Open Access Article
Open Access ArticleEvaluation of a novel low-carbon to nitrogen- and temperature-tolerant simultaneously nitrifying–denitrifying bacterium and its use in the treatment of river water
Peng Jin *a,
Yinyan Chenc,
Zhanwang Zhengbc and
Qizhen Du*a
*a,
Yinyan Chenc,
Zhanwang Zhengbc and
Qizhen Du*a
aThe College of Agricultural and Food Sciences, Zhejiang A & F University, Hangzhou, 311300, China. E-mail: jinpeng@zafu.edu.cn; qizhendu@163.com
bZhejiang Shuangliang Sunda Environment Co.,LTD, Hangzhou, 310000, China
cSchool of Environmental & Resource, Zhejiang A & F University, Hangzhou, 311300, China
First published on 1st August 2018
Abstract
In this study, a simultaneously heterotrophic nitrifying–aerobic denitrifying bacterium, designated KSND, was newly isolated from a lake wetland. Its removal efficiencies for 160 mg L−1 ammonium, 105 mg L−1 nitrate, and 8.39 mg L−1 nitrite were 86.56%, 74.52%, and 100% in 24 h, with removal rates of 5.77 mg L−1 h−1 for NH4+–N, 3.26 mg L−1 h−1 for NO3−–N, and 0.35 mg L−1 h−1 for NO2−–N. The bacterium retained ∼63% of its maximal removal rate at 10 °C and 56% of its maximal removal rate at a carbon to nitrogen (C/N) ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, with no nitrite accumulation. Gene-specific PCR indicated the absence of the key genes for nitrification and denitrification, encoding hydroxylamine oxidoreductase and nitrite reductase, respectively, suggesting that KSND achieves effective nitrogen removal by another pathway. KSND was used to treat river wastewater by culturing it in a floating bed bioreactor. Ammonia nitrogen decreased significantly from 8.76 mg L−1 initially to 1.87 mg L−1 in 90 days, with no NO3−–N or NO2−–N toxicants, indicating the great potential utility of KSND in future full-scale applications in the treatment of low-C/N wastewater.
1, with no nitrite accumulation. Gene-specific PCR indicated the absence of the key genes for nitrification and denitrification, encoding hydroxylamine oxidoreductase and nitrite reductase, respectively, suggesting that KSND achieves effective nitrogen removal by another pathway. KSND was used to treat river wastewater by culturing it in a floating bed bioreactor. Ammonia nitrogen decreased significantly from 8.76 mg L−1 initially to 1.87 mg L−1 in 90 days, with no NO3−–N or NO2−–N toxicants, indicating the great potential utility of KSND in future full-scale applications in the treatment of low-C/N wastewater.
1. Introduction
Human activities have caused an unprecedented increase in the levels of reactive nitrogen in the biosphere, with negative effects on human health, biodiversity, and water quality.1,2 The most common nitrogenous contaminants of groundwater are ammonium and nitrate, which are recognized as toxic to aquatic species and ecological health.3 Therefore, the control and elimination of NH4+–N and NO3−–N contamination are priorities in environmental water treatment strategies. Although bioremediation is recognized as the most economical and promising approach to nitrogen removal, traditional methods have many disadvantages,4 insofar as they involve nitrifiers and denitrifiers with distinct differences in their physiology and biochemistry,5 require specific conditions for nitrification (aerobic) and denitrification (anaerobic), and because the removal processes are inefficient and time-consuming.6To overcome such limitations, several novel nitrogen removal technologies have been developed, including anaerobic ammonium oxidation (anammox)7 and simultaneous nitrification and denitrification (SND).8 Unfortunately, anammox bacteria have long growth cycles and are not available in pure culture.7 Recently, a group of bacteria capable of simultaneous nitrification and denitrification were reported.9–11 These heterotrophic bacteria display higher growth rates than autotrophs and can use organic substrates as sources of carbon and energy to convert ammonium into nitrogenous gas under aerobic conditions.12 Importantly, these nitrogen removal processes could be carried out in the same reaction pool, and did not require the combined action of multiple classes of microorganisms. Although SND bacterial isolates have been reported,9,13 they pose great challenges because the removal efficiency of such bacteria is significantly inactivated in low-temperature or low-C/N wastewater environments, limiting their application. Therefore, further novel microbial resources with excellent tolerance of extreme environment must be exploited and characterized.
We report a novel bacterial isolate capable of simultaneous nitrification and denitrification that efficiently removes nitrate, nitrite, and ammonia. We determined the tolerance of this bacterium for high-strength inorganic nitrogen, low temperatures, and low-C/N wastewater. The functional biomarker genes involved in the known pathways of nitrification and denitrification were not detected with PCR. The performance of strain KSND as an inoculum for removing nitrogen from polluted rivers was examined.
2. Material and methods
2.1. Isolation of strain and culture media
The bacterium KSND was isolated from lake-wetland sediment in the city of Lin'an, Zhejiang Province, China. 1 mL sediment sample was added to 100 mL of sterile deionized water and incubated at 30 °C for 1 h. After four 10-fold serial dilutions, the sample was inoculated on MM plates with 2.0 g L−1 KNO3 and 5 g L−1 sodium citrate at 30 °C for 24 h. A bacterium with excellent nitrogen removal capability was obtained through shake flask experiments, and named KSND for further study.The modified medium MM (0.2 g L−1 MgSO4·7H2O, 1.0 g L−1 KH2PO4, 2.0 mL of trace element solution) was used in the nitrogen removal experiments. Ammonium chloride, ammonium nitrate, sodium nitrate, sodium nitrite and glucose were added as indicated. The trace element solution contained (per liter): 50 g of EDTA-2Na, 2.2 g of ZnSO4, 5.5 g of CaCl2, 5.06 g of MnCl2·4H2O, 5.0 g of FeSO4·7H2O, 1.1 g of (NH4)6MO7O24·4H2O, 1.57 g of CuSO4·5H2O, and 1.61 g of CoCl2·6H2O, at pH 7.0.
2.2. Assessment of nitrification and denitrification performance
KSND cells were incubated in MM with 5 g L−1 glucose, and then cultured at 37 °C with shaking at 200 rpm for 16 h as the seed culture (∼6.7 ± 0.4 × 107 colony-forming units per mL−1). Flasks (500 mL) containing 100 mL of MM were inoculated with 1 mL of the seed culture and incubated at 30 °C with shaking at 200 rpm. For the nitrogen removal analysis, either 160 mg L−1 NH4Cl, 105 mg L−1 KNO3, 8.39 mg L−1 KNO2, or 50 mg L−1 NH4NO3 was used as the sole nitrogen source. To evaluate the efficiency of high-strength ammonium removal, KSND was incubated in modified MM in the presence of NH4Cl (final concentration 20–1000 mg L−1) as the sole nitrogen source. Unless otherwise stated, all the experiments were conducted at C/N 8, initial pH 7.0, culturing temperature 35 °C and shaking speed 200 rpm. The optical density at 600 nm (OD600) was used to monitor cell growth. The concentrations of ammonium, nitrate, and nitrite were determined with Nessler's reagent spectrophotometry at 420 nm, N-(1-naphthalene)-diaminoethane spectrophotometry at 410 nm, and ultraviolet spectrophotometry at 540 nm, respectively. Total nitrogen (TN) was determined with the standard UV spectrophotometry method (DR6000, Hach, America). All the experiments were performed in triplicate.2.3. Degenerate primer design and PCR amplification
To design the degenerate primers, a neighbor-joining phylogenetic analysis was used to determine the evolutionary relationships of all hydroxylamine oxidoreductase and dissimilatory nitrite reductase sequences available in the National Center for Biotechnology Information nonredundant protein sequence database. The representtaive hydroxylamine oxidoreductase (Hao) or dissimilatory nitrite reductase (NirS) enzymes were used to construct multiple sequence alignments with the Clustal X program. Conserved regions were used to design the following degenerate primer pairs. JBNirS-1001F (CGTGGTGGGAAAYTAYTGGCCKCC)/JBNirS-1242R (CAYGAYGGHGGHTGGGAC) and JBHAO-170F (GTATGAVGCGYTGGTNAAGCGYTA)/JBHAO-939R (TGGAACTGGRAHGTHCVTCTCAAG) were used to identify nirS and Hao genes of KSND genome DNA by gene-specific PCR, respectively. The PCR reaction was performed with the following program: 3 min at 94 °C; 32 cycles of 30 s at 94 °C, 30 s at 50 °C and 60 s at 72 °C; 5 min at 72 °C and a hold period at 4 °C. Metagenomic DNA extracted from the lake sediments was used as the control template.2.4. Identification of strain KSND
The gene encoding 16S rDNA of KSND was amplified with PCR using the universal primers pair 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (TACGGCTACCTTGTTACGACTT). To determine the evolutionary relationship of this strain KSND with established denitrifying bacterium, 16S rRNA sequence of KSND was compared to 13 representative bacterial denitrifies from the non-redundant (NR) nucleic acid sequence database (NCBI), using neighbor-joining phylogenetic analysis. One thousand bootstrap replications were performed using the MEGA software (MEGA 5.0).2.5. Assessment of nitrogen removal in river ecological restoration
Xinqiao River, located in Hangzhou (Zhejiang, China), runs through a residential area, and the ammonia concentration in the river was ∼9 mg L−1 (chemical oxygen demand ∼127 mg L−1, pH 7.8, temperature 13–27 °C). KSND cells were cultured in MM medium at 37 °C with shaking at 200 rpm for 16 h as the seed cultures, and then stored at 4 °C. 20 L seed culture was mixed with 10 L MM medium and 10 L river water, with an aeration for 8 h. The mixtures were added into the bioreactor carried by floating-bed, and 950 L water and 10 L GT medium (glucose and trace element solution) were added with continuous aeration. The extensive culture of KSND was automatically controlled to rule out ∼500 L with the floating-bed moving in the whole river (786 m × 6 m), and then 500 L of water was replenished to the bioreactor every 24 h. The GT medium of 10 L was also automatically controlled to add into the bioreactor. The water was sampled at six equidistant positions in the river and tested every 7 days, as described above.2.6. Nucleotide sequence accession numbers
The nucleotide sequence has been deposited in the Genbank database under accession number MH155426.3. Results and discussion
3.1. Characterization of KSND's capacity to remove NH4+–N, NO3−–N, and NO2−–N
The characteristics of the nitrification and denitrification abilities of KSND are shown in Fig. 1. Cell growth showed similar trends on the three sole nitrogen sources, with the highest OD600 values of 2.65–2.84 at 24 h. The ammonium concentration decreased rapidly from 160 mg L−1 initially to 21.52 mg L−1, and the corresponding removal efficiency was 86.56 ± 1.12% after 24 h in culture (Fig. 1a). The nitrification rate was 5.77 mg L−1 h for NH4+–N, which was higher than that of Klebsiella pneumoniae CF-S9 (4.30 mg L−1 h−1).9 Analysis of the nitrogen balance showed that 31.1% of the initial NH4+–N was converted to intracellular nitrogen, and ∼68.9% was denitrified by KSND after 48 h in culture.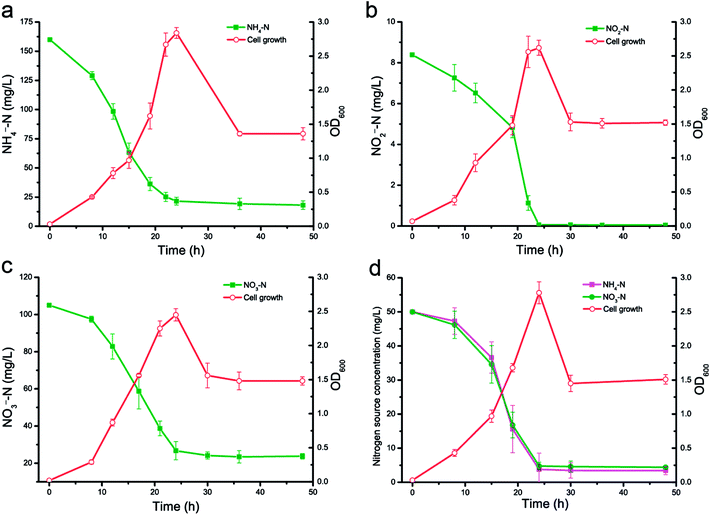 | ||
| Fig. 1 Nitrogen removal and cell growth of strain KSND using (a) NH4+–N, (b) NO2−–N, (c) NO3−–N and (d) NH4NO3–N as sole nitrogen source. | ||
With further denitrification by KSND, a significant reduction in NO2−–N, the sole nitrogen source, was observed at 24 h (Fig. 1b), and ∼100% of the initial nitrite (8.39 mg L−1) was removed, with an average removal rate of 0.35 mg L−1 h−1. When NO3−–N was the nitrogen source, it decreased significantly from 105 mg L−1 initially to 26.75 mg L−1 (Fig. 1c). Approximately 74.52% of the nitrate was removed and the corresponding maximum removal rate was 3.26 mg L−1 h−1, which was higher than that of Pseudomonas tolaasii strain Y-11 (1.99 mg L−1 h−1)13 and Rhodococcus sp. CPZ24 (0.93 mg L−1 h−1).14 Ammonium nitrate was used to investigate the simultaneous nitrification and denitrification capacity of KSND (Fig. 1d). The concentration of NH4+–N of 50 mg L−1 decreased to 3.82 mg L−1 after 24 h, with a removal efficiency of 92.36% and a removal rate of 1.92 mg L−1 h−1. Nitrate decreased from 50 to 4.80 mg L−1, with a removal efficiency of 90.40% and a removal rate of 1.88 mg L−1 h−1. These results indicate that KSND had no preference among the nitrogen sources and differs from P. tolaasii Y-11 and K. variicola sd-3 in that it more efficiently nitrifies ammonium than it denitrifies nitrate.10,13 Furthermore, the accumulation of nitrite was undetectable during the aerobic nitrogen removal process. This phenomenon may be attributable to the rapid removal by nitrite reductase of the trace nitrite produced by denitrification. Taken together, these results suggest that strain KSND has a broad substrate utilization capacity and efficient simultaneous nitrification and aerobic denitrification capabilities.
3.2. Factors affecting nitrogen removal by KSND
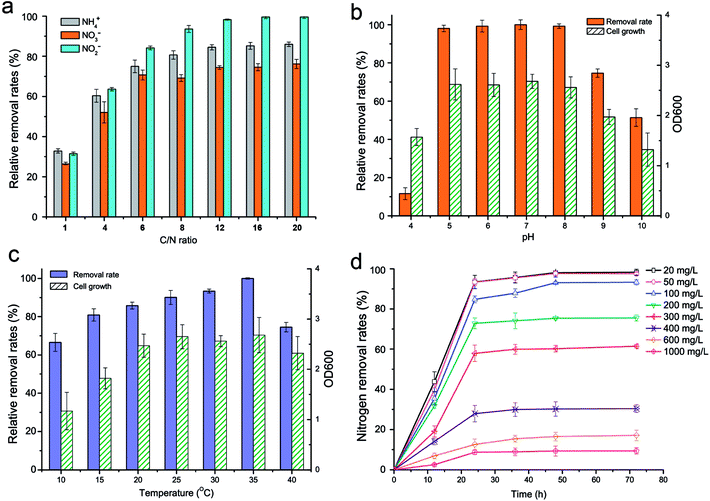
The effect of the C/N ratio (1–20) on the nitrogen removal efficiency of strain KSND is shown in Fig. 2a. The residual nitrogen removal activities were more than 35%, 56%, and 73% for C/N values of 1, 4, and 6, respectively. This result differs from that for Zobellella taiwanensis DN-7, in which higher C/N ratios (>12) resulted in a serious deterioration in ammonium removal.12 These results suggest that strain KSND is over excellent denitrifiers such as Bacillus strain N31 and B. methylotrophicus L7, even at high C/N ratios of 6–20.15,16 Therefore, strain KSND offers an alternative bacterium for the remediation of low-C/N domestic wastewater.KSND showed efficient nitrogen removal over a wide range of initial pHs (5.0–8.0), with approximate removal rates of 91–92%. More than 68% and 47% of this activity remained at pH 9.0 and pH 10.0, respectively (Fig. 2b). Strain KSND showed an optimal temperature for nitrogen removal of 35 °C. However, even at 10 °C, it maintained a removal rate of almost 63% (Fig. 2c). Previous reports have suggested that aerobic denitrification is seriously reduced below 10 °C because of the consequent delay in cell growth and nitrogen metabolism.17 Our results indicate that strain KSND can be used for low-temperature nitrification and denitrification. The temperature of most environmental water is <20 °C, suggesting the potential application of KSND to wastewater treatment in wintertime or in open reactors.
The effects of the ammonium concentration on cell growth, ammonium removal, and the nitrification products were evaluated. Higher NH4+–N concentrations resulted in slightly higher cell growth, indicating that KSND tolerates a broad range of ammonium concentrations. After culture for 24 h, ammonia was removed at average rates of 0.78, 1.94, 3.53, 6.07, 7.23, 4.65, 3.15, and 3.62 mg L−1 h−1 in NH4+–N concentrations ranging from 20 to 1000 mg L−1, respectively (Fig. 2d). No accumulation of the nitrification product NO2−–N was detectable (data not shown). The high-strength ammonium probably inhibited the enzyme activities involved in nitrification, which would account for the deterioration in the ammonium removal efficiency. By contrast, aquaculture water, effluent, and domestic wastewater have relatively low ammonium levels, ranging from 0.2 to 200 mg L−1.18,19 KSND removed 93.10% of the initial NH4+–N (100 mg L−1) within 24 h. This is highly significant for the practical application of KSND to most sewage treatments, including of aquaculture water, general effluent, domestic sewerage, and other wastewaters containing low levels of ammonia.
3.3. Determination of the genes involved in nitrogen metabolism
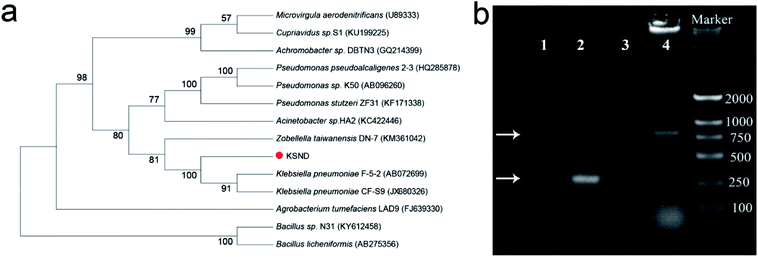
A phylogenetic analysis of 16S rRNA genes showed that the isolated strain KSND is affiliated with the genus Klebsiella (Fig. 3a). Genes encoding functional enzymes involved in the nitrification (Hao) and denitrification pathways (dissimilatory NirS) are often used as biomarkers to detect denitrifiers.20 Neither the hao nor nirS gene could be detected in the KSND strain with PCR and specific degenerate primers, whereas these primers highly efficiently amplified the hao and nirS genes from the environmental samples (Fig. 3b). These results are almost consistent with a previous report of K. pneumoniae EGD-HPI9-C.11 Therefore, the traditional nitrification and denitrification pathways are incomplete in Klebsiella sp. strain KSND, indicating a metabolic versatility in its nitrification and denitrification reactions. Recently, it is indicated that may be existed the other denitrification pathway via hydroxylamine intermediate.10,21,22 This matter requires further investigation to clarify the simultaneous nitrification and denitrification processes of this strain.3.4. Efficient nitrogen removal to a domestic sewerage system
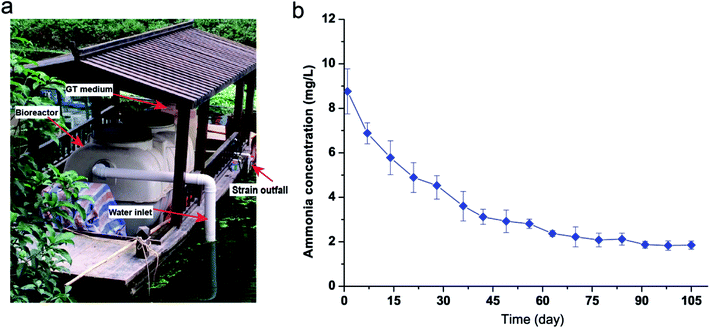
Strain KSND was used to remove ammonia nitrogen from a river, using a floating bed carrying a simple bioreactor (Fig. 4a). The cell growth of KSND was ∼OD600 = 1.7 (∼6.3 ± 0.5 × 107 colony-forming units per mL) after intense culture for 24 h in the bioreactor. NH4+–N (8.76 mg L−1) was present in the river, with undetectable amounts of NO3–N and NO2–N. After 14 days, the NH4+–N concentration was dramatically reduced to 5.78 mg L−1 (Fig. 4b). NO3–N and NO2–N were not detected during this time, which clearly differs from the results for Enterobacter cloacae CF-S27.22 Most studies have shown that nitrite accumulation is inevitable during the biological treatment of wastewater, which reduces the removal efficiency of TN. Nitrite nitrogen is a compound with particularly high toxicity to freshwater animals and environmental bacteria.23 Our results indicate that KSND caused no toxic compound pollution during the large-scale treatment of sewage. After 91 days, the NH4+–N concentration was further reduced to 1.87 mg L−1 (Fig. 4b), and this level was maintained throughout the experiment (180 days). Overall, our study shows the excellent performances of KSND as a nitrogen remediator and its great potential utility in the removal of nitrogen from domestic wastewater and in river restoration.4. Conclusions
In this study, a simultaneously nitrifying and denitrifying strain, Klebsiella sp. KSND, was successfully isolated, and demonstrated an excellent capacity for the simultaneous removal of high concentrations of ammonium, nitrite, and nitrate, with maximal removal rates of 5.77, 0.35, and 3.26 mg L−1 h−1, respectively. The average removal efficiency at 10 °C or in low-C/N wastewater was above 50%. This excellent performance makes KSND a promising candidate remediator of nitrogen pollution, with particular applications in nitrogen removal from low-C/N domestic wastewater.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant 31700078 and Grant 31270724), the Scientific Research Foundation for Talent program of Zhejiang Agricultural and Forestry University (W20170029), Distinguished Scholars of Zhejiang Agricultural and Forestry University (2014FR064).References
- J. N. Galloway, A. R. Townsend, J. W. Erisman, M. A. Bekunda, Z. Cai, J. R. Freney, L. A. Martinelli, S. P. Seitzinger and M. A. Sutton, Science, 2008, 320, 889–892 CrossRef PubMed.
- J. C. Finlay, G. E. Small and R. W. Sterner, Science, 2013, 342, 247–250 CrossRef PubMed.
- J. Granger and S. D. Wankel, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E6391–E6400 CrossRef PubMed.
- G. Zhu, Y. Peng, B. Li, J. Guo, Q. Yang and S. Wang, Rev. Environ. Contam. Toxicol., 2008, 192, 159–195 CrossRef.
- Y. Shi, S. Hu, J. Lou, P. Lu, J. Keller and Z. Yuan, Environ. Sci. Technol., 2013, 47, 11577–11583 CrossRef PubMed.
- Y. Wu, S. Shukal, M. Mukherjee and B. Cao, Environ. Sci. Technol., 2015, 49, 11551–11559 CrossRef PubMed.
- M. Strous, E. Pelletier, S. Mangenot, T. Rattei, A. Lehner, M. W. Taylor, M. Horn, H. Daims, D. Bartolmavel and P. Wincker, Nature, 2006, 440, 790–794 CrossRef PubMed.
- Z. Jing, L. Feng, G. Yang, D. Chen and J. Mu, Bioresour. Technol., 2017, 243, 800–809 CrossRef PubMed.
- S. K. Padhi, S. Tripathy, R. Sen, A. S. Mahapatra, S. Mohanty and N. K. Maiti, Int. Biodeterior. Biodegrad., 2013, 78, 67–73 CrossRef.
- Y. Feng, J. Feng and Q. Shu, J. Appl. Microbiol., 2018, 124, 1195–1211 CrossRef PubMed.
- R. R. Pal, A. A. Khardenavis and H. J. Purohit, Funct. Integr. Genomics, 2015, 15, 63–76 CrossRef PubMed.
- L. Yu, Y. Wang, H. Liu, C. Xi and L. Song, Appl. Microbiol. Biotechnol., 2016, 100, 4219–4229 CrossRef PubMed.
- H. Tengxia, L. Zhenlun, S. Quan, X. Yi and Y. Qing, Bioresour. Technol., 2016, 200, 493–499 CrossRef PubMed.
- P. Chen, J. Li, Q. X. Li, Y. Wang, S. Li, T. Ren and L. Wang, Bioresour. Technol., 2012, 116, 266–270 CrossRef PubMed.
- Q. L. Zhang, Y. Liu, G. M. Ai, L. L. Miao, H. Y. Zheng and Z. P. Liu, Bioresour. Technol., 2012, 108, 35–44 CrossRef PubMed.
- F. Huang, L. Pan, N. Lv and X. Tang, J. Biosci. Bioeng., 2017, 124, 564–571 CrossRef PubMed.
- S. Saleh-Lakha, K. E. Shannon, S. L. Henderson, C. Goyer, J. T. Trevors, B. J. Zebarth and D. L. Burton, Appl. Environ. Microbiol., 2009, 75, 3903–3911 CrossRef PubMed.
- S. Tsukuda, L. Christianson, A. Kolb, K. Saito and S. Summerfelt, Aquacult. Eng., 2015, 64, 49–59 CrossRef.
- R. Crab, Y. Avnimelech, T. Defoirdt, P. Bossier and W. Verstraete, Aquaculture, 2007, 270, 1–14 CrossRef.
- M. Li, T. Ford, X. Li and J. D. Gu, Environ. Sci. Technol., 2011, 45, 3547–3553 CrossRef PubMed.
- B. Zhao, Y. L. He, J. Hughes and X. F. Zhang, Bioresour. Technol., 2010, 101, 5194–5200 CrossRef PubMed.
- S. K. Padhi, S. Tripathy, S. Mohanty and N. K. Maiti, Bioresour. Technol., 2017, 232, 285–296 CrossRef PubMed.
- A. Alonso and J. A. Camargo, Ecotoxicol. Environ. Saf., 2009, 72, 2005–2008 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2018 |