Open Access Article
Open Access ArticleA high precision length-based carbon nanotube ladder
Zahra Borzooeian *a,
Mohammad E. Taslima,
Saina Rezvanib and
Giti Borzooeianc
*a,
Mohammad E. Taslima,
Saina Rezvanib and
Giti Borzooeianc
aDepartment of Mechanical and Industrial Engineering, College of Engineering, Northeastern University, Boston, MA, USA. E-mail: z.borzooeian@northeastern.edu; borzooeianm@gmail.com
bDepartment of Computer Science, Worcester Polytechnic Institute, Worcester, MA, USA
cDepartment of Biology, Payamnoor, University of Esfahan, Iran
First published on 23rd October 2018
Abstract
Today, carbon nanotubes manufacturers as well as users such as molecular electronics, nanomedicine, nano-biotechnology and similar industries are facing a major challenge: lack of length uniformity of carbon nanotubes in mass production. An effective solution to this major issue is the use of a length-based ladder. We are, for the first time, presenting such a valuable tool to determine the length purity. Our length-based carbon nanotubes ladder, containing a series of carbon nanotubes markers with different lengths, is made based on three combined techniques – bio-conjugation, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and silver staining. Creating an indicator using conjugation of a biomolecule with carbon nanotubes to make a carbon nanotubes ladder is a novel idea and a significant step forward for length-based carbon nanotubes separation. The very sensitive silver staining technique allows a precise visualization and quantification of the gel. This ladder with a pending patent by Northeastern University is an effective quality control tool when bulk quantities of nanotubes with a desirable length are manufactured.
Introduction
Carbon nanotubes (CNTs) of different lengths, diameters and structures are produced from different methods1–3 for different applications. These applications vary from nanoelectronics,4 semiconductors, probes and interconnects,5 nanosensors6,7 and nanotube transistors,8,9 to nanomaterials, energy storage/conversion devices, hydrogen storage10–12 and nanomedicine.13 Geometrical parameters are known to have a considerable impact on the properties (e.g., reactivity and conductivity) of carbon nanotubes.14 Thermal and electrical conductivities of carbon nanotubes are directly related to their length.15 CNT-based transistors performance,16 mechanical properties of epoxy composites17 and electromagnetic interference shielding are impacted by the nanotube length, to name a few. Therefore, a primary requirement for any technique that separates and purifies nanotubes in a scalable and reproducible manner is the ability to perform a precise measurement of the nanotubes length.18 To the best of these authors' knowledge, there exists no reliable quality control tool for precise and rapid measurement of nanotubes length and level of purity. Our proposed length-based ladder is intended for that purpose.The ladder is created through the covalent conjugation of lysozyme onto CNTs followed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Lysozyme was attached onto the carboxyl functionalized carbon nanotubes and horn sonication was used to create conjugated CNTs with different lengths. Creating an indicator for length-based separation of CNTs by the conjugation of biomolecules onto the nanotubes surfaces is a novel idea and a significant step forward in CNT-based science. Separation of bio-conjugated CNTs was performed using SDS-PAGE because of the charge- and size-dependent mobility of bio-conjugated CNTs under the influence of an applied electric field. Visualization with high resolution and quantification of nanotube fragments in the acrylamide gel, a challenge for a number of nano-tech researchers, was achieved using highly-sensitive silver staining. This ladder serves as a valuable quality-control tool when bulk quantities of CNTs with a specific length are produced.
Materials and methods
Conjugation of lysozyme onto SWCNTs
Details of covalent conjugation of lysozyme (lyophilized chicken egg white lysozyme, EC 3.2.1.17, Inovatech, Inc. Abbotsford, BC, Canada) to the functionalized single-walled carbon nanotubes (SWCNT–COOH) with outer diameter of 1–2 nm (MKnano, Canada) were given in our previous work.19,20 Conjugation was achieved using carbodiimide method.21 One mg of CNTs was dispersed in one mL of MES buffer, 50 mM, pH 6.2, and was added to an equal volume of 400 mM NHS in MES buffer. For coupling of NHS to the carboxylic groups on the surface of nanotubes, 20 mM EDC was added to the mixture. The mixture was then stirred at 200 rpm (30 min) followed by sonication (MSE Ultrasonic Disintegrators, 150 W, England) for 30 min and then centrifuged at 7000 rpm, three times, 15 min each to remove excess EDC and NHS. The enzyme solution which contained 10 mg of lysozyme in 1 mL of phosphate buffer (10 mM, pH 8) was then added to the nanotubes solution. The final mixture was sonicated for ca. 1 min to re-disperse the SWCNTs. The solution was shaken in an orbital shaker at 200 rpm at room temperature during the conjugation process. The conjugated lysozyme–SWCNTs solution was then centrifuged. To remove all nonspecifically adsorbed enzyme completely, the mixture was washed three times with triply distilled water and once with 1% (v/v) Tween-20. Control enzyme-nanotube conjugates were prepared using the same procedure, only without using EDC and NHS.Characterization of conjugated lysozyme–SWCNTs
The morphology of conjugated SWCNTs with lysozyme was compared with that of activated SWCNTs and pure lysozyme using scanning electron microscopy (SEM, S360 Oxford), TGA (TGA Q50), X-ray diffraction (XRD, D8, Advance, Bruker, axs) at λ = 0.1542 nm, and FTIR spectroscopy (Shimadzu FTIR 8300 spectrophotometer). To prepare three different ladders of SWCNTs, conjugated samples were sonicated for three time periods of 3, 7 and 10 minutes.22SDS-PAGE and silver staining
To prepare the gel stock solution (30%, m/v), acrylamide (29.2 g) and Bis (0.8 g) were dissolved in 100 mL of water and filtered. The separating gel solution was made up of 10.0 mL of the gel stock solution, 10.0 mL of Tris–HCl (1.5 mol L−1, pH 8.80), 200–800 μL of (NH4)2S2O8 (10% m/v) and 0.4 g of SDS, diluted with water to 40 mL. The stacking gel was prepared by mixing 1.33 mL of the gel stock solution with 2.5 mL Tris–HCl (0.5 mol L−1, pH 6.80) and 50 μL (NH4)2S2O8 (10%, m/v), and diluting with water to 10.0 mL. Then TEMED (10 μL) was added to the mixture. To remove any noncovalently-adsorbed enzyme, samples were washed several times with phosphate buffer (10 mM, pH 8). Electrophoresis buffer was made by dissolving Tris (15.14 g), glycine (72.05 g), and SDS (5 g) in 500 mL of distilled water. Solution's pH was adjusted to 8.30. The final gel which consisted of separating (10.0% m/v) and stacking (3.0%, m/v) gels was made in a vertical polyacrylamide gel system. Sample volumes of 15 μL were loaded on the gel. The silver staining procedure (Blum method23) consisted of several steps: fixation with methanol, acetic acid and paraformaldehyde solutions, washing with ethanol (50% and 30%) and ddH2O, sensitizing with Na2S2O3·5H2O, washing with ddH2O, impregnating with silver nitrate and paraformaldehyde solution, washing with ddH2O, developing with Na2CO3, paraformaldehyde and Na2S2O3·5H2O solution, washing with ddH2O, and ending reaction with a stop solution – methanol 50%, and acetic acid 12%.Image analysis techniques for length measurement
Two methods, a manual approach using ImageJ, a NIH approved software, and semi-automated method MATLAB used to analyse the gel images. Using the distance of each band, in a given lane, from the center (reference point) of the well and the electrical voltage applied to the gel, mobility and consequently length of the CNTs were calculated using Usrey's equation.24 A MATLAB code was developed to measure the intensities and the distances automatically. The input to this code was the 8 bit inverted image and the output was the distance of each band from the center of the well and the average intensity of the pixels of each band.Results and discussion
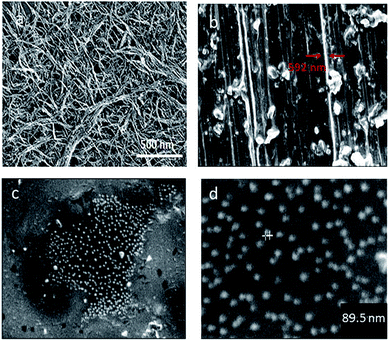
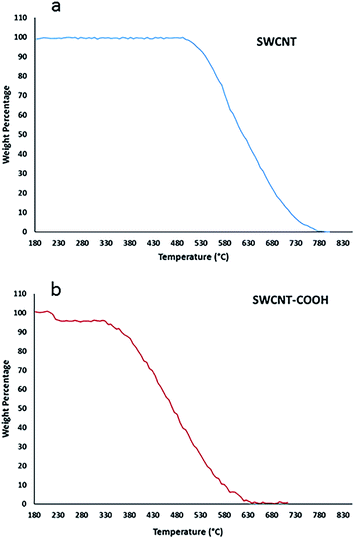
Carbodiimide method19,21,25 was used for bio-conjugation of lysozyme onto carboxylated carbon nanotubes surface. The results from SEM micrographs (Fig. 1a–d; before and after conjugation), TGA results (Fig. 2) used for evaluation of chemical functionalization of SWCNTs and XRD patterns (Fig. 3) and FTIR (Fig. 4) for evaluation of bio-conjugation and its chemical stability, confirmed the attachment of lysozyme onto the SWCNTs surface.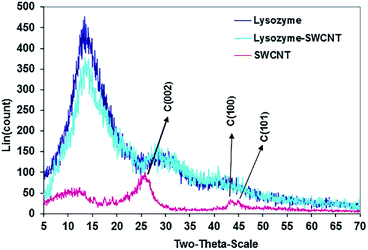 | ||
| Fig. 3 X-ray diffraction (XRD) patterns of SWCNTs (red), lysozyme (dark blue), and conjugated lysozyme–SWCNTs (light blue). | ||
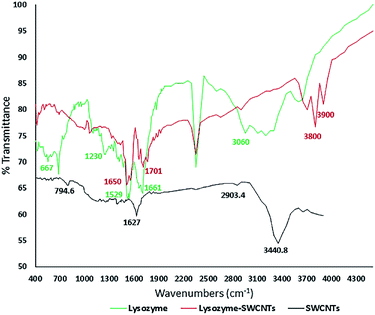 | ||
| Fig. 4 FTIR spectra showing amide bonds in the conjugated lysozyme–SWCNTs. FTIR spectrum for lysozyme (green), the SWCNTs (black) and conjugated lysozyme–SWCNT (red). | ||
The SEM micrographs (Fig. 1a–d) show the pre and post size and morphology of the conjugated SWCNTs. An increase of about 89.5–95 nm in the wall thickness of the conjugated nanotubes is an indication of a successful conjugation.
Thermograms of pure and functionalized SWNT with COOH are shown in Fig. 2a and b, respectively. Loss of weight with temperature is due to the burning of the attached chemical entities to the carbon nanotubes or nanotubes decomposition at elevated temperatures. Weight losses in two temperature ranges of 213–225 °C and 340–600 °C correspond to combustion of the covalently-linked –COOH and burning decomposition of the carbon nanotubes, respectively. As shown in part (a), SWCNT is thermally stable up to a temperature of about 500 °C indicating that it has no attached functional groups.
Detailed description of XRD results in Fig. 3 are presented in our earlier work.22 However, we shall refer to some important aspects of the results here. SWNCTs peak carbon planes are (002), (100) and (101). The Characteristic peaks of both free and conjugated lysozyme are the same and occur at 2θ positions 14.0, 30.0 and 42.0. The identical XRD patterns confirm the adsorption or absorption of lysozyme onto SWCNTs with no lysozyme phase change.
The results of XPS analysis of the atomic concentrations of oxygen and nitrogen in SWCNT, SWCNT–COOH, and lysozyme–SWCNT are presented in Table 1. The higher oxygen content for the SWCNT–COOH, compared with the pristine carbon nanotubes, confirms the presence of carboxyl groups on the surface of the nanotubes. The significant increase in both surface oxygen and nitrogen contents in conjugated lysozyme–SWCNTs indicates the covalent bonding between carboxyl-functionalized SWCNTs and lysozyme. The amount of immobilized enzyme was 1.1 mg mg−1 measured by elemental analysis of the activated carbon nanotubes and the lysozyme–SWNTs conjugates.
| Nanomaterial | Atomic concentration of oxygen (%) | Atomic concentration of nitrogen (%) |
|---|---|---|
| SWCNT | 0 | 0 |
| SWCNT–COOH | 12 | 0 |
| Lysozyme–SWCNT | 18 | 14 |
FTIR analyses
The amide linkages between the amino acid residues in polypeptides and proteins are detected in the FTIR diagrams.26 The covalent immobilization of polypeptides/proteins are studied by detecting the amide types I and II bands in the FTIR spectra which indicate the conformational changes in the protein secondary structure.27 Fig. 4 shows the FTIR spectra for pure lysozyme (green), pure SWCNTs (black), and lysozyme–SWCNT (red).Chemical functionalization of SWCNTs with –COOH groups are confirmed by the position of two absorption peaks at 1627.8 cm−1 and 3440.8 cm−1 (black curve). The wide and strong NH3 stretching band of 2950–2600 cm−1 in the enzyme spectrum is the amino acid characteristic. The plateau region near the band of 2222–2000 cm−1 corresponds to a combined bending vibration and torsional oscillation of the asymmetrical NH3+.28 A weak bending band of asymmetric NH3+ around 1661 cm−1 and a rather strong symmetric bending band around 1529 cm−1 are also observed. The 3600 cm−1 and 1230 cm−1 peaks represent the stretching of the N–H and C–N groups in the amine groups, respectively. Looking at the lysozyme–SWCNTs spectrum, the disappearance of the peaks is due to the formation of amide bonds between the carboxyl groups of functionalized SWCNTs and amine groups of the enzyme. Stretching vibration mode of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O creates the 1650 cm−1 peak and stretching of the N–H groups in the amide group creates the 3800 cm−1 and 1650 cm−1 peaks. In conclusion, the covalent bonding in the conjugation process was confirmed by the FTIR analyses, comparing the amide linkages between the free and conjugated lysozymes (peaks at 3800 cm−1 and 1650 cm−1).
O creates the 1650 cm−1 peak and stretching of the N–H groups in the amide group creates the 3800 cm−1 and 1650 cm−1 peaks. In conclusion, the covalent bonding in the conjugation process was confirmed by the FTIR analyses, comparing the amide linkages between the free and conjugated lysozymes (peaks at 3800 cm−1 and 1650 cm−1).
Precise visualization of nanotube fragments in the acrylamide gel which is a challenge for a number of nano-tech researchers, was achieved using silver staining. Fig. 5 shows the migration of lysozyme (a), SWCNTs (b) and conjugated lysozyme–SWCNTs (c) fragments across the gel.
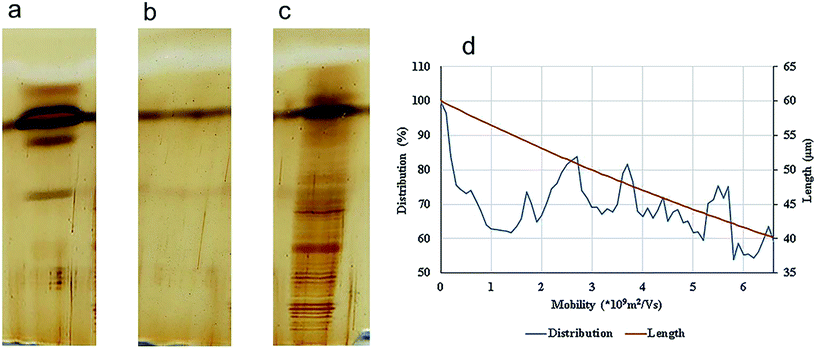 | ||
| Fig. 5 SDS-PAGE electrophoresis and silver staining of lysozyme. (a), SWCNT (b), and ladder/conjugated lysozyme–SWCNT (c). Distribution and the lengths of SWCNT fragments (d). | ||
Stability of conjugation and sensitivity of silver staining technique are the reasons for such sharp bands in Fig. 5c. The separation process of nano-carbon tubes in the gel based on their length is due to the following effects. Fragments of conjugated lysozyme–SWCNT with different lengths had different mobilities. Covalent attachment of lysozyme to carbon nanotubes gives rise to an intrinsic positive charge on any given individual nanotube or bundle affecting their mobilities. In other words, the degree of bioconjugation plays an important role in the separation process and net charge of fragments is directly proportional to the amount of conjugated lysozyme.
Depending on their length, each conjugated SWCNT moves differently through the gel matrix subjected to electrical field – small CNT fragments will experience less resistance when passing through the pores in the gel, while larger ones have more difficulty. Therefore, the CNTs migrate different distances based on their length. Smaller CNTs travel farther down the gel, while larger ones remain closer to the point of loading. The velocity (mobility) of the charged CNT fragments is directly proportional to the electrical field (E, volts per cm) and CNT fragments charge (q), and inversely proportional to the frictional coefficient of the mass and shape of the fragment (f).24
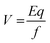 | (1) |
Since the gel acts like a sieve and retains the larger nanotubes while allowing the smaller ones pass through, the frictional coefficient is a representation of the level of resistance that the SWCNT fragments face as they pass through the pores of the gel. The SWCNT fragment length is also a key factor in its mobility in the gel matrix. In view of eqn (1), one will then have: mobility = (voltage)(charge)/(length).
To summarize, during gel electrophoresis, the mobility of a SWCNT fragment is primarily a function of its charge/length ratio.
Usrey's eqn (2), relating the fragment length to the intensity of the bands of the lanes is used for the calculation of the length distribution of the conjugated SWCNTs.
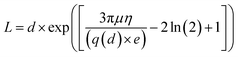 | (2) |
After an analysis of gel images using ImageJ, experimental data were obtained in the form of mobility distribution (number of nanotubes as a function of mobility). Fig. 5a–c show silver staining of free lysozyme (a), SWCNTs (b), and conjugated lysozyme–SWCNTs (c), and Fig. 5d shows that SWCNTs of various lengths are present in the population for each experimental electrophoretic mobility value.
To validate our method of creating CNT ladders, we made three different ladders of conjugated SWCNT fragments with different lengths, produced from sonication intervals of 3, 7, and 10 min (Fig. 6a–c, respectively). Conjugated lysozyme–SWCNT fragments of different lengths showed different mobilities.
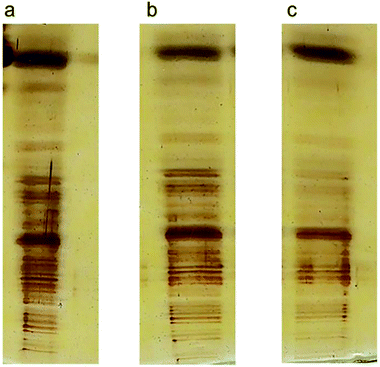 | ||
| Fig. 6 Three ladders produced from 3 (a), 7 (b), and 10 (c) min sonication time. It is clear the conjugated SWCNTs fragments are separated based on their lengths. | ||
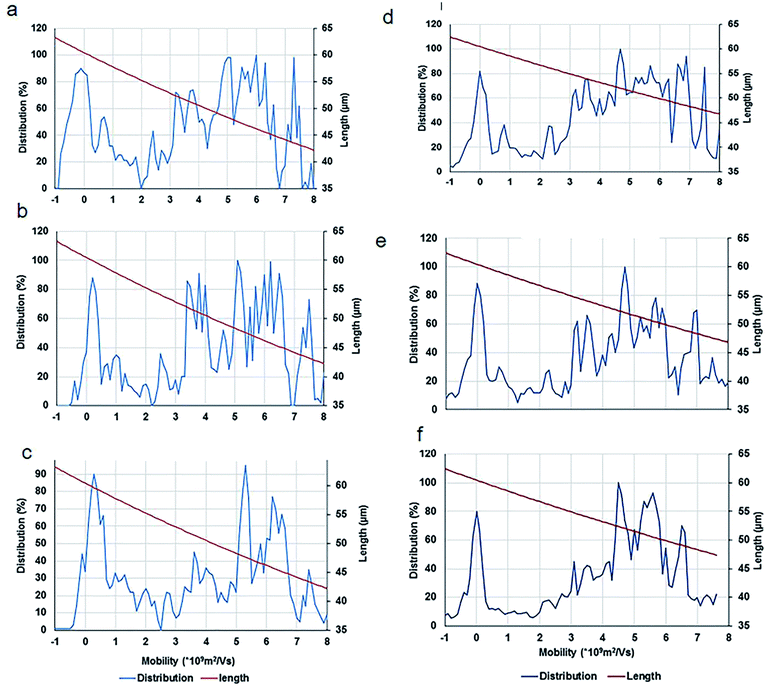
To calculate the length of CNTs, two methods were used. In the first method, a computer program was developed in MATLAB that subtracts the background, selects three lines on each lane of the gel and averages the signal (intensities of the bands) at each distance from the center of the wells. In the second method, ImageJ is used to calculate the same parameter using a narrow rectangle along each lane from the well to the bottom of the gel. These methods generated similar results (as shown in Fig. 7) that are in concordance with the visual evaluations.
After an analysis of gel images, experimental data were obtained in the form of mobility distribution (number of nanotubes as a function of mobility). The corresponding nanotube length of CNTs for every ladder were calculated from the Usray's formula (1). Fig. 7 shows the distribution and lengths of SWCNT fragments determined by two methods (ImageJ, Fig. 7a through Fig. 7c) & (MATLAB Fig. 7d through Fig. 7f) for sonication intervals of 3 min (Fig. 7a and 7d), 7 min (Fig. 7b and 7e), and 10 min (Fig. 7c and 7f). A key factor in our length-based separation technique is the change of surface charge density of nanotubes due to bio-conjugation and the number of attached biomolecules.
Fig. 8 shows the intensity of conjugated carbon nanotubes of different lengths for three different sonication intervals. Lengths are determine form the Usrey's eqn (1).
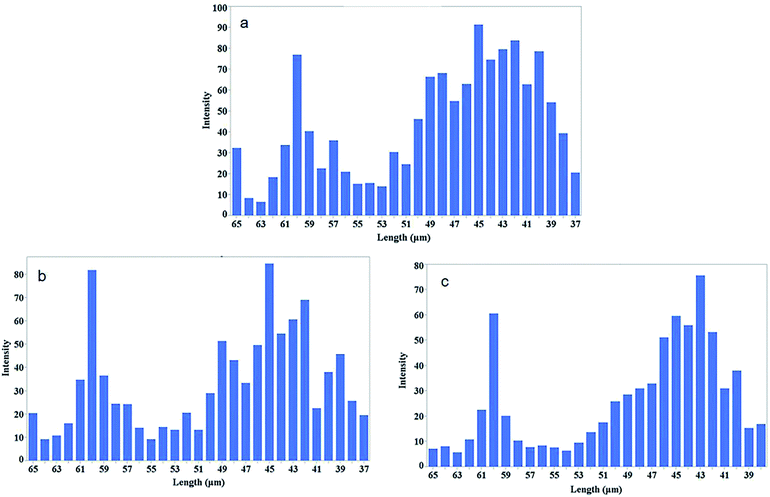 | ||
| Fig. 8 Length distribution of the conjugated SWCNTs after sonication time at 3 min (a), 7 min (b), and 10 min (c). The intensity of the CNTs at each ladder is plotted versus length of CNTs calculated from eqn (1). | ||
In conclusion, for the first time, taking advantage of conjugation of biomolecules onto CNT surfaces, a length-based CNT ladder is presented that can serve as a much-needed quality control tool for manufacturing bulk quantities of carbon nanotubes of specified lengths.
Conclusions
We have, for the first time, presented a valuable tool to determine the carbon nanotube length purity. Our length-based CNT ladder, containing a series of CNT markers with different lengths, is made by the combination of the following three techniques – bio-conjugation, SDS-PAGE, and silver staining. Creating an indicator using conjugation of a biomolecule with carbon nanotubes to make a CNT ladder is a novel idea and a significant step forward for length-based carbon nanotube separation. The very sensitive silver staining technique creates a high-resolution image, observable with naked eye and a precise quantification of the nanotubes. It further obviates the need for any further testing such as Raman spectroscopy. This ladder is an effective quality control tool when bulk quantities of nanotubes with a desirable length are manufactured.Conflicts of interest
There are no conflicts to declare.Abbreviations
| SWCNT | Single-walled carbon nanotubes |
| MES | 2-(N-Morpholino)ethane sulfonic acid |
| EDC | N-Ethyl-N′-(3-(dimethyl amino)propyl)carbodiimide hydrochloride |
| NHS | (N-Hydroxysuccinimide) |
| Tris | Tris-hydroxymethyl aminomethane |
| Bis | N,N-Methylenebisacrylamide |
| SDS | Sodium dodecyl sulfate |
| TEMED | Ammonium persulfate tetramethylethylenediamine |
| 2ME | 2-Mercaptoethanol |
| Bromophenol Blue | 3,3-5,5-Tetrabromophenolsulfonphthalein |
References
- E. G. Rakov, Russ. Chem. Rev., 2000, 69, 35–52 CrossRef CAS.
- F. Hennrich, S. Lebedkin, S. Malik, J. Tracy, M. Barczewski, H. Rösner and M. Kappes, Phys. Chem. Chem. Phys., 2002, 4, 2273–2277 RSC.
- S. Mohsenian, J. Fathi and B. Shokri, Appl. Phys. A: Mater. Sci. Process., 2018, 124, 546 CrossRef.
- S. Hermann, B. Pahl, R. Ecke, S. E. Schulz and T. Gessner, Microelectron. Eng., 2010, 87, 438–442 CrossRef CAS.
- R. H. Baughman, A. A. Zakhidov and W. A. de Heer, science, 2002, 297, 787–792 CrossRef CAS PubMed.
- M. S. Arnold, M. O. Guler, M. C. Hersam and S. I. Stupp, Langmuir, 2005, 21, 4705–4709 CrossRef CAS PubMed.
- J. Kong, N. R. Franklin, C. Zhou, M. G. Chapline, S. Peng, K. Cho and H. Dai, science, 2000, 287, 622–625 CrossRef CAS PubMed.
- A. Bachtold, P. Hadley, T. Nakanishi and C. Dekker, Science, 2001, 294, 1317–1320 CrossRef CAS PubMed.
- Z. Yao, H. W. C. Postma, L. Balents and C. Dekker, Nature, 1999, 402, 273–276 CrossRef CAS.
- A. Jones, T. Bekkedahl and C. Kiang, Nature, 1997, 386, 377 CrossRef.
- P. Chen, X. Wu, J. Lin and K. Tan, Science, 1999, 285, 91–93 CrossRef CAS PubMed.
- C. Liu, Y. Fan, M. Liu, H. Cong, H. Cheng and M. S. Dresselhaus, Science, 1999, 286, 1127–1129 CrossRef CAS PubMed.
- H. He, L. A. Pham-Huy, P. Dramou, D. Xiao, P. Zuo and C. Pham-Huy, BioMed Res. Int., 2013, 2013, 578290 Search PubMed.
- S. K. Doorn, R. E. Fields, H. Hu, M. A. Hamon, R. C. Haddon, J. P. Selegue and V. Majidi, J. Am. Chem. Soc., 2002, 124, 3169–3174 CrossRef CAS PubMed.
- X. Wang, Q. Jiang, W. Xu, W. Cai, Y. Inoue and Y. Zhu, Carbon, 2013, 53, 145–152 CrossRef CAS.
- A. D. Franklin and Z. Chen, Nat. Nanotechnol., 2010, 5, 858–862 CrossRef CAS PubMed.
- B. Singh, K. Saini, V. Choudhary, S. Teotia, S. Pande, P. Saini and R. Mathur, J. Nanopart. Res., 2014, 16, 1–11 Search PubMed.
- N. Komatsu and F. Wang, Materials, 2010, 3, 3818–3844 CrossRef CAS PubMed.
- Z. Borzooeian, A. Safavi, M. Hossain Sheikhi, M. Aminlari and M. Mahdi Doroodmand, J. Exp. Nanosci., 2010, 5, 536–547 CrossRef CAS.
- Z. Borzooeian, M. Taslim, G. Borzooeian, O. Ghasemi and M. Aminlari, RSC Adv., 2017, 7, 48692–48701 RSC.
- P. Asuri, S. S. Bale, R. C. Pangule, D. A. Shah, R. S. Kane and J. S. Dordick, Langmuir, 2007, 23, 12318–12321 CrossRef CAS PubMed.
- Z. Borzooeian, O. Ghasemi, S. Rezvani, G. Borzooeian and A. Nourbakhsh, PLoS One, 2018 DOI:10.1371/journal.pone.0197972.
- H. Blum, H. Beier and H. J. Gross, Electrophoresis, 1987, 8, 93–99 CrossRef CAS.
- M. L. Usrey, N. Nair, D. E. Agnew, C. F. Pina and M. S. Strano, Langmuir, 2007, 23, 7768–7776 CrossRef CAS PubMed.
- Z. Borzooeian, G. Borzooeian, O. Ghasemi and M. Aminlari, RSC Adv., 2017, 7, 48692 RSC.
- A. Dong, P. Huang and W. S. Caughey, Biochemistry, 1992, 31, 182–189 CrossRef CAS PubMed.
- A. Dong, B. Caughey, W. S. Caughey, K. S. Bhat and J. E. Coe, Biochemistry, 1992, 31, 9364–9370 CrossRef CAS PubMed.
- R. M. Silverstein, F. X. Webster, D. J. Kiemle and D. L. Bryce, Spectrometric identification of organic compounds, John wiley & sons, 2014 Search PubMed.
| This journal is © The Royal Society of Chemistry 2018 |