Open Access Article
Open Access ArticleBiosorption and bioaccumulation characteristics of cadmium by plant growth-promoting rhizobacteria†
Xingjie Li ,
Dongbo Li,
Zhenning Yan and
Yansong Ao*
,
Dongbo Li,
Zhenning Yan and
Yansong Ao*
School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China. E-mail: aoys@sjtu.edu.cn; Tel: +86-13916002737
First published on 3rd September 2018
Abstract
Plant growth-promoting rhizobacteria (PGPR) not only promote growth and heavy metal uptake by plants but are promising biosorbents for heavy metals remediation. However, there exist arguments over whether extracellular adsorption (biosorption) or intracellular accumulation (bioaccumulation) play dominant roles in Cd(II) adsorption. Therefore, three cadmium-resistant PGPR, Cupriavidus necator GX_5, Sphingomonas sp. GX_15, and Curtobacterium sp. GX_31 were used to study bioaccumulation and biosorption mechanisms under different initial Cd(II) concentrations, using batch adsorption experiments, desorption experiments, scanning electron microscopy coupled with energy dispersive X-ray (SEM-EDX) spectroscopy, transmission electron microscopy (TEM), and Fourier-transform infrared (FTIR) spectroscopy. In this study, with the increase of the initial Cd(II) concentrations, the removal efficiency of strains decreased and the adsorption capacity improved. The highest Cd(II) removal efficiency values were 25.05%, 53.88%, and 86.06% for GX_5, GX_15, and GX_31 with 20 mg l−1 of Cd(II), while the maximum adsorption capacity values were 7.97, 17.13, and 26.43 mg g−1 of GX_5, GX_15, and GX_31 with 100 mg l−1 of Cd(II). Meanwhile, the removal efficiency and adsorption capacity could be ordered as GX_31 > GX_15 > GX_5. The dominant adsorption mechanism for GX_5 was bioaccumulation (50.66–60.38%), while the dominant mechanisms for GX_15 and GX_31 were biosorptions (60.29–64.89% and 75.93–79.45%, respectively). The bioaccumulation and biosorption mechanisms were verified by SEM-EDX, TEM and FTIR spectroscopy. These investigations could provide a more comprehensive understanding of metal-bacteria sorption reactions as well as practical application in remediation of heavy metals.
1. Introduction
Remediation of heavy metal-contaminated soil has received much attention due to heavy metals' adverse effects on plants, animals, microorganisms, and humans. Among hazardous metals, cadmium (Cd) is of particular concern because it is difficult to degrade, accumulates easily, and is highly toxic.1 It has been demonstrated that a small amount of Cd(II) in the food chain can cause health risks in humans.2Physicochemical approaches such as filtration, ion exchange, chemical precipitation, and solvent extraction are widely used to remove heavy metals from the environment.3 However, these applications are mostly ineffective, expensive, and nonspecific, especially when concentrations of heavy metals are low.4 Therefore, it is imperative to find an efficient, cost-effective alternative. The use of microbiological biomass, including bacteria,5 fungi,6,7 and yeast,8 is increasingly accepted in metals removal due to the large and well defined surface area of biomass, its high binding affinity, its environmental friendliness, and its low cost.9
Microbial remediation takes place mainly through biosorption or bioaccumulation mechanisms.10 Biosorption is a passive-process, metabolism-independent extracellular adsorption, where heavy metal ions are passively adsorbed onto components of the cell surface.11 Generally, biosorption contains the following mechanisms: physical entrapment (physical adsorption), ion exchange, and complexation in functional groups,12,13 which may be independently or synergistically involved.14 Bioaccumulation, on the other hand, is an active-process, metabolism-dependent intracellular accumulation.15 It is a more complex process entailing many occurrences, including localization of the metal within specific organelles, metallothionein binding, and efflux pumping.14,16 Microorganisms show promise for the removal of heavy metals from polluted environments through both bioaccumulation and biosorption processes.
Bioaccumulation and biosorption have been extensively studied by some researchers.17–19 However, there have been arguments on whether bioaccumulation or biosorption plays a dominant role in Cd(II) adsorption.20–22 Little work has been done to investigate the predominant mechanisms (bioaccumulation and biosorption) involved in the reduction of Cd(II) toxicity of Gram-positive and Gram-negative bacteria at a time. Moreover, many researchers did not take metal concentrations into consideration during the adsorption process.23,24
The objectives of the present work were: (1) to investigate the capacities of Cd(II)-resistant PGPR, i.e., Cupriavidus necator GX_5 (CP002878), Sphingomonas sp. GX_15 (MF959440), and Curtobacterium sp. GX_31 (MF959445), for Cd(II) adsorption under the same experimental conditions; (2) to analyze surface interaction between static biomass and Cd(II) by means of SEM-EDX, TEM, and FTIR analysis; (3) to elucidate the main adsorption mechanism (bioaccumulation or biosorption) of bacteria for Cd(II) under different initial Cd(II) concentrations using batch adsorption experiments; and (4) to provide new insight into Cd-resistant PGPR's potential use for bioremediation of contaminated environments.
2. Materials and methods
2.1. Bacterial strains and culture medium
The PGPR used in this study were the Gram-negative strains Cupriavidus necator GX_5 (CP002878) and Sphingomonas sp. GX_15 (MF959440) and the Gram-positive strain Curtobacterium sp. GX_31 (MF959445), isolated in a 60 year-old lead-zinc core from rhizosphere soil of local (Guangxi, China) dominant plants, with an average Cd(II) concentration of 59.43 mg kg−1. Bacteria were cultured in Luria–Bertani (LB) broth medium and maintained at 28 °C.2.2. Preparation of the biosorbent
Strains were incubated at pH 6.0 in LB medium on a rotary shaker at 28 °C and 180 rpm. The cultural cells were harvested (exponential phase) and washed three times with sterile water, then separated by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min, collected, and lyophilized overnight in a Labconco freeze dryer.25 The dried cells were ground to fine powder and used as biosorbent.
000 × g for 10 min, collected, and lyophilized overnight in a Labconco freeze dryer.25 The dried cells were ground to fine powder and used as biosorbent.
2.3. Cd(II) solution
All chemicals used in this study were of analytical grade and solutions were prepared using ddH2O. Stock solutions of Cd(II) (1000 mg l−1) were prepared by dissolving CdCl2·2.5H2O in ddH2O. The initial pH of the working solutions was adjusted to 6.0 for Cd(II) binding experiments by the addition of 0.1 mol l−1 HCl and NaOH solution.2.4. Adsorption of Cd(II) by biosorbent
Adsorption experiments were conducted at 28 °C in batches with 0.02 g of resting cells in 50 ml plastic tubes containing 20 ml of 20, 50, and 100 mg l−1 working solution. The mixture was shaken for 2 h and then centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min. Then the concentration of Cd(II) in the supernatant was analysed via inductively coupled plasma. The difference between the initial Cd(II) concentration and the remaining ion concentration was assumed to have been adsorbed by cells. The removal efficiency value adsorption (%) for Cd(II) of the biomass was calculated using the following eqn (1):
000 × g for 10 min. Then the concentration of Cd(II) in the supernatant was analysed via inductively coupled plasma. The difference between the initial Cd(II) concentration and the remaining ion concentration was assumed to have been adsorbed by cells. The removal efficiency value adsorption (%) for Cd(II) of the biomass was calculated using the following eqn (1):
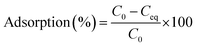 | (1) |
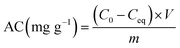 | (2) |
2.5. Desorption of Cd(II) by biosorbent
Desorption of Cd(II) from previously loaded resting cells was studied by using ddH2O, 1.0 mol l−1 of NH4NO3, and 0.1 mol l−1 of EDTA-Na2 as eluent.12,23 Briefly, biosorption can be described under three categories: (1) physical entrapment, which binds metals weakly to the cell surface and is easily susceptible to desorption by water; (2) ion exchange with K+, Ca2+, Na+, and Mg2+ on the cell wall, which can be desorbed using NH4NO3; and (3) complexes with functional groups, which can be desorbed via EDTA. Remaining metals are considered to be bioaccumulated and cannot be desorbed via EDTA.Desorption experiments were conducted in batches with 0.02 g Cd(II)-loaded cell biosorbent in 50 ml plastic tubes containing 20 ml of each eluent mentioned above. After 2 h of reaction, shaking at 28 °C, supernatants were analysed after centrifugation for Cd(II) concentration. Adsorption capacities for each of the above-mentioned categories was calculated from the amount of Cd(II) adsorbed in the strains and from the final Cd(II) concentration in the desorption solution. Meanwhile, bioaccumulation and biosorption capacity were also evaluated. The same procedure was repeated three times.
2.6. SEM-EDX observation
The surface characteristics of strains before and after interaction with Cd(II) (20, 50, and 100 mg l−1) were studied using a SEM (Sirion 200, USA). For SEM analysis, the prepared cell biosorbent was fixed with 2.5% glutaraldehyde at 4 °C overnight. The fixed sample was then smeared on the coverslip, air dried, dehydrated using a gradient series with ethanol, and sputter-coated with gold.26 Energy dispersive X-ray (EDX) of the samples was simultaneously analyzed.2.7. TEM observation
For transmission electron microscopy (TEM), strains were prepared without and with Cd(II) inoculations (20, 50, and 100 mg l−1). The prepared samples were fixed with 2.5% glutaraldehyde at 4 °C for 6 h, washed with 0.1 M PB solution, fixed in 1% osmium tetroxide for 2 h, dehydrated, and embedded in resin as described by Tyagi and Malik.27 The pre-treated samples were then cut into thin slices, stained, and observed using a TEM (Tecnai G2 spirit Biotwin, USA) at magnifications of 30k, 68k and 98k.2.8. Fourier transformed infrared spectroscopy observation
The functional groups of Cd(II)-loaded and unloaded cells were analysed using a Fourier-transform infrared (FTIR) spectrometer (Nicolet 6700, USA) at room temperature. All infrared spectra were recorded over the range of 4000–400 cm−1 with a resolution of 4 cm−1. Sample disks were made from 2 mg of cells encapsulated in 200 mg of KBr.2.9. Statistical analysis
Data were subjected to analysis of variance and represented as the mean ± standard deviation. Pairwise differences among treatments were tested using a mean separation test (least significant difference) at p = 0.05. All statistical analyses were performed using SAS8.1 software.3. Results and discussion
3.1. Cd(II) adsorption by biosorbent
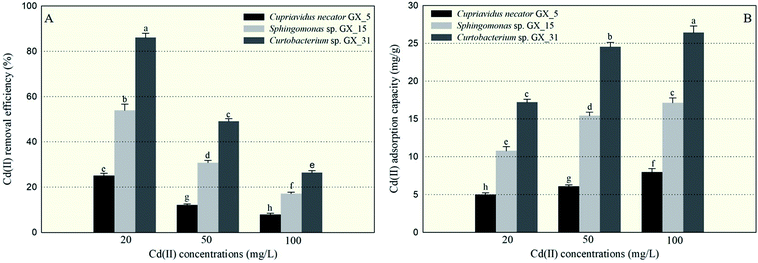
Under optimal conditions (pH: 6.0; reaction time: 6 h; biomass dosage: 1.0 g l−1) based on the preliminary data, the batch adsorption experiments were conducted with initial Cd(II) concentrations: 20, 50, and 100 mg l−1. The largest Cd(II) removal efficiency was 86.06%, for Curtobacterium sp. GX_31 under the initial concentration of 20 mg l−1; while the minimal removal efficiency was 7.98%, for Cupriavidus necator GX_5 with 100 mg l−1 Cd(II) treatment. It was obviously observed that a significant difference in Cd(II) adsorption existed among the three strains—more specifically, GX_31 > GX_15 > GX_5 (p < 0.05)—given the same amount of Cd(II) (Fig. 1A). In the study, biomass dosage (1.0 g l−1) was measured by weighing the freeze-dried fine powder of biosorbent, which was time-consuming but more accurate than methods in some previous studies.28–30 Meanwhile, the batch adsorption experiments were conducted using resting biomass rather than growing cells in medium. Considering the different growth rates of strains in medium, it is inappropriate to compare adsorption efficiency and capacity between strains. However, it makes sense that removal efficiency varied among strains based on their own adsorption or defence mechanisms.10In Fig. 1A, we also see that removal efficiency was significantly higher at lower concentrations than at higher concentrations for the same strains, which is in accordance with other studies.15,29,31 At low concentrations, the ratio of the moles of Cd(II) to the available surface area was low, leaving a large number of binding sites free for Cd(II) interactions and resulting in high adsorption efficiency.32 On the contrary, at high concentrations of Cd(II), a lack of sufficient free binding sites resulted in low removal efficiency.33
Similarly, the three strains showed a range of adsorption capacities when treated under the same Cd(II) concentrations: GX_31 > GX_15 > GX_5 (p < 0.05) (Fig. 1B). In considering identical strains, adsorption capacity was stronger at higher concentrations than that at lower concentrations (Fig. 1B). The strongest and weakest adsorption capacities were 26.43 mg g−1 for GX_31 at 100 mg l−1 of Cd(II) and 5.01 mg g−1 for GX_5 at 20 mg l−1, respectively. High initial concentrations could provide an effective force for driving metal ions to interact with finite metal binding sites, prompting adsorption by biomass strains.34 However, the adsorption capacity of the biosorbent would reach a saturation value with the increase of initial metal concentrations due to limited binding sites.26
3.2. Desorption of Cd(II) from loaded cell biosorbent
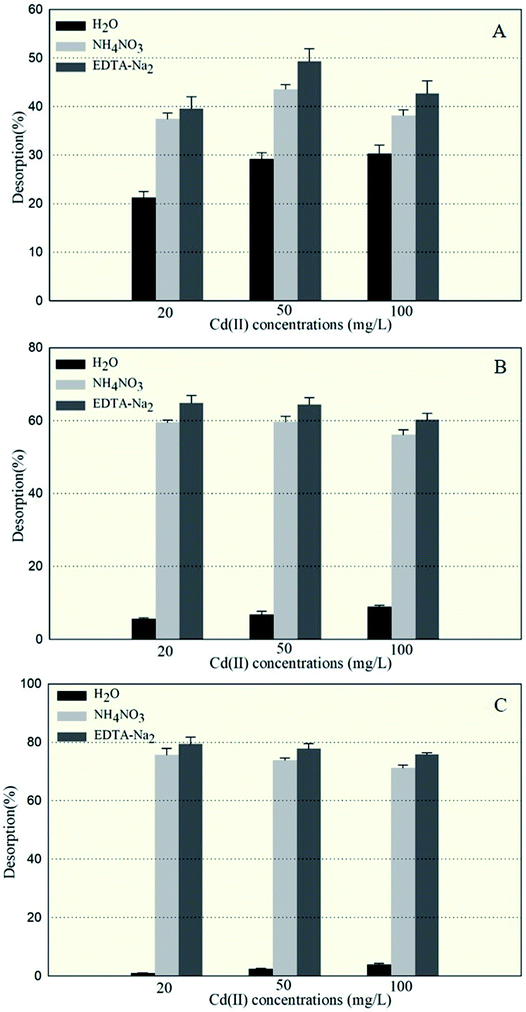
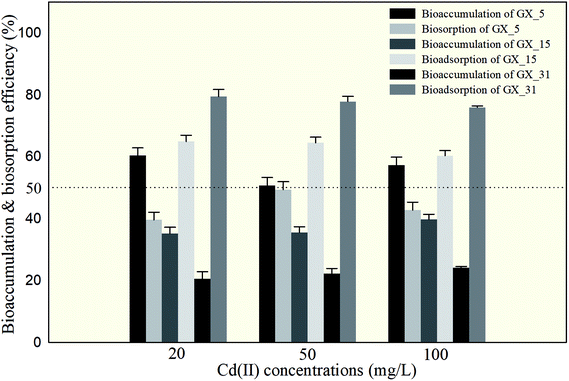
The amounts of Cd(II) desorbed from GX_5, GX_15, and GX_31 by water, NH4NO3, and EDTA-Na2 were investigated and displayed in Fig. 2, which shows that 5.67–8.95% and 0.97–3.93% of Cd(II) adsorbed by GX_15 and GX_31, respectively, was desorbed by water; 56.09–59.66% and 71.20–75.70%, respectively, was desorbed by NH4NO3; and 60.29–64.89% and 75.93–79.45% by EDTA-Na2 (Fig. 2B and C). In comparison, 21.21–30.34% of Cd(II) adsorbed by GX_5 was desorbed by water, while 37.48–43.58% and 39.62–49.34% was desorbed by NH4NO3 and EDTA-Na2, respectively (Fig. 2A).These results indicated that of the Cd(II) adsorbed by GX_5, 21.21–30.34% was physically entrapped, 7.82–16.27% was held by ion exchange, 2.14–5.76% was complexed in functional groups (Fig. 2A), and 50.66–60.38% was accumulated inside the cells (Fig. 3). On the contrary, for GX_15 and GX_31, 5.67–8.95% and 0.97–3.93% was physically entrapped. This suggested that the bound Cd(II) was not easily released and the contribution of physical adsorption was minor. According to Fang et al., only 3.6% of Cd(II) was adsorbed physically by Spirulina sp.23 A similar study was conducted by Chojnacka et al., who pointed out that the maximum contribution of physical adsorption by the blue-green algae Spirulina sp. was 3.7%.35 Desorption rates for Cd(II) held by ion exchange for strains GX_15 and GX_31 were 47.14–53.81% and 67.28–74.73%, respectively; rates for Cd(II) complexed in functional groups were 4.20–5.41% and 3.75–4.73%, respectively (Fig. 2B and C); 35.11–39.71% of Cd(II) was bioaccumulated in GX_5 and 20.55–24.07% in GX_15 (Fig. 3).
As these figures show, regardless of initial Cd(II) concentrations, the dominant mechanism for Cd(II) adsorption was bioaccumulation (intercellular accumulation) (50.66–60.38%) for GX_5, while the dominant adsorption mechanisms were both biosorptions (extracellular adsorptions) (60.29–64.89% for GX_15 and 75.93–79.45% for GX_31) (Fig. 3). It is obvious that the biosorption mechanism of GX_5 is more prone to physical entrapment (21.20–30.33%), while those of GX_15 and GX_31 tend toward ion exchange (47.14–53.81% and 67.28–74.73%, respectively). Adsorption mechanisms differed due to varying compositions and structures in bacterial cell walls.25 Another reason for adsorption mechanisms differing might be that exclusion mechanisms lead to Cd(II) being excreted from inside the cell and improving surface binding via metal-exporting proteins.36 Some researchers have concluded that the increase in surface adsorption might be a result of extracellular polymeric substances protecting cells from Cd(II) toxicity.21,37
The result was different from that obtained by Huang et al., who illustrated that intracellular accumulation is the main adsorption mechanism given lower metal concentrations and extracellular adsorption is the main adsorption mechanism at higher concentrations.11 Due to variance in experimental conditions and analytical methods, it is inappropriate to compare adsorption mechanisms between researchers.38–41 However, although study results may not be directly comparable, we can be sure that strains GX_5, GX_15, and GX_31 show different Cd(II) adsorption mechanisms and capacities. Meanwhile, bioaccumulation and biosorption were verified by SEM-EDX, TEM, and FTIR spectroscopy, which will be discussed in the following sections.
3.3. SEM-EDX and TEM analysis
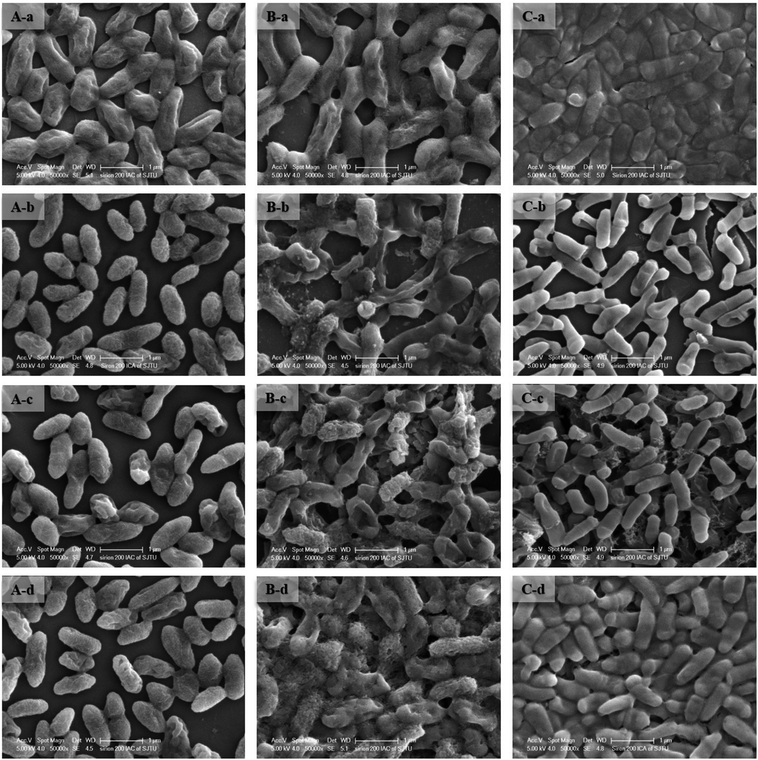
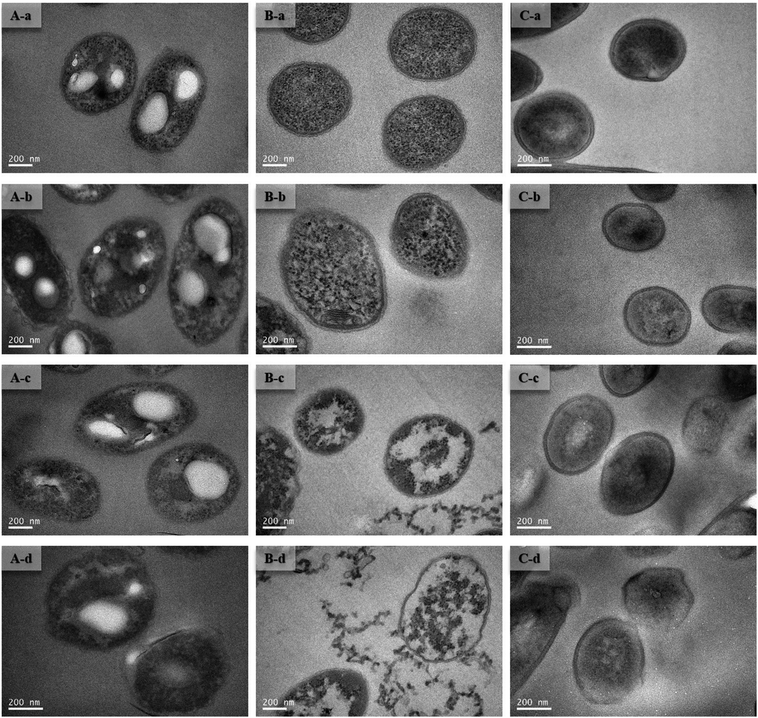
To improve understanding of the mechanisms of Cd(II) interactions with microbes, SEM-EDX and TEM were performed. Cell surfaces of GX_5, GX_15, and GX_31 were all observed to be rod shapes with clear boundary before adsorption (Fig. 4A-a, B-a and C-a). There were no obvious changes in morphology of these strains after interaction with Cd(II) at a concentration of 20 mg l−1 (Fig. 4A-b, B-b and C-b). However, their surfaces became rough and were covered by sediments after the reaction; this effect was yet more evident when bacterial cells were exposed to 100 mg l−1 Cd(II) (Fig. 4A-d, B-d and C-d). Changes in cell morphology could be explained as a protective mechanism responding to a stressful environment, which has previously been reported.42,43 Moreover, for strain GX_15, cells appeared to aggregate after reaction with Cd(II), an effect which Fig. 4B-c and B-d show to be especially pronounced. Some floccus precipitation was found on the surface of GX_15 (Fig. 4B-b, B-c and B-d). Aggregation and precipitation might be caused by extracellular polymeric substances, which had an important role in binding heavy metals.30 EDX is a useful tool for chemical and elemental analysis of biosorbents and has been extensively applied.44 EDX spectra recorded the signals of carbon, nitrogen, and oxygen, which were likely in polysaccharides and proteins of the biosorbents (Fig. S1†). Unloaded biomass showed no Cd(II) signals in the EDX spectra, but signals could be observed after Cd(II) exposure, revealing the presence of Cd(II) in the cell after adsorption (Fig. S1†). However, EDX spectra could only determine the presence or absence of Cd(II) on the biomass qualitatively, not quantitatively.Although most heavy metals are not essential to bacteria, some of them can cross the cell membrane and enter the cells via a range of processes.45 Therefore, TEM analysis of strains was conducted to intuitively show the effects of metal concentrations on cells. As shown in Fig. 5A-a, B-a and C-a, the cells were intact, the contents were identically dispersed in the cells, and the cell wall could be clearly distinguished from cytoplasm. With increased initial Cd(II) concentrations, the cell walls became unclear and vague and it was hard to tell the cell wall from cytoplasm (Fig. 5A-b, A-c, B-b, B-c, C-b and C-c). This phenomenon was especially evident given a Cd(II) concentration of 100 mg l−1 (Fig. 5A-d, B-d and C-d). Fig. 5B-c and B-d show that, under concentrations of 50 and 100 mg l−1 of Cd(II), some contents flowed out of the cells. This indicates that the cell walls of GX_15 were destroyed by high Cd(II) concentrations, and thus that GX_15 is more sensitive to Cd(II) than GX_5 and GX_31.
3.4. FTIR spectra study
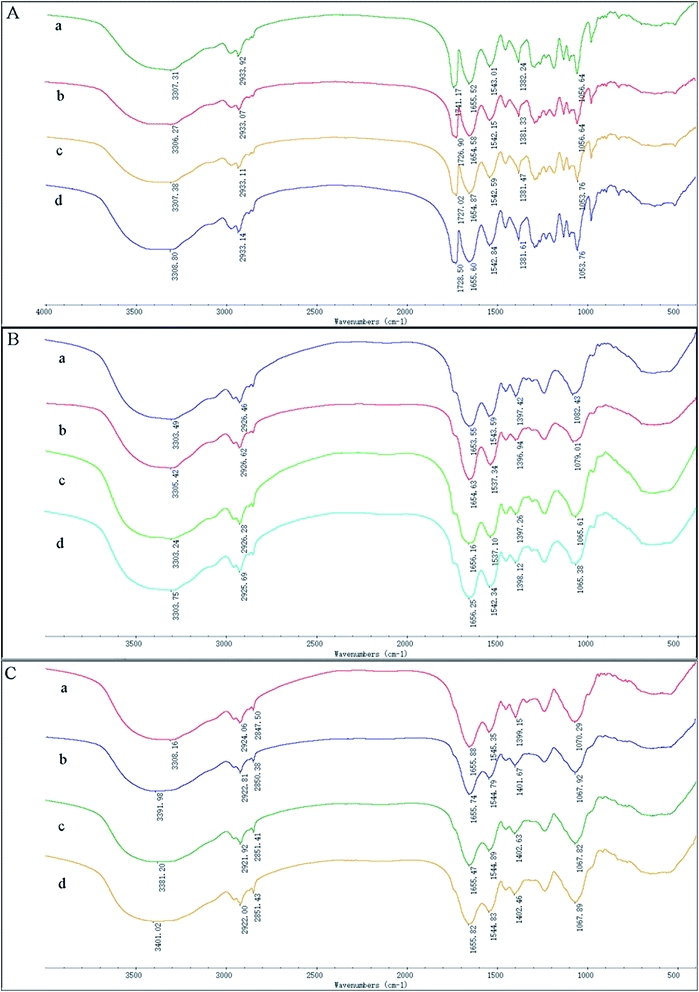
To investigate possible interactions between Cd(II) and functional groups on the cell walls, the FTIR spectra of GX_5, GX_15, and GX_31 were recorded before and after Cd(II) adsorption. The pre-adsorption FTIR spectra revealed the presence of many functional groups on the cell surface, indicating the complex nature of the strains (Fig. 6A-a, B-a and C-a). Meanwhile, the IR spectra of GX_15 and GX_31 were similar to each other, but different from that of GX_5 (Fig. 6A–C).Broad spectra bands were observed in the range of 3300–3500 cm−1, representing the stretching bond of the –NH from an amino group and a bonded hydroxyl group.46 After contact with Cd(II), the spectra had the tendency to shift to lower frequencies (Fig. 6A–C), an effect that was more evident for GX_31 (Fig. 6C).
The band around 2930 cm−1 corresponded to symmetrical –CH– vibration of –CH2 and –CH3 in lipids,47 which showed subtle changes after adsorption (Fig. 6). For strains GX_5 and GX_15, there were no changes at the band of 2850 cm−1, which corresponded to asymmetrical –CH– vibration in lipids.34 However, the adsorption peaks at 2850 cm−1 for GX_31 shifted from 2847.50 cm−1 to 2850.38 cm−1 (20 mg l−1 Cd(II)-loaded), 2851.41 cm−1 (50 mg l−1 Cd(II)-loaded), and 2851.43 cm−1 (100 mg l−1 Cd(II)-loaded) (Fig. 6C).
Carbonyl groups stretching vibration was prominent at 1741.17 cm−1 for strain GX_5 (Fig. 6A).48 After interaction with 20, 50, and 100 mg l−1 concentrations of Cd(II), the spectra of Cd(II)-loaded biomass demonstrated a clear shift of this peak to 1726.90 cm−1, 1727.02 cm−1, and 1728.50 cm−1, respectively. Strains GX_15 and GX_31 did not show any peak at this band (Fig. 6B and C).
The peaks between 1650 cm−1 and 1540 cm−1 could be assigned to amide groups in proteins.49 The typical amide I (–CO–) appeared at 1655.52 cm−1, 1653.55 cm−1, and 1655.88 cm−1, respectively, for GX_5, GX_15 and GX_31; while the peaks at 1543.01 cm−1, 1543.59 cm−1, and 1545.35 cm−1 were considered to be amide II (–NH–). The spectra showed a minor shift of these two bands to 1654.58 cm−1 and 1655.47 cm−1 for GX_5 and 1542.15 cm−1 and 1544.79 cm−1 for GX_31 (Fig. 6A and C); for GX_15, the two bands shifted to 1656.25 cm−1 and 1537.34 cm−1 (Fig. 6B).
A minor peak shift at 1397.42 cm−1 to 1398.12 cm−1 for GX_15 (Fig. 6B), 1399.15 cm−1 to 1402.46 cm−1 for GX_31 (Fig. 6C), and 1382.24 cm−1 to 1381.33 cm−1 for GX_5 (Fig. 6A) indicated the role of carboxyl groups in Cd(II) binding.31,50 For GX_15, there existed a significant shift from 1082.43 cm−1 to 1065.38 cm−1, corresponding to the –CO– group vibration in the cyclic structure of carbohydrates.26 Meanwhile, a band at 1070.29 cm−1 shifted to 1067.82 cm−1 for GX_31, representing the –CO– groups as well.20 In the control spectra, the adsorption peak at 1056.64 cm−1 due to the phosphate groups was observed,38 and a shift of this peak to 1053.76 cm−1 (50 and 100 mg l−1 Cd(II)-loaded) (Fig. 6A) suggested the interaction of bound metals with phosphates.
After Cd(II) adsorption occurred, the overall IR spectra analysis indicated the involvement of functional groups such as hydroxyl, carbonyl, and carboxyl groups of saccharides; amino and amide groups of proteins; phosphate groups; and –COC– groups of carbohydrates in the interaction of Cd(II) with bacteria. Moreover, with increased initial metal concentrations, the differences between IR spectra for Cd(II)-free and for Cd(II)-loaded cells was more distinct.
4. Conclusions
Three cadmium-resistant PGPR, Cupriavidus necator GX_5, Sphingomonas sp. GX_15, and Curtobacterium sp. GX_31, were used to study bioaccumulation and biosorption mechanisms under different initial Cd(II) concentrations. Removal efficiency and adsorption capacity of the assessed PGPR can be ordered as GX_31 > GX_15 > GX_5. Strain GX_15 showed high potential (86.06%) for Cd(II) remediation. Physical entrapment, ion exchange, and complexation were involved in biosorption processes. The dominant adsorption mechanism for GX_5 was bioaccumulation, while the dominant mechanisms for GX_15 and GX_31 were both biosorptions. The elucidation of the binding mechanisms could provide new perspectives of strains in practical bioremediation applications for heavy metals. However, more strains from different genera or even phyla are needed to be assessed for biosorption and bioaccumulation mechanisms under different metal concentrations and using various analysis methods.Conflicts of interest
There is no conflict of interest and all the authors are interested to publish the manuscript.Acknowledgements
This work was supported by Agriculture Committee of Shanghai (2014/5–2). We are thankful to MS Yao Han for SEM and EDX analysis, MS Yanhua Zhu and Ge Wang for TEM analysis, and Prof. Bangshang Zhu for FTIR spectra analysis (Instrumental Analysis Center of Shanghai Jiao Tong University, Shanghai, China).References
- Y. Wang, J. Shi, H. Wang, Q. Lin, X. Chen and Y. Chen, Ecotoxicol. Environ. Saf., 2007, 67, 75–81 CrossRef PubMed.
- C. Xu, S. He, Y. Liu, W. Zhang and D. Lu, Chemosphere, 2017, 173, 622–629 CrossRef PubMed.
- C. N. Mulligan, R. N. Yong and B. F. Gibbs, J. Hazard. Mater., 2001, 85, 145–163 CrossRef PubMed.
- J. Wang and C. Chen, Biotechnol. Adv., 2009, 27, 195–226 CrossRef PubMed.
- D. Prithviraj, K. Deboleena, N. Neelu, N. Noor, R. Aminur, K. Balasaheb and M. Abul, Ecotoxicol. Environ. Saf., 2014, 107, 260–268 CrossRef PubMed.
- X. Xu, Z. Zhang, Q. Huang and W. Chen, Geomicrobiol. J., 2018, 35, 40–49 CrossRef.
- B. Ye, Y. Luo, J. He, L. Sun, B. Long, Q. Liu, X. Yuan, P. Dai and J. Shi, Bioresour. Technol., 2018, 264, 206–210 CrossRef PubMed.
- Y. Zhang, W. Liu, M. Xu, F. Zheng and M. Zhao, J. Hazard. Mater., 2010, 178, 1085–1093 CrossRef PubMed.
- A. S. Ayangbenro and O. O. Babalola, Int. J. Environ. Res. Public Health, 2017, 14, 94 CrossRef PubMed.
- T. Limcharoensuk, N. Sooksawat, A. Sumarnrote, T. Awutpet, M. Kruatrachue, P. Pokethitiyook and C. Auesukaree, Ecotoxicol. Environ. Saf., 2015, 122, 322–330 CrossRef PubMed.
- F. Huang, C. Guo, G. Lu, X. Yi, L. Zhu and Z. Dang, Chemosphere, 2014, 109, 134–142 CrossRef PubMed.
- J. Bai, X. Yang, R. Du, Y. Chen, S. Wang and R. Qiu, J. Environ. Sci., 2014, 26, 2056–2064 CrossRef PubMed.
- Y. Li, Q. Yue and B. Gao, J. Hazard. Mater., 2010, 178, 455–461 CrossRef PubMed.
- T. Srinath, T. Verma, P. Ramteke and S. Garg, Chemosphere, 2002, 48, 427–435 CrossRef PubMed.
- M. Pérez-Rama, E. Torres, C. Suárez, C. Herrero and J. Abalde, J. Environ. Manage., 2010, 91, 2045–2050 CrossRef PubMed.
- G. M. Gadd, Experientia, 1990, 46, 834–840 CrossRef.
- R. Chakravarty and P. C. Banerjee, Bioresour. Technol., 2012, 108, 176–183 CrossRef PubMed.
- J. E. McLean, M. W. Pabst, C. D. Miller, C. O. Dimkpa and A. J. Anderson, Chemosphere, 2013, 91, 374–382 CrossRef PubMed.
- V. I. Slaveykova, N. Parthasarathy, K. Dedieu and D. Toescher, Environ. Pollut., 2010, 158, 2561–2565 CrossRef PubMed.
- Y. Lin, X. Wang, B. Wang, O. Mohamad and G. Wei, Ecotoxicol. Environ. Saf., 2012, 77, 7–17 CrossRef PubMed.
- M. W. Pabst, C. D. Miller, C. O. Dimkpa, A. J. Anderson and J. E. McLean, Chemosphere, 2010, 81, 904–910 CrossRef PubMed.
- M. Vargas-García, M. López, F. Suárez-Estrella and J. Moreno, Sci. Total Environ., 2012, 431, 62–67 CrossRef PubMed.
- L. Fang, C. Zhou, P. Cai, W. Chen, X. Rong, K. Dai, W. Liang, J.-D. Gu and Q. Huang, J. Hazard. Mater., 2011, 190, 810–815 CrossRef PubMed.
- V. Rajesh, A. S. K. Kumar and N. Rajesh, Chem. Eng. J., 2014, 235, 176–185 CrossRef.
- L. Fang, P. Cai, W. Chen, W. Liang, Z. Hong and Q. Huang, Colloids Surf., A, 2009, 347, 50–55 CrossRef.
- E. Khadivinia, H. Sharafi, F. Hadi, H. S. Zahiri, S. Modiri, A. Tohidi, A. Mousavi, A. H. Salmanian and K. A. Noghabi, J. Ind. Eng. Chem., 2014, 20, 4304–4310 CrossRef.
- A. K. Tyagi and A. Malik, Micron, 2010, 41, 797–805 CrossRef PubMed.
- Y. Jin, X. Wang, T. Zang, Y. Hu, X. Hu, G. Ren, X. Xu and J. Qu, J. Microbiol. Biotechnol., 2016, 26, 1428–1438 CrossRef PubMed.
- C. Sukumar, V. Janaki, S. Kamala-Kannan and K. Shanthi, Clean Technol. Environ. Policy, 2014, 16, 405–413 CrossRef.
- Y. Yang, M. Hu, D. Zhou, W. Fan, X. Wang and M. Huo, RSC Adv., 2017, 7, 18793–18802 RSC.
- P. Arivalagan, D. Singaraj, V. Haridass and T. Kaliannan, Ecol. Eng., 2014, 71, 728–735 CrossRef.
- M. Oves, M. S. Khan and A. Zaidi, Saudi J. Biol. Sci., 2013, 20, 121–129 CrossRef PubMed.
- P. Chakravarty, N. S. Sarma and H. Sarma, Chem. Eng. J., 2010, 162, 949–955 CrossRef.
- N. Masoudzadeh, F. Zakeri, T. bagheri Lotfabad, H. Sharafi, F. Masoomi, H. S. Zahiri, G. Ahmadian and K. A. Noghabi, J. Hazard. Mater., 2011, 197, 190–198 CrossRef PubMed.
- K. Chojnacka, A. Chojnacki and H. Gorecka, Chemosphere, 2005, 59, 75–84 CrossRef PubMed.
- D. H. Nies, FEMS Microbiol. Rev., 2003, 27, 313–339 CrossRef PubMed.
- M. Ueshima, B. R. Ginn, E. A. Haack, J. E. Szymanowski and J. B. Fein, Geochim. Cosmochim. Acta, 2008, 72, 5885–5895 CrossRef.
- X. Li, C. Ding, J. Liao, T. Lan, F. Li, D. Zhang, J. Yang, Y. Yang, S. Luo and J. Tang, J. Environ. Radioact., 2014, 135, 6–12 CrossRef PubMed.
- G. Ren, Y. Jin, C. Zhang, H. Gu and J. Qu, Ecotoxicol. Environ. Saf., 2015, 117, 141–148 CrossRef PubMed.
- D. R. Shaw and J. Dussan, Water, Air, Soil Pollut., 2015, 226, 112 CrossRef.
- H. Wang, A. McCarthney, X. Qiu and R. Zhao, Geomicrobiol. J., 2012, 29, 199–205 CrossRef.
- R. Chakravarty and P. C. Banerjee, Extremophiles, 2008, 12, 279–284 CrossRef PubMed.
- C. Nithya, B. Gnanalakshmi and S. K. Pandian, Mar. Environ. Res., 2011, 71, 283–294 CrossRef PubMed.
- Q. Zhai, F. Tian, G. Wang, J. Zhao, X. Liu, K. Cross, H. Zhang, A. Narbad and W. Chen, RSC Adv., 2016, 6, 5990–5998 RSC.
- T. Lan, Y. Feng, J. Liao, X. Li, C. Ding, D. Zhang, J. Yang, J. Zeng, Y. Yang and J. Tang, J. Environ. Radioact., 2014, 134, 6–13 CrossRef PubMed.
- L. Fan, Z. Ma, J. Liang, H. Li, E. Wang and G. Wei, Bioresour. Technol., 2011, 102, 703–709 CrossRef PubMed.
- D. Das, B. B. Salgaonkar, K. Mani and J. M. Braganca, Chemosphere, 2014, 112, 385–392 CrossRef PubMed.
- T. Wang and H. Sun, Environ. Sci. Pollut. Res., 2013, 20, 7450–7463 CrossRef PubMed.
- F. Masoumi, E. Khadivinia, L. Alidoust, Z. Mansourinejad, S. Shahryari, M. Safaei, A. Mousavi, A.-H. Salmanian, H. S. Zahiri and H. Vali, J. Environ. Chem. Eng., 2016, 4, 950–957 CrossRef.
- S. S. Zamil, M. H. Choi, J. H. Song, H. Park, J. Xu, K.-W. Chi and S. C. Yoon, Appl. Microbiol. Biotechnol., 2008, 80, 531–544 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra06270f |
| This journal is © The Royal Society of Chemistry 2018 |