Open Access Article
Open Access ArticleImmunomagnetic separation and size-based detection of Escherichia coli O157 at the meniscus of a membrane strip†
Hyeonjeong Lee,
Jeongin Hwang,
Yunsung Park ,
Donghoon Kwon,
Sanghee Lee,
Inseok Kang and
Sangmin Jeon
,
Donghoon Kwon,
Sanghee Lee,
Inseok Kang and
Sangmin Jeon *
*
Department of Chemical Engineering, Pohang University of Science and Technology (POSTECH), 77 Cheongam-Ro, Nam-Gu, Pohang, Gyeongbuk, Republic of Korea. E-mail: jeons@postech.ac.kr
First published on 23rd July 2018
Abstract
We developed a facile method for the detection of pathogenic bacteria using gold-coated magnetic nanoparticle clusters (Au@MNCs) and porous nitrocellulose strips. Au@MNCs were synthesized and functionalized with half-fragments of Escherichia coli O157 antibodies. After the nanoparticles were used to capture E. coli O157 in milk and dispersed in a buffer solution, one end of a test strip was dipped into the solution. Due to the size difference between the E. coli–Au@MNC complexes (approximately 1 μm) and free Au@MNCs (approximately 180 nm), only E. coli–Au@MNC complexes accumulated at the meniscus of the test strip and induced a color change. The color intensity of the meniscus was proportional to the E. coli concentration, and the detection limit for E. coli in milk was 103 CFU mL−1 by the naked eye. The presence of E. coli–Au@MNC complexes at the meniscus was confirmed using a real-time PCR assay. The developed method was highly selective for E. coli when compared with Salmonella typhimurium, Listeria monocytogenes, and Staphylococcus aureus.
Introduction
Foodborne illnesses that are frequently caused by pathogenic bacteria are a major public health concern worldwide. Since microorganisms can contaminate foods at any point during handling, prevention of foodborne illness requires rigorous inspection. Considering the short time before each meal, conventional cultivation techniques that require a lengthy period of time for bacterial cell culture are not suitable for field tests. Instrument-related methods that do not require bacterial cell culture have been developed and include quartz crystal microbalance,1,2 surface plasmon resonance,3,4 surface-enhanced Raman scattering,5,6 fluorescence spectroscopy,7,8 electrochemical sensors,9 and polymerase chain reaction.10–12 Although these techniques enable the detection of bacteria within a few hours, they require expensive instruments and experienced technicians.In contrast, the lateral flow immunoassay (LFIA) uses a series of membrane pads, such as a sample pad, a conjugation pad containing antibody-functionalized gold nanoparticles, a nitrocellulose (NC) membrane with antibodies immobilized at test and control lines, and an absorbent pad. The presence of target bacteria is determined by observing a color change at the test and control lines caused by the accumulation of the Au nanoparticles.13–17 The LFIA is economical, disposable and easy to use.16,18–22 However, the presence of high concentrations of food debris in food samples impedes sensitive measurement.
The sensitive and selective detection of foodborne bacteria may be achieved by adopting antibody-conjugated magnetic nanoparticles to separate target bacteria from food debris. After magnetic separation of target bacteria from a sample solution, the concentration of target bacteria can be determined either by a size sorting method,23,24 an electrochemical method,25,26 or by a sandwich assay using probes to which the specific detection antibody has been conjugated, such as fluorescence dyes and catalytic enzymes.27–30 Since the use of additional probes for a sandwich assay is a time-consuming and expensive process, a size sorting method is preferred for screening purposes.
Conventional size sorting methods utilize membrane filters to separate large bacteria-magnetic nanoparticle complexes from small free magnetic nanoparticles. The combination of membrane filters and magnetic nanoparticles offers an easy and cost-effective method of detection of foodborne bacteria.24 However, it has a critical drawback. The sample solution dropped on a membrane filter flows down and spreads laterally, which increases the contact area with the solution and degrades the detection sensitivity. The contact area can be reduced by dispensing the solution using a fine nozzle or making the membrane surface hydrophobic. However, the use of fine nozzles increases the analysis time and the hydrophobic coating increases nonspecific binding.
We overcame these limitations by separating bacteria–magnetic nanoparticle complexes and free magnetic nanoparticles at the meniscus of a test strip. The test strip was prepared by simply overlapping an absorbent pad on an NC membrane with appropriate pore size. Half-antibody fragment-conjugated, Au-coated magnetic nanoparticle clusters (Au@MNCs) were used to capture Escherichia coli O157 in milk, followed by magnetic separation and dispersal in phosphate buffer (PB). The immersion of one end of the test strip in the solution containing the free Au@MNCs and Au@MNC–bacteria complexes induced a color change at the meniscus due to the accumulation of the Au@MNC–bacteria complexes. The detection limit for E. coli in milk was 103 colony forming units (CFU) mL−1 with the naked eye, which was superior to the typical LFIA (105 CFU mL−1).31–34
Experimental
Materials and reagents
Sodium citrate tribasic dihydrate, iron(III) chloride hexahydrate (FeCl3·6H2O), urea, gold(III) chloride trihydrate (HAuCl4·3H2O), sodium borohydride (NaBH4), polyacrylamide, (3-aminopropyl)triethoxysilane (APTES), hydroxylamine hydrochloride (NH2OH·HCl), tris(2-carboxyethyl)phosphine (TCEP), potassium phosphate monobasic (KH2PO4), potassium phosphate dibasic (K2HPO4), bovine serum albumin (BSA), and Tween 20 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Anti-E. coli O157:H7 antibody was purchased from KPL (Gaithersburg, MD, USA). Absorbent pads, NC membranes, and Amicon ultra-0.5 centrifugal filters were purchased from Millipore (Billerica, MA, USA). Real-time polymerase chain reaction (PCR) experiments were carried out using PowerChek™ EHEC real-time PCR kit (KogeneBiotech, Seoul, Korea) and analyzed using a QuantStudio™ 3 Real-Time PCR system (Applied Biosystems, Franklin Lakes, NJ, USA).Preparation of antibody-conjugated Au@MNCs
Gold-coated superparamagnetic Fe3O4 nanoparticle clusters were synthesized as described elsewhere.35–37 In brief, 4 mmol of FeCl3·6H2O, 12 mmol of urea, 8 mmol of sodium citrate tribasic dehydrate, and 0.4 g polyacrylamide were dissolved in 80 mL deionized (DI) water. After the solution was heated in a Teflon-lined autoclave at 200 °C for 10 h, the precipitate (MNC) was magnetically collected and rinsed several times with DI water. To coat the gold nanoparticles onto an MNC, amine groups were produced on the surface of the MNC by incubating 8 mg of MNC overnight in 40 mL of 1% APTES in ethanol. After washing the MNCs with ethanol and DI water, they were added to a solution of 14 nm gold nanoparticles and shaken for 5 h. Half fragments of anti-E. coli antibody were obtained by adding 40 μg of the antibody to 400 μL of PB containing 0.125 mM of TCEP for 1 h. The direct conjugation of half-antibody fragments to Au@MNCs via Au–S bonds provided the desired orientation of the antibody to efficiently capture target bacteria.38,39Results and discussion
Antibody-conjugated Au@MNCs
Fig. 1(a) presents a representative scanning electron microscopy (SEM) image of Au@MNCs. The bright spots correspond to Au nanoparticles. The mean diameter of Au@MNCs was 180 nm, and each MNC comprised hundreds of 14 nm magnetic nanoparticles. Fe3O4 nanoparticles larger than 30 nm exhibit ferromagnetism. Therefore, clusters of 14 nm Fe3O4 nanoparticles were used to maintain the paramagnetic property and allow the efficient magnetic separation of the nanoparticles. Fig. 1(b) displays a representative transmission electron microscopy image of E. coli covered with many Au@MNCs. The E. coli–Au@MNC complex was one order of magnitude larger than the Au@MNCs, which made it possible to separate the E. coli–Au@MNC complexes from the free Au@MNCs.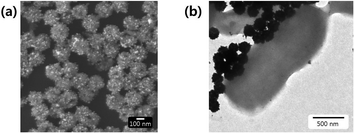 | ||
| Fig. 1 (a) SEM image of Au@MNCs, wherein 14 nm gold nanoparticles appear as bright spots. (b) TEM image of E. coli–Au@MNC complexes. | ||
Detection of E. coli-Au@MNCs at the meniscus of the test strip
Fig. 2 illustrates the structure of the test strip, which comprises an absorbent pad and NC membrane. The NC membrane was pressed to generate a control line with considerably smaller pores compared to the initial membrane (see SEM images). As a result, the Au@MNC–bacteria complexes accumulated at the meniscus (i.e., the test line) upon immersion of the test strip into a sample solution containing free Au@MNCs and E. coli–Au@MNC complexes, whereas the free Au@MNCs moved up and accumulated at the control line, with the smaller pores. The immersion of the test strip into a sample solution resulted in the formation of a meniscus line.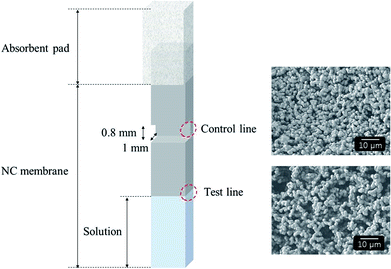 | ||
| Fig. 2 Schematic of test strip with an NC membrane overlapped by an absorbent pad (left) and SEM images (right) showing the pores at the control line (upper) and test line (lower). | ||
The line's shape changed depending on the width of the test strip. To examine the meniscus shape, a test strip with an NC membrane (HF180, Millipore) was dipped into a solution containing Au@MNCs. The meniscus became more curved and blurred as the width increased, which degraded the detection sensitivity (Fig. 3). Since handling the 2 mm-wide test strip was not convenient, the 3 mm-wide test strip was selected as the optimal membrane and was used throughout the experiments.
 | ||
| Fig. 3 Optical images of test strips after immersion in Au@MNC solution for 3 min. As the width of the test strip increases, the meniscus becomes more curved and blurred. | ||
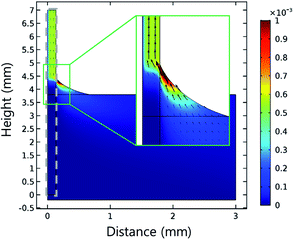
Interestingly, most E. coli–Au@MNC complexes accumulate at the meniscus and rarely accumulate below the meniscus. Numerical simulations were conducted using the COMSOL Multiphysics package to examine the accumulation of E. coli–Au@MNC complexes at the meniscus line. The permeability of the NC membrane was assumed to be 10−10 m2. The solution flowed toward the meniscus of the NC membrane and the flow velocity reached a maximum at the meniscus (Fig. 4). Thus, most E. coli–Au@MNC complexes in the solution would accumulate at the meniscus.
Detection of E. coli in milk
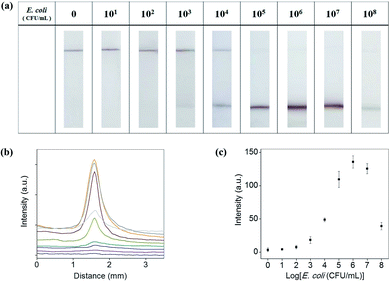
Antibody-conjugated Au@MNCs were incubated in 15 mL of E. coli O157-spiked milk at various concentrations (0 to 108 CFU mL−1) and magnetically separated and dispersed in 200 μL of 0.1% Tween 20. The immersion of a test strip into the solution induced the absorption of free Au@MNCs and E. coli–Au@MNC complexes on the membrane. Fig. 5(a) shows optical images of the test strips after immersion for 3 min. The meniscus became darker with increasing E. coli concentration because of the increase in the number of E. coli–Au@MNC complexes in the solution. In contrast, the control line having a reduced pore size became paler with increasing E. coli concentration because of the decrease in the number of free Au@MNCs. Of note, the accumulation of E. coli–Au@MNC complexes occurred at the meniscus even though the test strip below the meniscus was fully immersed in the solution, which increased the assay sensitivity.For quantitative analysis, the colors at the meniscus and control line were converted to a gray-scale value using ImageJ software (NIH, Bethesda, MD, USA). Fig. 5(b) shows the variation in the intensities at the meniscus along the flow direction and Fig. 5(c) shows the maximum intensities, both as a function of E. coli concentration. The intensity at the meniscus increased with increasing E. coli concentration and showed a maximum value at 106 CFU mL−1. The decrease in intensity at concentrations above 106 CFU mL−1 may be attributed to the relative decrease (compared with E. coli) in the number of Au@MNCs (∼108 mL−1). At E. coli concentrations above 106 CFU mL−1, a lower number of Au@MNCs bound to the surface of each E. coli bacterium, and the color at the meniscus became paler despite the accumulation of a greater number of complexes. The detection limit was determined to be 103 CFU mL−1 of E. coli in milk after 3 min of immersion, which is higher than the sensitivity of typical LFIA (105 CFU mL−1).31–34 Table 1 shows that the sensitivity of our method is superior to other LFIAs for detection of E. coli O157.31–33,40–44
E. coli detection using a real-time PCR assay
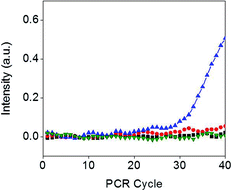
The presence of E. coli strains at the meniscus line was confirmed using a real-time PCR (RT-PCR) assay. Two test strips in Fig. 5(a) with 0 and 103 CFU mL−1 E. coli were used for the RT-PCR measurement. After carefully cutting 2 mm of the test strip containing the meniscus line or the control line, each sample was immersed in 200 μL of DI water and sonicated for 10 min. Then, 5 μL of each solution was mixed with 15 μL of RT-PCR premix solution, and RT-PCR was conducted for 1.5 h. Amplification of VT2 genes in E. coli was observed only for samples containing the meniscus line with 103 CFU mL−1 E. coli (Fig. 6), indicating that the test strip successfully separated E. coli–Au@MNC complexes from free Au@MNCs.Selectivity of the assay
To evaluate the assay selectivity, a control experiment was conducted. E. coli antibody-functionalized Au@MNCs were incubated in 15 mL of milk with 105 CFU mL−1 Listeria monocytogenes, Salmonella typhimurium, and Staphylococcus aureus. After the magnetic separation of the bacteria–Au@MNC complexes, they were dispersed in 200 μL of 0.1% Tween 20 and transferred into a 96-well plate. Fig. 7 shows optical images of the test strips after immersion in each solution for 3 min. A dark meniscus was observed for the test strip dipped into the E. coli-spiked sample, while a pale meniscus was observed for those treated with S. typhimurium, L. monocytogenes, and Staph. aureus, demonstrating the selectivity of this method.Conclusions
In this study, we developed a novel method for the size-based detection of E. coli in milk. After immunomagnetic separation of E. coli, the immersion of the test strip in a solution containing free Au@MNCs and E. coli–Au@MNC complexes induced a color change at the meniscus due to the accumulation of the complexes. The accumulation at the meniscus occurred even though the test strip below the meniscus was fully immersed in the solution, which increased the assay sensitivity. The color intensity of the meniscus was proportional to the bacterial concentration (up to 106 CFU mL−1), and the detection limit was 103 CFU mL−1 for E. coli O157 in milk with the naked eye. The developed method has great potential to assess the presence of hazardous bacteria or heavy metal ions in drinking water simply by changing probe molecules.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grants from the POSCO Research Fund (Project No. 2017Y018) and the National Research Foundation of Korea (NRF-2016R1A4A1010735).Notes and references
- Z. Farka, D. Kovář and P. Skládal, Sensors, 2015, 15, 79 CrossRef PubMed.
- Z. Shen, M. Huang, C. Xiao, Y. Zhang, X. Zeng and P. G. Wang, Anal. Chem., 2007, 79, 2312–2319 CrossRef PubMed.
- Y. Wang, W. Knoll and J. Dostalek, Anal. Chem., 2012, 84, 8345–8350 CrossRef PubMed.
- F. C. Dudak and İ. H. Boyacı, Biotechnol. J., 2009, 4, 1003–1011 CrossRef PubMed.
- C. Fan, Z. Hu, A. Mustapha and M. Lin, Appl. Microbiol. Biotechnol., 2011, 92, 1053–1061 CrossRef PubMed.
- H. Zhou, D. Yang, N. E. Mircescu, N. P. Ivleva, K. Schwarzmeier, A. Wieser, S. Schubert, R. Niessner and C. Haisch, Microchim. Acta, 2015, 182, 2259–2266 CrossRef.
- F. Mouffouk, A. M. Rosa da Costa, J. Martins, M. Zourob, K. M. Abu-Salah and S. A. Alrokayan, Biosens. Bioelectron., 2011, 26, 3517–3523 CrossRef PubMed.
- L. Wang, W. Zhao, M. B. O'Donoghu and W. Tan, Bioconjugate Chem., 2007, 18, 297–301 CrossRef PubMed.
- C.-X. Zhou, R.-J. Mo, Z.-M. Chen, J. Wang, G.-Z. Shen, Y.-P. Li, Q.-G. Quan, Y. Liu and C.-Y. Li, ACS Sens., 2016, 1, 965–969 CrossRef.
- P. de Boer, H. Rahaoui, R. J. Leer, R. C. Montijn and J. M. B. M. van der Vossen, Food Microbiol., 2015, 51, 96–100 CrossRef PubMed.
- Y. Zeng, Y. Wan and D. Zhang, Microchim. Acta, 2016, 183, 741–748 CrossRef.
- Y. Yang, F. Xu, H. Xu, Z. P. Aguilar, R. Niu, Y. Yuan, J. Sun, X. You, W. Lai, Y. Xiong, C. Wan and H. Wei, Food Microbiol., 2013, 34, 418–424 CrossRef PubMed.
- J. E. Schonhorn, S. C. Fernandes, A. Rajaratnam, R. N. Deraney, J. P. Rolland and C. R. Mace, Lab Chip, 2014, 14, 4653–4658 RSC.
- A. K. Yetisen, M. S. Akram and C. R. Lowe, Lab Chip, 2013, 13, 2210–2251 RSC.
- D. H. Choi, S. K. Lee, Y. K. Oh, B. W. Bae, S. D. Lee, S. Kim, Y.-B. Shin and M.-G. Kim, Biosens. Bioelectron., 2010, 25, 1999–2002 CrossRef PubMed.
- X. Mao, Y. Ma, A. Zhang, L. Zhang, L. Zeng and G. Liu, Anal. Chem., 2009, 81, 1660–1668 CrossRef PubMed.
- S. Rong-Hwa, T. Shiao-Shek, C. Der-Jiang and H. Yao-Wen, Food Chem., 2010, 118, 462–466 CrossRef.
- Y. Zhang, J. Bai and J. Y. Ying, Lab Chip, 2015, 15, 1465–1471 RSC.
- D. J. You, T. S. Park and J.-Y. Yoon, Biosens. Bioelectron., 2013, 40, 180–185 CrossRef PubMed.
- G. A. Posthuma-Trumpie, J. Korf and A. van Amerongen, Anal. Bioanal. Chem., 2009, 393, 569–582 CrossRef PubMed.
- J. Yan, Y. Liu, Y. Wang, X. Xu, Y. Lu, Y. Pan, F. Guo and D. Shi, Sens. Actuators, B, 2014, 197, 129–136 CrossRef.
- J. Hu, J. R. Choi, S. Wang, Y. Gong, S. Feng, B. Pingguan-Murphy, T. J. Lu and F. Xu, Sens. Actuators, B, 2017, 243, 484–488 CrossRef.
- H. Lee, T.-J. Yoon and R. Weissleder, Angew. Chem., Int. Ed. Engl., 2009, 48, 5657–5660 CrossRef PubMed.
- W.-B. Shim, C.-W. Lee, M.-G. Kim and D.-H. Chung, Anal. Methods, 2014, 6, 9129–9135 RSC.
- C. Lulu, Z. Qi, D. Huang, F. Yingchun and L. Yanbin, Electroanalysis, 2018, 30, 517–524 CrossRef.
- Q. Zhang, L. Li, Z. Qiao, C. Lei, Y. Fu, Q. Xie, S. Yao, Y. Li and Y. Ying, Anal. Chem., 2017, 89, 12145–12151 CrossRef PubMed.
- Y. Che, Y. Xu, R. Wang and L. Chen, Anal. Bioanal. Chem., 2017, 409, 4709–4718 CrossRef PubMed.
- X.-L. Su and Y. Li, Anal. Chem., 2004, 76, 4806–4810 CrossRef PubMed.
- Y. Zhao, M. Ye, Q. Chao, N. Jia, Y. Ge and H. Shen, J. Agric. Food Chem., 2009, 57, 517–524 CrossRef PubMed.
- J. Joo, D. Kwon, H. H. Shin, K.-H. Park, H. J. Cha and S. Jeon, Sens. Actuators, B, 2013, 188, 1250–1254 CrossRef.
- C. Song, C. Liu, S. Wu, H. Li, H. Guo, B. Yang, S. Qiu, J. Li, L. Liu, H. Zeng, X. Zhai and Q. Liu, Food Control, 2016, 59, 345–351 CrossRef.
- M. S. Suria, A. T. Mohd Afendy, M. Noor Azlina and I. Zamri, Int. Food Res. J., 2015, 22, 2587–2593 Search PubMed.
- J. Wang, R. Katani, L. Li, N. Hegde, E. L. Roberts, V. Kapur and C. DebRoy, Toxins, 2016, 8, 92 CrossRef PubMed.
- P. Noguera, G. A. Posthuma-Trumpie, M. van Tuil, F. J. van der Wal, A. de Boer, A. P. H. A. Moers and A. van Amerongen, Anal. Bioanal. Chem., 2011, 399, 831–838 CrossRef PubMed.
- W. Cheng, K. Tang, Y. Qi, J. Sheng and Z. Liu, J. Mater. Chem., 2010, 20, 1799–1805 RSC.
- L. Wang, J. Bai, Y. Li and Y. Huang, Angew. Chem., Int. Ed., 2008, 47, 2439–2442 CrossRef PubMed.
- J. Hwang, D. Kwon, S. Lee and S. Jeon, RSC Adv., 2016, 6, 48445–48448 RSC.
- H. Sharma and R. Mutharasan, Anal. Chem., 2013, 85, 2472–2477 CrossRef PubMed.
- D. Kwon, H. Yoo, H. Lee and S. Jeon, Sens. Actuators, B, 2018, 255, 552–556 CrossRef.
- Y. Zhao, H. Wang, P. Zhang, C. Sun, X. Wang, X. Wang, R. Yang, C. Wang and L. Zhou, Sci. Rep., 2016, 6, 21342 CrossRef PubMed.
- C. Song, J. Liu, J. Li and Q. Liu, Biosens. Bioelectron., 2016, 85, 734–739 CrossRef PubMed.
- I.-H. Cho, A. Bhunia and J. Irudayaraj, Int. J. Food Microbiol., 2015, 206, 60–66 CrossRef PubMed.
- J. Han, L. Zhang, L. Hu, K. Xing, X. Lu, Y. Huang, J. Zhang, W. Lai and T. Chen, J. Dairy Sci., 2018, 101, 5770–5779 CrossRef PubMed.
- M. Chen, Z. Yu, D. Liu, T. Peng, K. Liu, S. Wang, Y. Xiong, H. Wei, H. Xu and W. Lai, Anal. Chim. Acta, 2015, 876, 71–76 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra04739a |
| This journal is © The Royal Society of Chemistry 2018 |