncoTargets Ther., 2015, 6(42), 44061–44071 Search PubMed.
- M. Ono and M. Kuwano, Molecular mechanisms of epidermal growth factor receptor (EGFR) activation and response to gefitinib and other EGFR-targeting drugs, Clin. Cancer Res., 2006, 12(24), 7242–7251 CrossRef CAS PubMed.
- C. Kasap, O. Elemento and T. M. Kapoor, DrugTargetSeqR: a genomics- and CRISPR-Cas9-based method to analyze drug targets, Nat. Chem. Biol., 2014, 10(8), 626–628 CrossRef CAS PubMed.
- J. L. Raoul, Definitive chemoradiotherapy with FOLFOX versus fluorouracil and cisplatin in patients with oesophageal cancer (PRODIGE5/ACCORD17): final results of a randomised, phase 2/3 trial, Lancet Oncol., 2014, 15(3), 305–314 CrossRef.
- D. S. H. Liu, M. Read and C. Cullinane, et al., APR-246 potently inhibits tumour growth and overcomes chemoresistance in preclinical models of oesophageal adenocarcinoma, Gut, 2015, 64(10), 1506–1516 CrossRef CAS PubMed.
- K. M. Bompiani, C. Y. Tsai and F. P. Achatz, et al., Copper transporters and chaperones CTR1, CTR2, ATOX1, and CCS as determinants of cisplatin sensitivity, Metallomics, 2016, 8(9), 951–962 RSC.
- J. U. Marquardt and S. S. Thorgeirsson, Linking MLL and the HGF-MET signaling pathway in liver cancer, J. Clin. Invest., 2013, 123(7), 2780–2783 CrossRef CAS PubMed.
- S. Wu, Z. Yang and R. Ye, et al., ovel variants in MLL confer to bladder cancer recurrence identified by whole-exome sequencing, OncoTargets Ther., 2016, 7(3), 2629–2645 Search PubMed.
- Y. Liu, X. Wu and Y. Sun, et al., Silencing of X-linked inhibitor of apoptosis decreases resistance to cisplatin and paclitaxel but not gemcitabine in non-small cell lung cancer, J. Int. Med. Res., 2011, 39(5), 1682–1692 CrossRef CAS PubMed.
- J. J. Sheu, B. Guan and J. H. Choi, et al., Rsf-1, a Chromatin Remodeling Protein, Induces DNA Damage and Promotes Genomic Instability, J. Biol. Chem., 2010, 285(49), 38260–38269 CrossRef CAS PubMed.
- J. H. Choi, J. J. Sheu and B. Guan, et al., Functional analysis of 11q13.5 amplicon identifies Rsf-1 (HBXAP) as a gene involved in paclitaxel resistance in ovarian cancer, Cancer Res., 2009, 69(4), 1407–1415 CrossRef CAS PubMed.
- X. Chen, X. Sun and J. Guan, et al., Rsf-1 Influences the Sensitivity of Non-Small Cell Lung Cancer to Paclitaxel by Regulating NF-κB Pathway and Its Downstream Proteins, Cell. Physiol. Biochem., 2017, 44(6), 2322 CrossRef CAS PubMed.
- N. Zhang and M. J. Bevan, CD8(+) T cells: foot soldiers of the immune system, Immunity, 2011, 35(2), 161–168 CrossRef CAS PubMed.
- P. Sharma, S. Hu-Lieskovan, J. A. Wargo and A. Ribas, Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy, Cell, 2017, 168(4), 707–723 CrossRef CAS PubMed.
- D. Pan, A. Kobayashi and P. Jiang, et al., A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing, Science, 2018, 359(6377), 770–775 CrossRef CAS PubMed.
- V. Pattanayak, S. Lin and J. P. Guilinger, et al., High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity, Nat. Biotechnol., 2013, 31(9), 839–843 CrossRef CAS PubMed.
- D. Baltimore, P. Berg and M. Botchan, et al., A prudent path forward for genomic engineering and germline gene modification, Science, 2015, 348(6230), 36 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh and J. W. Lee, et al., Nucleic acid detection with CRISPR-Cas13a/C2c2, Science, 2017, 356(6336), 438–442 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh and J. W. Lee, et al., Nucleic acid detection with CRISPR-Cas13a/C2c2, Science, 2017, 356(6336), 438–442 CrossRef CAS PubMed.
- D. A. Scott and F. Zhang, Implications of human genetic variation in CRISPR-based therapeutic genome editing, Nat. Med., 2017, 23(9), 1095 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |