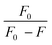Open Access Article
Open Access ArticleMolecular design and synthesis of new dithiocarbazate complexes; crystal structure, bioactivities and nano studies†
Zahra Yekke-ghasemia,
Reza Takjoo *a,
Mohammad Ramezanib and
Joel T. Maguec
*a,
Mohammad Ramezanib and
Joel T. Maguec
aDepartment of Chemistry, Faculty of Science, Ferdowsi University of Mashhad, Mashhad, Iran. E-mail: r.takjoo@um.ac.ir; rezatakjoo@yahoo.com; Tel: +98 513 880 5536
bPharmaceutical Research Center, School of Pharmacy, Mashhad University of Medical Sciences, Mashhad, Iran
cDepartment of Chemistry, Tulane University, New Orleans, LA 70118, USA
First published on 14th December 2018
Abstract
The syntheses of a new set of metal complexes MoO2L′(CH3OH), VOL′(CH3O)(CH3OH), 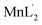 ,
, 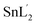 , SnL′Cl2 and SnL′I2 with a new ligand (L = (2,2′(disulfanediylbis((ethylthio)methylene)bis(hydrazin-2-yl-1-ylidene)bis(methanylylidene)) diphenol; L′ = S-ethyl-3-(2-hydroxyphenyl)methylenedithiocarbazate are described along with characterization by elemental analysis, mass spectrometry, spectroscopic (IR, 1H- and 13C-NMR) and TGA techniques. The crystal structures of compounds were determined by single crystal X-ray diffraction analysis and compared to powder X-ray diffraction (PXRD) patterns of the nano complexes obtained using ultrasonic methods. The PXRD results indicate that the compounds synthesized by ultrasonic methods have high crystallinity. The compounds were evaluated in an in vitro cytotoxicity study with two human cancer cell lines. The results of this study revealed that all complexes exhibit good cytotoxic activity when compared to the clinical drug, cisplatin. Interaction of the samples with human serum albumin (HSA) was investigated using fluorescence spectrophotometric methods and the Stern–Volmer quenching constant (KSV) and free energy changes (ΔG) were calculated at 298 K. The fluorescence quenching method is used to determine the number of binding sites (n) and association constants (Ka) at the same temperatures.
, SnL′Cl2 and SnL′I2 with a new ligand (L = (2,2′(disulfanediylbis((ethylthio)methylene)bis(hydrazin-2-yl-1-ylidene)bis(methanylylidene)) diphenol; L′ = S-ethyl-3-(2-hydroxyphenyl)methylenedithiocarbazate are described along with characterization by elemental analysis, mass spectrometry, spectroscopic (IR, 1H- and 13C-NMR) and TGA techniques. The crystal structures of compounds were determined by single crystal X-ray diffraction analysis and compared to powder X-ray diffraction (PXRD) patterns of the nano complexes obtained using ultrasonic methods. The PXRD results indicate that the compounds synthesized by ultrasonic methods have high crystallinity. The compounds were evaluated in an in vitro cytotoxicity study with two human cancer cell lines. The results of this study revealed that all complexes exhibit good cytotoxic activity when compared to the clinical drug, cisplatin. Interaction of the samples with human serum albumin (HSA) was investigated using fluorescence spectrophotometric methods and the Stern–Volmer quenching constant (KSV) and free energy changes (ΔG) were calculated at 298 K. The fluorescence quenching method is used to determine the number of binding sites (n) and association constants (Ka) at the same temperatures.
1. Introduction
S-Alkyl/aryl dithiocarbazates as sulfur–nitrogen chelating agents are interesting in the context of coordination chemistry. Owing to hard nitrogen and soft sulfur donor atoms, formation of five-membered chelate rings can stabilize transition and non-transition metals in various oxidation states.1,2 Through reacting various aldehydes and ketones with dithiocarbazates multi-dentate ligands will form for which, depending on the different structures obtained upon coordination with metal ions, different biological applications including antibacterial, antifungal and anticancer activity could be obtained.3 A variety of coordination complexes and cytotoxic compounds have been investigated for the treatment of human cancer in recent years4,5 with cisplatin as the most widely used drug but its use is limited in medicine due to the side effects such as nephrotoxicity and drug resistance.6,7 Other studies have suggested that dithiocarbazates, due to the sulfur–nitrogen chelating functionality, can reduce these side effects.8 Thus complexation of tridentate ONS-chelating derivatives of dithiocarbazates with metals such as Mn,9 Sn,10 V11 and Mo12 should be interesting in chemotherapeutics especially with regard to antitumor effects.13 These metals are also known to accelerate drug action and play major roles in biological systems. For example, vanadium is present in the two enzymes of haloperoxidase and nitrogenase and also plays a role in the regulation of phosphate metabolism.14 Manganese exists at the catalytic center of the three enzymes manganese catalase, ribonucleotide reductase of certain bacteria and the photosystem II of green plants to facilitate the metabolism of the O2n− unit15 and molybdenum is at the active sites of molybdoenzymes.16Investigation of protein–ligand interactions helps us to understand the antitumor effects of metal complexes.17,18 Albumins as a major transporter of the proteins in blood plasma are good candidates for studying these interactions.19 HSA is responsible for carrying substances in the blood and is also able to carry pharmaceutical compounds with different chemical structures.20 Six linkages are identified for ligand binding on HSA.21,22 From these six sites, two major sites are responsible for binding with metals23 with the linkage being either covalent or non-covalent.24 In general, evaluation of drug–protein interaction in order to achieve optimal therapeutic dose and the awareness of drug–protein binding capacity is essential.25
Therefore, in the present article, the new complexes of dithiocarbazates are designed, synthesized and characterized in order to examine their antitumor properties. The cytotoxic test of the compounds using MTT assay was performed against the Hela and MCF-7 cells. Also, the effects of dithiocarbazates in the presence of different metal ions on HSA are studied from a molecular point of view.
2. Experimental
2.1. Materials and methods
Human serum albumin (HSA 99%, fatty acid free) was purchased from Sigma Chemical Company and RPMI-1640 medium and fetal bovine serum (FBS) from GIBCO (Gaithersburg, USA). Penicillin and streptomycin were purchased from Biochrom AG (Berlin, Germany).All other material and solvents were purchased and used without purification. IR spectra in the region 600–4000 cm−1 were recorded with a Buck 500 Scientific. Elemental analyses (CHNS) were performed with a Thermo Finnigan Flash 1112EA elemental analyzer. The EI-mass spectra were carried out using a Varian CH-7 instrument at 70 eV. Melting points were determined with an electrothermal digital melting point apparatus. 1H and 13C NMR spectra of compounds were recorded on a Bruker FUM-300 spectrometer using DMSO-d6 as solvent. TG analysis was performed with a TGA-50 SHIMADZU instrument at a heating rate of 10 °C min−1 under air atmosphere from ambient temperature to 950 °C. X-ray powder diffraction (XRD) measurements were recorded on a Philips diffractometer manufactured by X'pert with graphite monochromatized Cu-Kα radiation. Simulated XRD powder patterns were calculated by using MERCURY based on the single crystal data. The size and morphology of nanoparticles of CP have been studied by SEM, a Leo 1450 VP, Germany.
2.2. Synthesis of (2,2′-(disulfanediylbis((ethylthio)methylene))bis(hydrazin-2-yl-1-ylidene)bis(methanylylidene))diphenol (L)
Carbon disulfide (7.75 g, 102 mmol) was added dropwise to a solution of hydrazine hydrate (5.00 g, 100 mmol), potassium hydroxide (6.00 g, 107 mmol), water (6 mL) and 25 mL of ethanol with stirring and cooling in an ice-salt bath for one hour. Ethyl bromide (10.35 g, 94.98 mmol) was added to the above mixture over 30 minutes. The reaction mixture was heated for 30 minutes to 25 °C and then cooled down to 10 °C. Cold distilled water (60 mL) was added to the above solution and a two-phase mixture was obtained. Salicylaldehyde (5.20 g, 24.58 mmol) solution in 30 mL of 75% ethanol, at room temperature, was added to the separated organic phase. The resulting yellow precipitate was filtered off, washed with water and dried in a vacuum desiccator over silica gel.Colorless column-like crystals were obtained after recrystallization from ethanol, yield: 5.79 g, 14.5% (based on the mass of the oily organic phase), mp: 151 °C. Anal. calc. for C20H22N4O2S4 (478.67 g mol−1): C, 49.98; H, 5.03; N, 11.66; S, 26.68. Found: C, 50.07; H, 5.10; N, 11.36; S, 27.30%. IR (KBr), cm−1: ν(OH) 3108 s, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1612 s, ν(N-CSS) 1526 m, ν(C–O) 1262 m, ν(N–N) 1028 s. Mass spectrometry, m/z (%): 239 (100) {M/2 = L′}. 1H-NMR (300 MHz, DMSO-d6, 295 K) δ: 1.298 (t, 3H, J = 7.35 Hz, CH3), 3.202 (q, 2H, J = 7.35 Hz, CH2), 6.919 (m, 2H, H-3 and H-5), 7.316 (td, J = 7.80 Hz, 1H, H-4), 7.669 (dd, J = 7.77, 1.69 Hz, 1H, H-2), 10.259 (s, 1H, H-7). 13C-NMR (75.6 MHz, DMSO-d6, 296 K) δ ppm: 14.37 (C10); 27.88 (C9); 127.87 (C1); 120.08 (C2); 132.65 (C3); 116.86 (C4); 157.70 (C5); 119.52 (C6); 145.15 (C7); 196.59 (C8).
N) 1612 s, ν(N-CSS) 1526 m, ν(C–O) 1262 m, ν(N–N) 1028 s. Mass spectrometry, m/z (%): 239 (100) {M/2 = L′}. 1H-NMR (300 MHz, DMSO-d6, 295 K) δ: 1.298 (t, 3H, J = 7.35 Hz, CH3), 3.202 (q, 2H, J = 7.35 Hz, CH2), 6.919 (m, 2H, H-3 and H-5), 7.316 (td, J = 7.80 Hz, 1H, H-4), 7.669 (dd, J = 7.77, 1.69 Hz, 1H, H-2), 10.259 (s, 1H, H-7). 13C-NMR (75.6 MHz, DMSO-d6, 296 K) δ ppm: 14.37 (C10); 27.88 (C9); 127.87 (C1); 120.08 (C2); 132.65 (C3); 116.86 (C4); 157.70 (C5); 119.52 (C6); 145.15 (C7); 196.59 (C8).
2.3. Synthesis of the metal complexes
Orange block-like crystal, yield: 0.1 g, 68% (based on the metal salt). Mp: 230 °C. Anal. calc. for C11H14MoN2O4S2 (398.32 g mol−1): C, 33.17; H, 3.54; N, 7.03; S, 16.10. Found: C, 32.00; H, 3.42; N, 6.76; S, 15.93%. IR (KBr), cm−1: ν(OH) 3418 w, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1592 s, ν(C–O) 1548 m, ν(N–N) 1012 s. Mass spectrometry, m/z (%): 366 (98) {M-CH3OH}. 1H-NMR (300 MHz, DMSO-d6) δ: 1.356 (t, 3H, J = 7.31 Hz, CH3), 3.180 (m, 3H, OCH3), 4.131 (q, 2H, J = 5.24 Hz, CH2), 6.970 (d, 1H, J = 8.27 Hz, H-2), 7.086 (m, 1H, H-4), 7.575 (m, 1H, H-3), 7.789 (dd, 1H, J = 1.75 and 7.86 Hz, H-5), 8.957 (S, 1H, H-7). 13C-NMR (75.6 MHz, DMSO-d6, 296 K) δ ppm: 15.04 (C10); 28.27 (C9); 49.08 (C11); 136.21 (C1); 120.14 (C2); 135.68 (C3); 118.84 (C4); 121.77 (C5); 171.00 (C6); 160.15 (C7).
N) 1592 s, ν(C–O) 1548 m, ν(N–N) 1012 s. Mass spectrometry, m/z (%): 366 (98) {M-CH3OH}. 1H-NMR (300 MHz, DMSO-d6) δ: 1.356 (t, 3H, J = 7.31 Hz, CH3), 3.180 (m, 3H, OCH3), 4.131 (q, 2H, J = 5.24 Hz, CH2), 6.970 (d, 1H, J = 8.27 Hz, H-2), 7.086 (m, 1H, H-4), 7.575 (m, 1H, H-3), 7.789 (dd, 1H, J = 1.75 and 7.86 Hz, H-5), 8.957 (S, 1H, H-7). 13C-NMR (75.6 MHz, DMSO-d6, 296 K) δ ppm: 15.04 (C10); 28.27 (C9); 49.08 (C11); 136.21 (C1); 120.14 (C2); 135.68 (C3); 118.84 (C4); 121.77 (C5); 171.00 (C6); 160.15 (C7).
Dark yellow-brown thick plate-like crystal, yield: 0.1 g, 67% (based on the metal salt). Mp: 83 °C. Anal. calc. for C12H17N2O4S2V (368.34 g mol−1): C, 39.13; H, 4.65; N, 7.61; S, 17.41. Found: C, 39.29; H, 3.84; N, 8.20; S, 19.75%. IR (KBr), cm−1: ν(OH) 3425 w, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1596 s, ν(C–O) 1543 m, ν(N–N) 1028 s. Mass spectrometry, m/z (%): 336 (54) {M-CH3OH}.
N) 1596 s, ν(C–O) 1543 m, ν(N–N) 1028 s. Mass spectrometry, m/z (%): 336 (54) {M-CH3OH}.
Intense green plate-like crystals, yield: 0.030 g, 67% (based on the metal salt). Mp: 182 °C. Anal. calc. for C20H20MnN4O2S4 (530.98 g mol−1): C, 45.19; H, 3.97; N, 10.54; S, 24.12. Found: C, 46.33; H, 4.00; N, 10.43; S, 27.14%. IR (KBr), cm−1: ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1596 s, ν(C–O) 1572 m, ν(N–N) 1015 s, mass spectrometry, m/z (%): 530 (<1) {M}.
N) 1596 s, ν(C–O) 1572 m, ν(N–N) 1015 s, mass spectrometry, m/z (%): 530 (<1) {M}.
Light yellow plate-like crystals, Yield: 0.1 g, 85% (based on metal salt). Mp: 255 °C. Anal. calc. for C20H20N4O2S4Sn (595.36 g mol−1): C, 40.35; H, 3.39; N, 9.41; S, 21.54. Found: C, 40.10; H, 3.43; N, 9.49; S, 22.37%. IR (KBr), cm−1: ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1604 s, ν(C
N) 1604 s, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1464 s, ν(C–O) 1284 m, ν(N–N) 968 s. Mass spectrometry, m/z (%): 595 (<1) {M}. 1HNMR (300 MHz, DMSO-d6) δ ppm: 1.344 (t, 3H, J = 7.32 Hz, CH3), 3.217 (q, 2H, J = 7.31 Hz, CH2), 6.740 (d, 1H, H-2), 6.928 (m, 1H, H-4), 7.473 (m, 1H, H-3), 7.665 (dd, 1H, J = 1.76 and 8.01 Hz, H-5), 9.306 (S, 1H, H-7). 13C-NMR (75.6 MHz, DMSO-d6, 296 K) δ ppm: 14.92 (C10); 25.80 (C9); 137.55 (C1); 119.21 (C2); 136.71 (C3); 116.55 (C4); 122.11 (C5); 167.64 (C6); 165.19 (C7); 169.28 (C8).
C) 1464 s, ν(C–O) 1284 m, ν(N–N) 968 s. Mass spectrometry, m/z (%): 595 (<1) {M}. 1HNMR (300 MHz, DMSO-d6) δ ppm: 1.344 (t, 3H, J = 7.32 Hz, CH3), 3.217 (q, 2H, J = 7.31 Hz, CH2), 6.740 (d, 1H, H-2), 6.928 (m, 1H, H-4), 7.473 (m, 1H, H-3), 7.665 (dd, 1H, J = 1.76 and 8.01 Hz, H-5), 9.306 (S, 1H, H-7). 13C-NMR (75.6 MHz, DMSO-d6, 296 K) δ ppm: 14.92 (C10); 25.80 (C9); 137.55 (C1); 119.21 (C2); 136.71 (C3); 116.55 (C4); 122.11 (C5); 167.64 (C6); 165.19 (C7); 169.28 (C8).
Orange plate-like crystals, yield: 0.04 g, 37% (based on metal salt). Mp: 160 °C. Anal. calc. for C13H17I2N3O2S2Sn (683.94 g mol−1): C, 22.83; H, 2.51; N, 6.14; S, 9.38. Found: C, 23.35; H, 2.49; N, 6.17; S, 9.17%. IR (KBr), cm−1: ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1647 s, ν(C–O) 1284 m, ν(N–N) 10
N) 1647 s, ν(C–O) 1284 m, ν(N–N) 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 230 w. Mass spectrometry, m/z (%): 683 (<1) {M}. 1H-NMR (300 MHz, DMSO-d6) δ ppm: 1.346 (t, 3H, J = 7.32 Hz, C10H3), 3.220 (q, 2H, J = 7.32 Hz, C9H2), 6.739 (dd, 1H, J = 8.51 and 1.01, H2), 6.932 (ddd, 1H, J = 8.02, 7.08 and 1.11, H4), 7.476 (m, 1H, J = 7.04 and 1.82, H3), 7.671 (dd, 1H, J = 7.96 and 1.77 Hz, H5), 9.314 (s, 1H, H7).
230 w. Mass spectrometry, m/z (%): 683 (<1) {M}. 1H-NMR (300 MHz, DMSO-d6) δ ppm: 1.346 (t, 3H, J = 7.32 Hz, C10H3), 3.220 (q, 2H, J = 7.32 Hz, C9H2), 6.739 (dd, 1H, J = 8.51 and 1.01, H2), 6.932 (ddd, 1H, J = 8.02, 7.08 and 1.11, H4), 7.476 (m, 1H, J = 7.04 and 1.82, H3), 7.671 (dd, 1H, J = 7.96 and 1.77 Hz, H5), 9.314 (s, 1H, H7).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) was added to the L (0.500 g, 2.083 mmol) in the same solvent (6 mL). The resulting yellow solution was refluxed for 2 h on a water bath. Yellow crystals were obtained from slow evaporation of the solvent in a refrigerator over 10 days.
1 v/v) was added to the L (0.500 g, 2.083 mmol) in the same solvent (6 mL). The resulting yellow solution was refluxed for 2 h on a water bath. Yellow crystals were obtained from slow evaporation of the solvent in a refrigerator over 10 days.Light-yellow tablet crystals, yield: 0.91 g, 87% (based on metal salt). Mp: 195 °C. Anal. calc. for C12H16Sn N2O2S3Cl2 (506.6 g mol−1): C, 28.48; H, 3.19; N, 5.54; S, 19.01. Found: C, 28.88; H, 3.16; N, 5.98; S, 19.51%. IR (KBr), cm−1: ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1600 s, ν(C–O) 15
N) 1600 s, ν(C–O) 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340 m, ν(N–N) 1030 w. Mass spectrometry, m/z (%): 506.8 (<1). 1H-NMR (300 MHz, DMSO-d6) δ ppm: 1.372 (t, 3H, J = 7.28 Hz, CH3), 2.554 (S, 6H, CH3), 3.234 (q, 2H, J = 7.08 Hz, CH2), 6.982 (m, 1H, H-4), 7.007 (d, 1H, J = 1.08, H-2), 7.551 (td, 1H, H-3), 7.677 (dd, 1H, J = 1.87 and 7.71 Hz, H-5), 9.117 (S, 1H, H-7). 13C-NMR (75.6 MHz, DMSO-d6, 296 K) δ ppm: 14.99 (C10); 25.63 (C9); 40.90 (C11, C12); 136.98 (C1); 119.55 (C2); 136.58 (C3); 116.96 (C4); 122.31 (C5); 164.64 (C6); 164.85 (C7); 169.26 (C8).
340 m, ν(N–N) 1030 w. Mass spectrometry, m/z (%): 506.8 (<1). 1H-NMR (300 MHz, DMSO-d6) δ ppm: 1.372 (t, 3H, J = 7.28 Hz, CH3), 2.554 (S, 6H, CH3), 3.234 (q, 2H, J = 7.08 Hz, CH2), 6.982 (m, 1H, H-4), 7.007 (d, 1H, J = 1.08, H-2), 7.551 (td, 1H, H-3), 7.677 (dd, 1H, J = 1.87 and 7.71 Hz, H-5), 9.117 (S, 1H, H-7). 13C-NMR (75.6 MHz, DMSO-d6, 296 K) δ ppm: 14.99 (C10); 25.63 (C9); 40.90 (C11, C12); 136.98 (C1); 119.55 (C2); 136.58 (C3); 116.96 (C4); 122.31 (C5); 164.64 (C6); 164.85 (C7); 169.26 (C8).
2.4. X-ray crystallography
Suitable crystals of L and 1–6 were mounted on polymer loops and placed in a cold nitrogen stream on a Bruker Smart APEX CCD diffractometer. Full spheres of data were collected under control of the APE×3 program suite.26 The raw data were converted to F2 values with SAINT26 and empirical absorption corrections as well as merging of equivalent reflections were performed with SADABS.26 The structures were solved by direct methods (SHELXT27) and refined by full-matrix, least-squares procedures (SHELXL28). Hydrogen atoms attached to carbon were placed in idealized positions while those attached to oxygen were placed in locations derived from difference maps and their coordinates adjusted to give O–H = 0.87 Å. All were included as riding contributions with isotropic displacement parameters tied to those of the attached atoms.2.5. Biology studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 M−1 cm−1. Samples were prepared by dissolving each compound in DMSO with gradual increase in concentration (0, 1, 2 and 13 μM) at room temperature. Appropriate concentrations of drug–protein solutions were prepared by mixing protein solutions with different concentrations of drug solutions. The final concentration of DMSO after addition of the drug solution to the protein solution was 1% of the total volume. For fluorescence determination, spectra (λex = 280 nm) in the wavelength range of 285–500 nm were recorded at 298 K and the slit width was 5/5. Absorption titration experiments with the same concentrations for protein and compounds were also done.
700 M−1 cm−1. Samples were prepared by dissolving each compound in DMSO with gradual increase in concentration (0, 1, 2 and 13 μM) at room temperature. Appropriate concentrations of drug–protein solutions were prepared by mixing protein solutions with different concentrations of drug solutions. The final concentration of DMSO after addition of the drug solution to the protein solution was 1% of the total volume. For fluorescence determination, spectra (λex = 280 nm) in the wavelength range of 285–500 nm were recorded at 298 K and the slit width was 5/5. Absorption titration experiments with the same concentrations for protein and compounds were also done.2.6. X-ray powder diffraction studies
To prepare the nano-particles, 10 mL of a 0.01 M solution of the metal salt was positioned in an ultrasonic probe with a maximum power output of 200 W. Into this solution, 10 mL of a 0.01 M solution of the L was added dropwise. The precipitate obtained was filtered off and dried in air. The X-ray diffraction pattern of the complexes was determined for a 2θ range of 6.000° to 79.990° using the unit cell data from the single crystal determinations. The morphology and size of the nanoparticles were investigated by scanning electron microscopy (SEM).3. Results and discussion
3.1. IR and NMR studies
In the infrared spectrum of L (Fig. SI 1a†), a broad, medium intensity band in the region 3200–3400 cm−1 is assigned to the OH stretching vibration. This band disappears after complexation and indicates the coordination takes place through the deprotonated phenolic oxygen atom.29–31 Also the medium intensity ν(C–O) band at ∼1260 cm−1 in the L undergoes a blue shift in the complexes (Fig. SI 1b–g†) which provides further evidence of involving the phenolic oxygen atom in coordination.32The bands at 1307 and 3108 cm−1 are assigned to ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) and ν(N–H) vibrations respectively.33,34 Disappearance of these bands in the spectra of the complexes suggest the coordination of the S atom to the metal ion. Also, the absence of a ν(S–H) band at approximately 2750 cm−1 emphasizes the existence of the thione tautomeric form in the solid state.35 A strong ν(C
S) and ν(N–H) vibrations respectively.33,34 Disappearance of these bands in the spectra of the complexes suggest the coordination of the S atom to the metal ion. Also, the absence of a ν(S–H) band at approximately 2750 cm−1 emphasizes the existence of the thione tautomeric form in the solid state.35 A strong ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) band is observed at 1613 cm−1 in the L spectrum which shows a red shift in the metal complexes and confirms coordination of the azomethine nitrogen atom to the metal ion.10,34 Finally, νsy(cis-MoO2), νasy(cis-MoO2)36 and ν(V
N) band is observed at 1613 cm−1 in the L spectrum which shows a red shift in the metal complexes and confirms coordination of the azomethine nitrogen atom to the metal ion.10,34 Finally, νsy(cis-MoO2), νasy(cis-MoO2)36 and ν(V![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)37,38 frequencies appear at 842, 929 and 990 cm−1 respectively.39
O)37,38 frequencies appear at 842, 929 and 990 cm−1 respectively.39
In the 1HNMR spectra of compounds (Fig. SI 2a–e†), the aromatic protons are observed as multiplet signals in the range 6.5–7.9 ppm.40 The triplet and quartet signals which are, respectively, observed in the ranges 1.2–1.4 and 3.2–4.2 ppm correspond to the ethyl protons.41 In the spectrum of the ligand (L), however, the sharp signal at 10.26 ppm is assigned to the phenolic proton as it disappears upon D2O addition (Fig. SI 2f†).42 The absence of this signal in the spectra of the complexes confirms that coordination occurs through the phenolic oxygen.43 Also the singlet signal assigned to the imine hydrogen in the L spectrum (8.54 ppm) shifts down field after complexation44,45 while satellites appear around this signal in compounds 4–6 which correspond to coupling of this hydrogen with 117Sn and 119Sn isotopes,46 all of which indicate coordination of this nitrogen atom.
In the 13C-NMR spectra of the compounds (Fig. SI 3a–e†), signals of the aromatic carbons are in the range 116–164 pm. The signals of the thioamide and azomethine carbons in the L spectrum are observed at 196 and 145 ppm respectively and are shifted upfield after complexation.47 This indicates that the thioamide and azomethine carbons are close to complexation sites (see Experimental section for more details about other carbons).48
3.2. Thermogravimetric analysis
TGA and DTG curves of complexes 1–6 (Fig. SI 4a–f†) were carried out over a temperature range from room temperature up to 950 °C. The thermal studies using TGA methods show a multistep decomposition pattern in the thermograms of the metal complexes under investigation.The data from thermogravimetric analysis show that decomposition of compound 1 occurs in four steps. The degradation starts over the temperature range 28–145 °C with a mass loss of 7.90% (calc. 8.04%), consistent with removal of the coordinated methanol molecule. Next, weight losses of 26.79% (calc. 26.40%) and 28.83% (calc. 29.40%) are associated with removal of the coordinated L′ which occurs in two consecutive steps over the temperature range 224–719 °C. The remaining residue (35.22% of the initial mass) is formulated as MoO3 (calc. 36.14%).12,49
The vanadium complex (2) also, in the first stage of its thermal decomposition, involves loss of the coordinated methanol molecule (8.63% mass loss; calc. 8.69%) over the temperature range of 21–113 °C. In the second step, the methoxide ligand is removed by further heating to 217 °C (8.30% mass loss; calc. 8.42%). Then, the L′ ligand is removed in three steps with a total mass loss of 60.39% (calc. 60.35%) over the temperature range 178–743 °C and VO2 remains as the final product with a mass of 26.60% (calc. 22.51%).49
Complex 3 decomposes in three steps. No weight loss is seen up to 150 °C. The coordinated L′ in two consecutive steps are eliminated over the temperature range 153–570 °C with a mass loss of 22.76% (calc. 23.02%) for release of S-ethyl groups and a mass loss of 32.96% (calc. 33.61%) for release of another part of the L′ ligand. The residue at the temperature of 570 °C is a mixture of metal oxides comprising 25.59% of the initial mass which at 822 °C converts to Mn2O3 with a mass of 19.21% (calc. 21.40%).50–52
TGA and DTG curves of 4, 5 and 6 shows two thermal decomposition stages and for all the three complexes, SnO2 remains as the final product.46
In 4, the first step consists of the loss of iodine groups and coordinated dimethyl formamide molecule with a mass of 42.88% (calc. 47.80%) at temperature range of 100–370 °C. The second step indicates the decomposition of the organic part (L′) up to 590 °C with mass loss of 30.52% (calc. 32.50%) and formation of SnO2.
Likewise, in 5, the first step involves the removal of dimethyl sulfoxide molecule, chlorine atoms and CS2–ethyl group with a mass of 46.19% (calc. 50.25%) at temperature range of 21–357 °C. By increasing of temperature up to 725 °C, the residual organic part decomposes with a loss of 26.36% (calc. 26.13%) of the complex mass, and the SnO2 with mass of 26.40% (calc. 29.79%) remains as the final product.
Finally, compound 6 is stable up to 195 °C. The L′ removes during two steps at temperature range of 195–580 °C and the SnO2 with mass loss of 22.7% (calc. 25.31%) remains as the final product.
3.3. X-ray crystal structures
Molecular structures were determined by single crystal X-ray diffraction for the L and all six complexes and ORTEP drawings are shown in Fig. 1. Crystal data and refinement results for the compounds are listed in Table 1.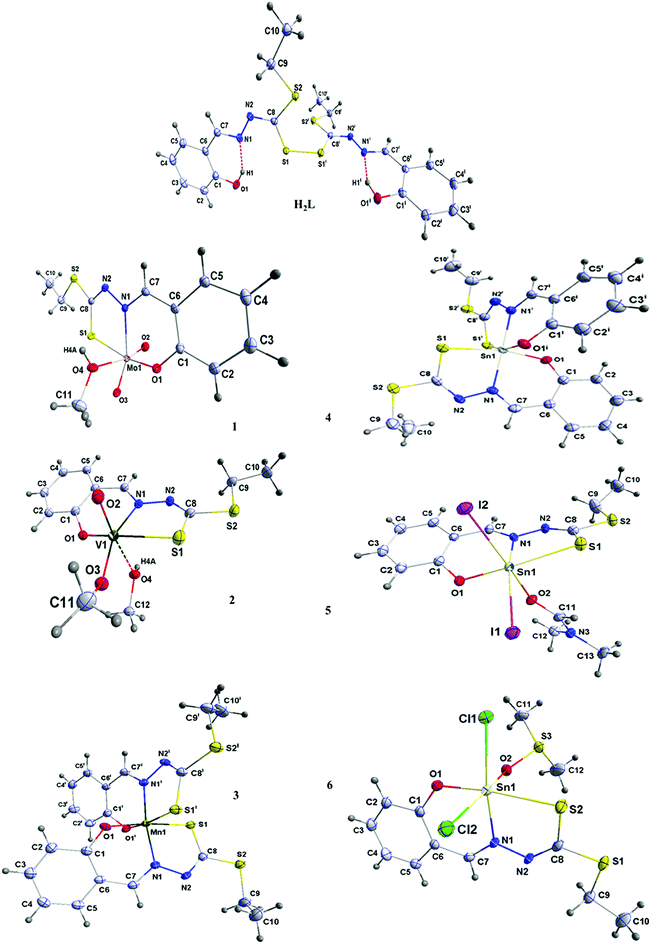 | ||
| Fig. 1 Views of L ligand and complexes showing the atomic numbering and 50% probability displacement ellipsoids. | ||
| L | 1 | 2 | 3 | 4 | 5 | 6 | |
| Chemical formula | C20H22N4O2S4 | C11H14MoN2O4S2 | C12H17N2O4S2V | C20H20MnN4O2S4 | C20H20N4O2S4Sn | C13H17I2N3O2S2Sn | C12H16Cl2N2O2S3Sn |
| Mr | 478.65 | 389.30 | 368.33 | 531.58 | 595.33 | 683.90 | 506.04 |
| Crystal system | Monoclinic | Monoclinic | Triclinic | Monoclinic | Monoclinic | Orthorhombic | Triclinic |
| Space group | C2/c | P21/c | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
C2/c | C2/c | Pbca | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
| Unit cell (Å, °) | |||||||
| a | 23.712(2) | 10.0788(8) | 8.0281(17) | 7.7764(10) | 7.7957(7) | 18.999(11) | 8.5293(4) |
| b | 9.1025(8) | 9.8639(8) | 10.127(2) | 24.056(3) | 24.197(2) | 8.978(5) | 9.3173(5) |
| c | 10.4517(9) | 14.4725(11) | 11.104(2) | 12.1308(15) | 12.3003(10) | 23.512(14) | 12.6067(6) |
| α | 90 | 90 | 110.554(3) | 90 | 90 | 90 | 100.635(1) |
| β | 105.353(1) | 90.690(1) | 110.250(3) | 104.462(2) | 101.157(1) | 90 | 108.831(1) |
| γ | 90 | 90 | 94.785(3) | 90 | 90 | 90 | 94.334(1) |
| V (Å3) | 2175.3(3) | 1438.7(2) | 771.2(3) | 2197.4(5) | 2276.3(3) | 4011(4) | 922.06(8) |
| Z | 4 | 4 | 2 | 4 | 4 | 8 | 2 |
| Radiation type | Mo/Kα (λ = 0.71075 Å) | Mo/Kα (λ = 0.71075 Å) | Mo/Kα (λ = 0.71075 Å) | Mo/Kα (λ = 0.71075 Å) | Mo/Kα (λ = 0.71075 Å) | Mo/Kα (λ = 0.71075 Å) | Mo/Kα (λ = 0.71075 Å) |
| ρ (g cm−1) | 1.462 | 1.839 | 1.586 | 1.607 | 1.737 | 2.265 | 1.823 |
| μ (mm−1) | 0.462 | 1.215 | 0.929 | 1.007 | 1.515 | 4.571 | 2.020 |
| Crystal size (mm) | 0.060 × 0.080 × 0.200 | 0.090 × 0.130 × 0.160 | 0.120 × 0.210 × 0.240 | 0.033 × 0.085 × 0.214 | 0.060 × 0.140 × 0.240 | 0.040 × 0.100 × 0.280 | 0.060 × 0.110 × 0.290 |
| T (K) | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| F(000) | 1000 | 800 | 380 | 1092 | 1192 | 2560 | 500 |
| No. of refls. | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 130 130 |
27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 178 178 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 305 305 |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 217 217 |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 190 190 |
72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 505 505 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 013 013 |
| R1 [I ≥ 2σ(I)] | 0.0378 | 0.0278 | 0.464 | 0.0399 | 0.0351 | 0.0362 | 0.0291 |
| R1 (all data) | 0.0507 | 0.0341 | 0.634 | 0.0520 | 0.0413 | 0.0497 | 0.0369 |
| wR2 | 0.0970 | 0.0752 | 0.1311 | 0.1104 | 0.1032 | 0.0884 | 0.0632 |
| GOF on F2 | 1.079 | 1.083 | 1.026 | 1.099 | 1.146 | 1.081 | 0.917 |
| Largest diff. Peak/hole (e Å−3) | 0.497, −0.239 | 1.604, −0.355 | 1.149, −0.380 | 0.915, −0.285 | 1.624, −0.580 | 1.909, −0.869 | 1.247, −0.871 |
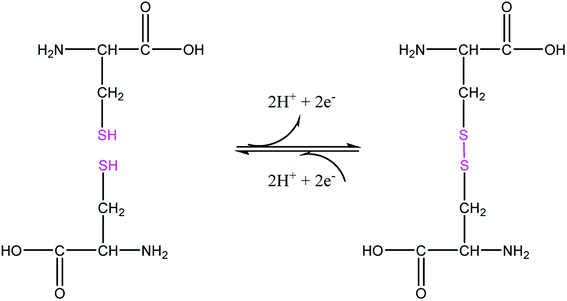
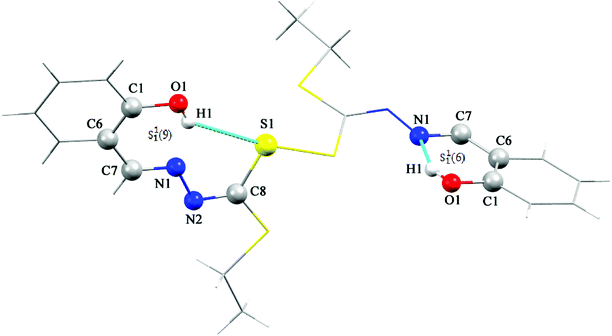
The sample of the L ligand material used for the X-ray structure was found to be a dimer with an S–S bond. Formation of the S–S bond is a consequence of S–H oxidation which is a phenomenon which frequently happen in the amino acid cysteine (Scheme 1).45
The S–S bond length in hydrogen disulfide is 2.06 Å, however, formation of this bond with a length of 2.045 Å in the L is close to the length of the bond in other compounds.53 The torsion angle S1′–S1–C8–S2 (−8.86(8) Å) shows that the molecule is not planar and is twisted. In fact, the two equal fragments are perpendicular to each other in the molecule. The intramolecular O1–H1⋯N1 hydrogen bonding creates a S11(6) graph set. Also, another intramolecular interaction between the S1 and O1 atoms leads to formation of a nine-membered ring (S1/C8/N2/N1/C7/C6/C1/O1/H1) with S11(9) graph set (Fig. 2).
Adjacent molecules connect to each other by C10–H10A⋯O1i (2.644 Å) (symm. code i: 1 − x, 1 − y, − z) and C7–H7⋯O1 (2.692 Å) hydrogen bonds along the c axis. Additionally, the C5–H5⋯Cgii (Cg: C1–C6) (3.715 Å) (symm. code ii: 1 − x, 1 − y, 1 − z) interaction plays a decisive role in the development of these chains. These chains expand through C2–H2⋯Cgiii (Cg: C1–C6) (2.865 Å) (symm. code iii: 1 − x, 1 − y, 1 − z) interaction in the bc plane and complete second and third dimensions (Fig. SI 5a and b†).
In the complexation the S–S bond of the L is broken and two L′ is formed (Scheme 1). The L′ ligand acts as a tridentate chelate and connects to the metal ion through the deprotonated phenolic oxygen, the azomethine nitrogen and the thioamide sulfur atom in 1–6. All the complexes are six coordinated and exhibit a distorted octahedral configuration. The asymmetric units of 1, 3, 4 and 5 are composed of one complete molecule and for 2 and 6 only half of the molecule lies in the asymmetric unit. Selected bond lengths and angles are listed in Table 2.
| L | 1 | 2 | 3 | 4 | 5 | 6 | |
| C7–N1 | 1.2864(18) | 1.300(2) | 1.301(3) | 1.298(3) | 1.290(3) | 1.301(6) | 1.286(3) |
| C8–N2 | 1.2859(17) | 1.300(3) | 1.311(3) | 1.296(3) | 1.302(4) | 1.301(6) | 1.289(3) |
| S1–C8 | 1.7917(13) | 1.729(2) | 1.719(2) | 1.735(2) | 1.741(3) | 1.753(5) | 1.739(3) |
| S2–C8 | 1.7413(13) | 1.742(2) | 1.746(2) | 1.747(2) | 1.754(3) | 1.754(5) | 1.760(3) |
| C1–C6 | 1.4106(19) | 1.408(3) | 1.416(3) | 1.420(3) | 1.418(4) | 1.423(6) | 1.426(4) |
| C1–O1 | 1.3516(17) | 1.349(2) | 1.340(3) | 1.330(2) | 1.339(3) | 1.343(5) | 1.338(3) |
| N1–N2 | 1.4050(16) | 1.397(2) | 1.414(3) | 1.410(2) | 1.395(3) | 1.401(5) | 1.406(3) |
| S2–C9 | 1.8201(14) | 1.807(2) | 1.818(3) | 1.822(2) | 1.821(3) | 1.820(5) | 1.818(3) |
| M1–N1 | — | 2.2821(16) | 2.152(2) | 1.9836(16) | 2.173(2) | 2.203(4) | 2.183(2) |
| M1–S1 | — | 2.4568(5) | 2.4183(9) | 2.3196(6) | 2.4827(7) | 2.4910(16) | 2.4624(7) |
| M1–O1 | — | 1.9284(14) | 1.8699(17) | 1.9036(15) | 2.045(2) | 2.040(3) | 2.0109(18) |
| M1–O2 | — | 1.7017(16) | 1.5904(18) | — | — | 2.220(3) | 2.1662(17) |
| S1–C8–S2 | 103.83(4) | 119.81(12) | 126.61(19) | 113.70(12) | 111.35(15) | 111.6(2) | 111.13(15) |
| N1–N2–C8 | 111.96(11) | 115.03(16) | 112.12(19) | 111.69(16) | 114.2(2) | 114.5(4) | 115.6(2) |
| O1–C1–C6 | 122.28(12) | 121.64(18) | 120.5(2) | 122.81(18) | 124.1(2) | 123.5(4) | 124.7(2) |
| C7–N1–N2 | 113.72(11) | 113.30(16) | 113.57(19) | 115.13(16) | 116.3(2) | 114.8(4) | 115.8(2) |
| C8–S2–C9 | 99.47(6) | 104.36(10) | 104.66(12) | 103.08(10) | 103.69(14) | 102.8(2) | 102.32(13) |
| N2–C8–S2 | 121.76(10) | 113.75(14) | 120.26(18) | 120.37(15) | 119.4(2) | 118.7(3) | 120.0(2) |
| O2–M1–O1 | — | 100.02(7) | 99.83(8) | — | — | 82.32(13) | 81.51(7) |
| O2–M1–N1 | — | 93.99(6) | 94.44(8) | — | — | 79.97(13) | 83.41(7) |
| O1–M1–N1 | — | 82.42(6) | 84.55(8) | 91.67(6) | 86.46(8) | 84.26(13) | 87.84(7) |
| N1–M1–S1 | — | 76.14(4) | 77.54(6) | 82.07(5) | 79.25(6) | 79.24(10) | 80.32(6) |
| O1–M1–S1 | — | 154.98(5) | 156.89(6) | 91.46(5) | 165.58(6) | 161.79(10) | 165.95(5) |
| O2–M1–S1 | — | 94.24(6) | 96.04(7) | — | — | 87.24(10) | 89.59(5) |
| O2–M1–O3 | — | 105.46(7) | 102.95(9) | — | — | — | — |
| C8–S1–M1 | — | 100.08(7) | 98.73(8) | 94.10(7) | 92.60(9) | 93.97(15) | 102.32(13) |
In all these complexes, the coordination of the L′ to the central metal forms one five- and one six-membered chelate ring. These two rings are not coplanar and therefore it can be concluded that the L′2− is coordinated to the central metal with steric strain. Moreover, the greater electronegativity of oxygen atom over sulfur atom causes shortening of the oxygen–metal bond which leads to an out-of-plane orientation of the oxygen atom within the six-membered ring. Among these complexes, compound 5 has the longest M–S bond (2.4910(16) Å) which causes the greatest amount of oxygen atom deviation (0.278 Å) from the M, N1, C7, C6, C1, O1 mean plane and compound 2 has the shortest M–O bond (1.869(17) Å). In 1, the O1 atom bears such a strain that it is 0.16 Å off the mean plane of the six-membered ring (Mo1, N1, C7, C6, C1, O1). The Mo1–O1 and Mo1–S1 bond lengths are 1.9284(14) and 2.4568(5) Å respectively. However, the Mo1–O2 (1.702(2) Å) and Mo1–O3 (1.703(1) Å) bonds are approximately the same length, which confirms the equal distribution of electron density of oxido atoms around the central metal. Oxido atoms, O2 and O3, are oriented cis to each other with an angle of 105.46(7)°. All lengths and angles around the central atom are almost equal to those of the similar compounds reported elsewhere.54–56 The L′ and O3 atom occupy the equatorial positions while methanol solvent and the O2 atom are in axial positions. The O2 and O4 atoms are trans relative to each other with angle of 169.59(7)°. In this compound, the adjacent molecules form a dimer with an R22(10) graph set through pairwise hydrogen bond of O4–H4A⋯N2i (1.913(2) Å) (symm. code i: 1 − x, 1 − y, 1 − z). These dimers connect to each other through the hydrogen bond C9ii–H9Aii⋯O2 (2.543 Å) (symm. code ii: x, 1.5 − y, 1/2 + z) and form a one-dimensional chain in the direction of the (−567) plane. Likewise, through C9–H9A⋯O2iii and C4–H4A⋯S1iv (2.961 Å) (symm. code iii and iv: 1 − x, 1/2 + y, 1/2 − z and x, −1 + y, z) hydrogen bonds the second dimension forms along the b axis. The planes thus created connect to each other through C2–H2⋯ O3v (2.606 Å) (symm. code v: x, 1.5 − y, 1/2 + z) hydrogen bonds along the c direction to generate the three dimensional structure (Fig. SI 5c–f†).
In complex 2, also due to steric strain the vanadium center moves from the mean coordination plane toward the axial position by about 0.303(3) Å. The axial positions are occupied by the methanol and O2 atom and the L′ ligand and methoxy groups fill the equatorial positions. The angle between the O2 and O4 atoms (172.98(8) °) is higher than compound 1. In this crystalline structure, just like 1, adjacent molecules form dimers and create ten member graph set R22(10) through pairwise hydrogen bonds O4–H4A⋯N2i (1.963 Å) (symm. code i: 1 − x, 1 − y, 1 − z). The preformed dimers form one-dimensional chains in the direction of the c axis through the hydrogen bonds C9–H9A⋯Cg, C12–H12C⋯O2ii (2.681 Å) and C5i–H5i⋯O2iii (2.665 Å) (symm. code ii: 2 − x, 1 − y, 1 − z, iii: −1 + x, y, z). Moreover, the C2–H2⋯O1iv (symm. code iv: 1 − x, 1 − y, −z) hydrogen bond is responsible for the formation of the second dimension along the ac plane and the last dimension is formed through C4–H4⋯Sv and C5–H5⋯C11vi (symm. code v: x, −1 + y, −1 + z, vi: x, −1 + y, z) interactions along the b axis (Fig. SI 5g–j†).
In compounds 3 and 4, the central metal in its +IV oxidation state surrounded by two tridentate chelate ligands (L′). In 3 and 4, respectively, the two L′ coordinate to the manganese and tin ions through the deprotonated phenol oxygen, the azomethine nitrogen and the thioamide sulfur donor atoms. In the coordination sphere, the two phenolic oxygen atoms are cis to one another as are the thioamide sulfur atoms with the two azomethine nitrogen atoms disposed trans to one another. The O1 atom deviation from the coordination plane is 0.217 Å and 0.227 Å in 3 and 4 respectively. It seems that steric strain is higher in 4. However, in both compounds the C4–H4⋯S1i (2.920 Å for 3 and 2.883 Å for 4) (symm. code i: −1/2 + x, 1/2 + y, 1/2 + z) interaction leads to formation of one-dimensional chains in the −a + c direction. These chains within the (−10−1) plane form 2D sheets through chalcogen–chalcogen S2⋯S2ii (ii: −1 + x, 1 − y, 1/2 + z) (3.403 Å) interactions and finally the planes expand by C9–H9B⋯C9iii (iii: −1 + x, y, −1 + z) interactions to construct the 3D supramolecular network (Fig. SI 5k–m†).
In the structure of 5, the deprotonated L′ and one iodine atom are located in equatorial positions and the other iodine atom and the DMF solvent are placed in the axial position. Iodine atoms are positioned cis relative to each other (97.84(2)°) which angle is 7° higher than the idealized 90° for a regular octahedron geometry. Interactions such as C9–H9A⋯N2i, C13–H13C⋯I1ii (3.047 Å) and C7–H7⋯Cgi (symm. code i: 1.5 − x, −1/2 + y, 1 − z, ii: x, −1 + y, z) creates chains along the b axis direction. The hydrogen bonding C13–H13B⋯S2iii (2.985 Å), C12–H12A⋯O1vi (2.451 Å) and C11–H11⋯S2iii (2.975 Å) (symm. code iii: 1 − x, −y, 1 − z, vi: 1/2 + x, −1 + y, 1/2 − z) forms the second dimension along the –a + c direction. The last dimension is formed along the –a −c direction through C12v–H12Av⋯O1, C11vi –H11vi⋯S2vi and C13vi–H13Bvi⋯S2vi (symm. code v: 1 − x, −1.5 + y, 1/2 + z, vi: 1/2 + x, 1/2 − y, 1 − z) hydrogen bonds (Fig. SI 5n–p†).
The crystalline structure of 6 is similar to that of 5, in which the two chlorine atoms are arranged cis to each other (96.81(3)°) and the angle is 6° higher than the idealized 90° for a regular octahedron geometry and 2° higher than the reported similar structure by Souse et al.,57 Therefore, the polyhedron around the Sn(IV) atom is best described as a distorted octahedral. The deprotonated L′ and one chlorine atom are located at the equatorial positions and the other chlorine atom and the DMSO solvent are in the axial positions.
The three-dimensional structure is formed according to the following interactions. The C11–H11A⋯C10i (2.899 Å), C7–H7⋯Cl2ii (2.819 Å), C10–H10C⋯Cl2ii (3.397 Å) and H12B–C12⋯Cl2 (2.868 Å) interactions (symm. code i: x, y, −1 + z, ii: 2 − x, 1 − y, 1 − z) form chains along the c axis direction. The molecules form the second dimension along the a axis direction via the hydrogen bonds C11–H11A··· Cl2iii, C12–H12C⋯Cl1iii and C11–H11B⋯Cgiv (symm. code iii: −1 − x, y, z, iv: 1 − x, 1 − y, −z). The C4–H4C⋯Cl2v (symm. code v: x, 1 + y, z) interaction along the b axis forms the third dimension (Fig. SI 5q–s†).
All bond lengths in these compounds are similar to those observed in reported compounds.15,58,59 The S1–C8 and S2–C8 bond distances are in the range 1.729(2) Å to 1.792(13) Å for the new compounds which suggest the S–C bond is very close to a single bond. The N1–C8 and N2–C8 bond lengths are between 1.285(17) Å and 1.311(3) Å consistent with substantial double bond character. The observed values for the N1–N2 bond (1.395(3) Å to 1.414(3) Å) shows that the bond is shorter than a single N–N bond (1.44 Å). This variation in bond lengths shows that a significant π-charge delocalization occurs within the SSCNNC fragment.
3.4. In vitro cytotoxicity study
To use a substance as a drug, a cytotoxicity test is required. If it does not show cytotoxicity or just reveals a lower level than the determined standards and if other tests are passed it could enter the pharmaceutical marketing. One of the most important methods to measure cytotoxicity is the MTT assay.60 MTT is a compound that reduces in the presence of mitochondria dehydrogenase enzyme and its yellow color changes to violet. Color intensity is proportional to the number of live cells.61 In this method, the cell is exposed to various concentrations of the target material and then evaluated for cell death indices.62,63The in vitro cytotoxicity evaluation results in three cell lines for the L and complexes 1–6 are summarized in Table 3 (see graphs in Fig. SI 6a–c†).
| Cell lines | IC50 ± SD (μM)a | ||
|---|---|---|---|
| Hela | MCF-7 | CHO | |
| a Data are presented as mean ± SD (standard deviation). All experiments were independently performed at least four times. | |||
| L | 0.1766 ± 0.022 | 0.4762 ± 0.0261 | 0.4698 ± 0.0236 |
| 1 | 0.846 ± 0.023 | 1.067 ± 0.0223 | 0.6628 ± 0.224 |
| 2 | 0.0506 ± 0.0226 | 0.6692 ± 0.0243 | 0.9544 ± 0.0186 |
| 3 | 0.3415 ± 0.0232 | 0.7031 ± 0.0241 | 0.8267 ± 0.025 |
| 4 | 0.6842 ± 0.012 | 0.5862 ± 0.0244 | 1.456 ± 0.0246 |
| 5 | 0.6842 ± 0.02 | 1.048 ± 0.0247 | 0.3179 ± 0.0254 |
| 6 | 0.05065 ± 0.0226 | 0.7018 ± 0.0247 | 1.236 ± 0.0262 |
| Cisplatin | 0.324 ± 0.023 | 0.67430 ± 0.023 | 1.347 ± 0.025 |
Cytotoxicity evaluation results reveal that all complexes exhibit good cytotoxic activity. L and complexes 2 and 6 showed IC50 values lower than cisplatin against Hela cell and likewise complexe 4 and L against MCF-7 cell which indicating a greater cytotoxicity and anticancer activity of these compounds. So, complex 2 and L show better in vitro therapeutic index than cisplatin against all the two cell lines (see the pictures of the color change of compounds during the MTT experiment in Fig. SI 6d–f†).
3.5. Interaction of compounds with HSA using fluorescence spectroscopy
The L effects on the fluorescence intensity of HSA is shown in Fig. 3. With increasing concentration of L at 298 K there is a decrease in the fluorescence intensity of HSA. The results show that the L quenches the fluorescence peak of HSA significantly even before its concentrations become equal to that of HSA.
 | ||
| Fig. 3 Fluorescence quenching spectra of HSA in the presence of increasing concentration of L. [HSA] = 15 μM, [L] = 0–14 μM, λex = 280 nm, T = 298 K. | ||
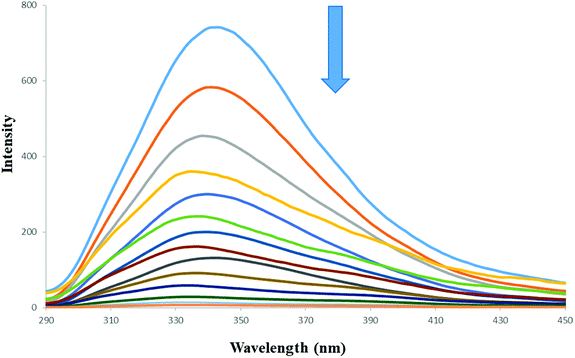
Fluorescence quenching is described by the Stern–Volmer equation:70
 | (1) |
Fluorescence quenching occurs by two mechanisms: dynamic and static quenching. Dynamic quenching is due to the collision of the quencher molecules with the desired protein, which reduces fluorescence and converts the energy into heat. Static quenching takes place by forming a complex of protein-quencher molecules.25 The formation of this complex produces a new species that has no emission in the range of the previous wavelength. In fact, dynamic quenching and static quenching are caused by diffusion and ground–state complex formation, respectively.73
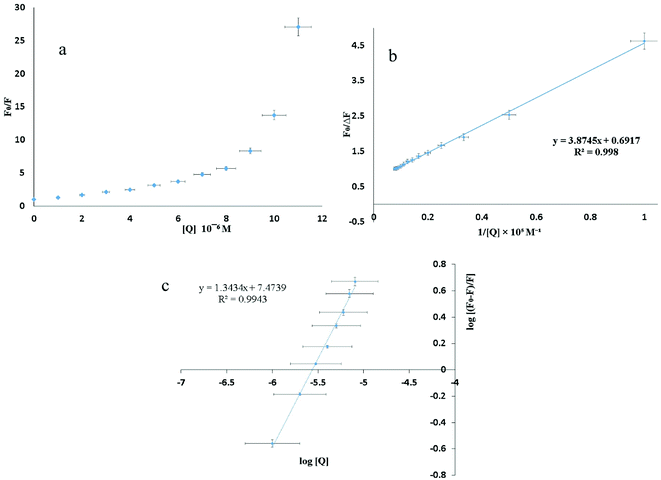
The shape of the SV plot gives information on the quenching mechanism involved. The Stern–Volmer diagram is not linear and is curved (Fig. 4a), indicating that the quencher has created dynamic and static quenching (Fig. SI 7g–l†).
For the positive deviation cases a modified SV equation accounting for simultaneous static and dynamic mechanisms was used. For negative deviation cases the following equation was used:
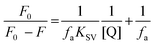 | (2) |
The graph is linear (Fig. 4b) and from this one can get fa and KSV. According to the literature, if Kq is more than 2 × 1010 M−1 S−1 this indicates that static quenching has a major role in quenching process and the role of dynamic quenching is almost insignificant (Fig. SI 7m–r†).75 The KSV values, calculated from eqn (2) for compounds 1–6 are collected in Table 4.
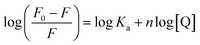 | (3) |
From the plot of 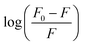 versus log[Q] (Fig. 4c), the number of binding sites (n) and association constant (Ka) values were calculated for L ligand and complexes (Fig. SI 7s–x†) from the slope and the intercept on the Y-axis respectively (Table 5).76
versus log[Q] (Fig. 4c), the number of binding sites (n) and association constant (Ka) values were calculated for L ligand and complexes (Fig. SI 7s–x†) from the slope and the intercept on the Y-axis respectively (Table 5).76
| a Data are presented as mean ± SD (standard deviation). All experiments were independently performed three times. | |||||||
|---|---|---|---|---|---|---|---|
| L | 1 | 2 | 3 | 4 | 5 | 6 | |
| Ka (×10−5 M−1) ± SDa | 2.98 ± 0.09 | 1.76 ± 0.1 | 3.3 ± 0.05 | 0.8 ± 0.06 | 1.01 ± 0.03 | 3.4 ± 0.1 | 1.8 ± 0.07 |
| n | 1.34 | 1.29 | 1.29 | 1.25 | 1.2 | 1.3 | 1.3 |
| ΔG | −41.6 | −41.3 | −42.8 | −39.4 | −40 | −43 | −41.5 |
The thermodynamic parameters for reaction between a ligand and a protein are the main evidence for confirming the binding forces. The four types of non-covalent interactions include hydrogen bonds, van der Waals forces, electrostatic and hydrophobic bond interactions which play a key role in ligand binding to proteins.77 The value of the free energy change (ΔG) was calculated using van't Hoff equation (eqn (4))
ΔG = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka Ka
| (4) |
3.6. Powder X-ray diffraction studies accompanied by SEM and TEM images
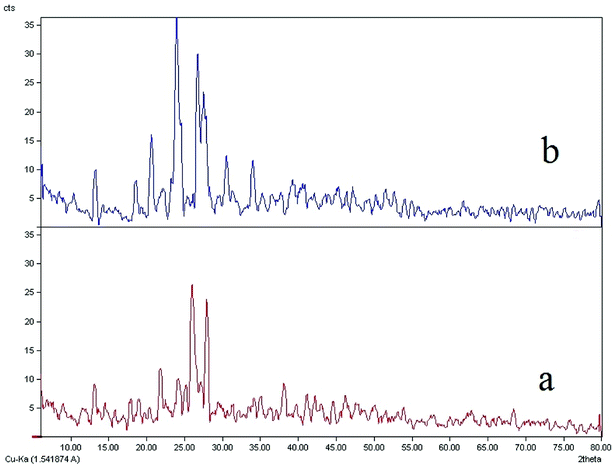
Comparisons of the XRD patterns for the nanoparticulate materials obtained by the ultrasonic method show the experimental data are in good agreement with the simulated XRD patterns obtained from single crystal data which also confirm the phase purity of these compounds (Fig. SI 8a–d†). The sharp bands of the samples indicate that the nanoparticles are well crystallized under these synthetic conditions and due to absence of a characteristic bar of impurities in the crystalline phase, the product is highly pure.In Fig. 5b, it is clear that the bands 20, 22, 23, 26 and 27 have much higher intensity which is due to the crystallization method of the sample. The nanoparticle size is estimated by the Debye–Scherrer formula:79
dRX = kλ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ θ
| (5) |
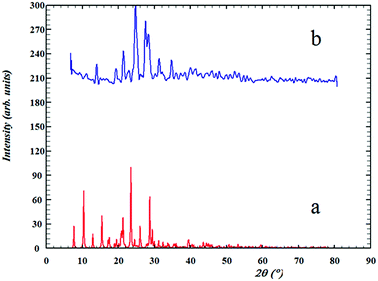 | ||
| Fig. 5 The XRD patterns of (a) computed from single-crystal X-ray data of ligand (L) and (b) nanostructure of L ligand. | ||
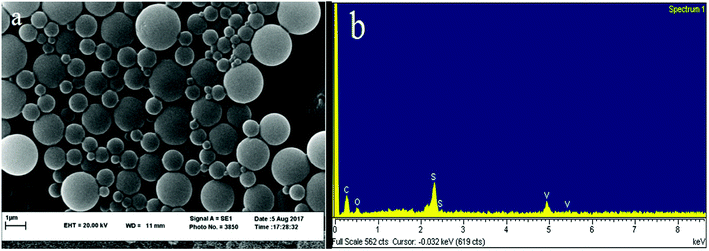
The XRD spectrum of the ligand is compared to the XRD spectrum of the complexes. Fig. 6 illustrates that in the complex spectrum of 2, some bands are removed or displaced regards to the ligand spectrum, indicating a reaction between the ligand and the metal. EDX analyses indicate that compound 2 is successfully obtained via the current synthetic route (Fig. 7b).
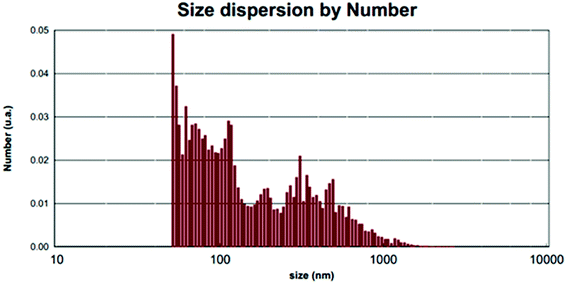
The average nanoparticle size for bands no. 21, 24, 25 and 27 is 90.4 nm by the Debye–Scherrer formula which is also confirmed by SEM images (Fig. 7a) and measurements based on PLS (Fig. 8). See SEM and TEM images, EDS analyses and PLS measurements for samples in SI 8e–o Section.†
4. Conclusion
In this work, we synthesized and characterized one novel L ligand and six metal complexes from the dithiocarbazate family. Structural analysis of compounds shows that the L ligand crystallizes as a dimer with S–S bond while in the complexation the S–S bond of the ligand is broken and L′ is formed. L′ act as a tridentate ONS donor ligand. All the complexes are six coordinated with distorted octahedral configuration. These compounds are designed to be able to bind to HSA protein and to have anticancer properties. Studying the interaction of compounds with HSA using fluorescence spectroscopy reveals that with increasing concentration of compounds at 298 K the fluorescence intensity of HAS decreases and also these compounds quench the fluorescence peak of HSA significantly even before their concentrations become equal to that of HSA. KSV values indicate that static quenching plays a major role in the quenching process and the role of dynamic quenching is almost insignificant. Moreover, the cellular toxicity of the compounds was tested against the Hela and MCF-7 cell lines. All compounds exhibit good cytotoxic activity when compared to the clinical drug, cisplatin. Even complexes 2 and 6 show better in vitro therapeutic index than cisplatin against Hela and MCF-7 cell lines. Furthermore, nanoparticles of the complexes are obtained by ultrasonic methods. XRD spectra of the samples confirm that the products are highly pure. The nanoparticle size is estimated by PLS analysis.Conflicts of interest
There are no conflict to declare.Acknowledgements
This work was supported by the Ferdowsi University of Mashhad (3/39699-1394/11/11) and Buali Research Institute. Authors thank Tulane University for support of the Tulane Crystallography Laboratory.References
- R. Takjoo, S. S. Hayatolgheibi and H. Amiri Rudbari, Inorg. Chim. Acta, 2016, 447, 52–58 CrossRef CAS.
- R. Centore, R. Takjoo, A. Capobianco and A. Peluso, Inorg. Chim. Acta, 2013, 404, 29–33 CrossRef CAS.
- M. Hakimi, K. Moeini, Z. Mardani and R. Takjoo, Phosphorus, Sulfur Silicon Relat. Elem., 2014, 189, 596–605 CrossRef CAS.
- T.-J. Khoo, M. K. b. Break, K. A. Crouse, M. I. M. Tahir, A. M. Ali, A. R. Cowley, D. J. Watkin and M. T. H. Tarafder, Inorg. Chim. Acta, 2014, 413, 68–76 CrossRef CAS.
- M. Eslami Moghadam, M. Saidifar, A. Divsalar, H. Mansouri-Torshizi, A. A. Saboury, H. Farhangian and M. Ghadamgahi, J. Biomol. Struct. Dyn., 2016, 34, 206–222 CrossRef CAS PubMed.
- Z. H. Siddik, Oncogene, 2003, 22, 7265–7279 CrossRef CAS PubMed.
- S. H. Lai, G. B. Jiang, J. H. Yao, W. Li, B. J. Han, C. Zhang, C. C. Zeng and Y. J. Liu, J. Inorg. Biochem., 2015, 152, 1–9 CrossRef CAS PubMed.
- M. Kudrat-E-Zahan and M. S. Islam, Russ. J. Gen. Chem., 2015, 85, 979–983 CrossRef CAS.
- R. Takjoo, R. Centore, M. Hakimi, S. Ali Beyramabadi and A. Morsali, Inorg. Chim. Acta, 2011, 371, 36–41 CrossRef CAS.
- A. Alim, Sci. J. Chem., 2015, 3, 35 CrossRef CAS.
- A. Taha, A. A. Emara, M. M. Mashaly and O. M. Adly, Spectrochim. Acta, Part A, 2014, 130, 429–439 CrossRef CAS PubMed.
- M. R. Maurya, S. Dhaka and F. Avecilla, Polyhedron, 2014, 81, 154–167 CrossRef CAS.
- E. Zangrando, M. T. Islam, M. A.-A. A. A. Islam, M. C. Sheikh, M. T. H. Tarafder, R. Miyatake, R. Zahan and M. A. Hossain, Inorg. Chim. Acta, 2015, 427, 278–284 CrossRef CAS.
- S. Kundu, D. Mondal, K. Bhattacharya, A. Endo, D. Sanna, E. Garribba and M. Chaudhury, Inorg. Chem., 2015, 54, 6203–6215 CrossRef CAS PubMed.
- B.-K. Koo and U. Lee, Bull. Korean Chem. Soc., 2002, 23, 613–616 CrossRef CAS.
- D.-W. KIM, U. LEE and B.-K. KOO, Bull. Korean Chem. Soc., 2004, 25, 1071–1074 CrossRef CAS.
- T. S. Morais, F. C. Santos, L. Corte-Real and M. H. Garcia, J. Inorg. Biochem., 2013, 129, 94–101 CrossRef CAS PubMed.
- A. Bijelic, S. Theiner, B. K. Keppler and A. Rompel, J. Med. Chem., 2016, 59, 5894–5903 CrossRef CAS PubMed.
- J. Toneatto and G. A. Arguello, J. Inorg. Biochem., 2011, 105, 645–651 CrossRef CAS PubMed.
- X. B. Fu, Z. H. Lin, H. F. Liu and X. Y. Le, Spectrochim. Acta, Part A, 2014, 122, 22–33 CrossRef CAS PubMed.
- T. Chatterjee, A. Pal, S. Dey, B. K. Chatterjee and P. Chakrabarti, PLoS One, 2012, 7, 37468 CrossRef PubMed.
- Y. Wang, X. Wang, J. Wang, Y. Zhao, W. He and Z. Guo, Inorg. Chem., 2011, 50, 12661–12668 CrossRef CAS PubMed.
- N. Shahabadi, A. Khorshidi and N. H. Moghadam, Spectrochim. Acta, Part A, 2013, 114, 627–632 CrossRef CAS PubMed.
- Y. Gou, Y. Zhang, J. Qi, Z. Zhou, F. Yang and H. Liang, J. Inorg. Biochem., 2015, 144, 47–55 CrossRef CAS PubMed.
- F. Faridbod, M. R. Ganjali, B. Larijani, S. Riahi, A. A. Saboury, M. Hosseini, P. Norouzi and C. Pillip, Spectrochim. Acta, Part A, 2011, 78, 96–101 CrossRef PubMed.
- Bruker, APE×3 and SADABS, Bruker AXS Inc., Madison, Wisconsin, USA, 2016 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3–8 CrossRef PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- K. Mohanan, R. Aswathy, L. P. Nitha, N. E. Mathews and B. S. Kumari, J. Rare Earths, 2014, 32, 379–388 CrossRef CAS.
- F. R. Pavan, P. I. d. S. Maia, S. R. A. Leite, V. M. Deflon, A. A. Batista, D. N. Sato, S. G. Franzblau and C. Q. F. Leite, Eur. J. Med. Chem., 2010, 45, 1898–1905 CrossRef CAS PubMed.
- H. L. Singh and A. K. Varshney, Bioinorg. Chem. Appl., 2006, 2006, 1–7 CrossRef PubMed.
- L. Ronconi, C. Maccato, D. Barreca, R. Saini, M. Zancato and D. Fregona, Polyhedron, 2005, 24, 521–531 CrossRef CAS.
- Y. Zhang, X. Wang, W. Fang, X. Cai, F. Chu, X. Liao and J. Lu, Bioinorg. Chem. Appl., 2013, 2013, 437134 Search PubMed.
- N. R. Pramanik, M. Chakraborty, D. Biswal, S. S. Mandal, S. Ghosh, S. Chakrabarti, W. S. Sheldrick, M. G. Drew, T. K. Mondal and D. Sarkar, Polyhedron, 2015, 85, 196–207 CrossRef CAS.
- N. Gandhi, A. Kumar, C. Kumar, N. Mishra, P. Chaudhary, N. K. Kaushik and R. Singh, Main Group Chem., 2015, 15, 35–46 CAS.
- J. U. Mondal, J. G. Zamora, M. D. Kinon and F. A. Schultz, Inorg. Chim. Acta, 2000, 309, 147–150 CrossRef CAS.
- C. Das, P. Adak, S. Mondal, R. Sekiya, R. Kuroda, S. I. Gorelsky and S. K. Chattopadhyay, Inorg. Chem., 2014, 53, 11426–11437 CrossRef CAS PubMed.
- A. Mathavan, A. Ramdass and S. Rajagopal, Transition Met. Chem., 2015, 40, 355–362 CrossRef CAS.
- K. Bhattacharya, M. Maity, S. M. T. Abtab, M. C. Majee and M. Chaudhury, Inorg. Chem., 2013, 52, 9597–9605 CrossRef CAS PubMed.
- S. Zaidi, A. K. Chaturvedi, N. Singh and D. Chaturvedi, Curr. Chem. Lett., 2017, 6, 143–150 CrossRef.
- E. Zangrando, M. S. Begum, M. C. Sheikh, R. Miyatake, M. M. Hossain, M. M. Alam, M. A. Hasnat, M. A. Halim, S. Ahmed, M. N. Rahman and A. Ghosh, Arabian J. Chem., 2017, 10, 172–184 CrossRef CAS.
- D. A. Abdel-Latif, H. M. Youssef and Y. G. Abou El Reash, J. Mol. Liq., 2017, 241, 456–468 CrossRef CAS.
- M. Yazdanbakhsh and R. Takjoo, Struct. Chem., 2008, 19, 895–903 CrossRef CAS.
- R. Takjoo, R. Takjoo, M. Yazdanbakhsh, A. Aghaei kaju and Y. Chen, Chin. J. Chem., 2010, 28, 221–228 CrossRef CAS.
- M. Amirnasr, M. Bagheri, H. Farrokhpour, K. J. Schenk, K. Mereiter and P. C. Ford, Polyhedron, 2014, 71, 1–7 CrossRef CAS.
- A. Kumar, P. Chaudhary, R. Singh and N. Kaushik, Main Group Chem., 2016, 15, 163–178 CAS.
- A. A. Alshaheri, M. I. M. Tahir, M. B. A. Rahman, T. B. S. A. Ravoof and T. A. Saleh, Chem. Eng. J., 2017, 327, 423–430 CrossRef CAS.
- N. Nanjundan, R. Narayanasamy, R. J. Butcher, J. P. Jasinski, K. Velmurugan, R. Nandhakumar, M. D. Balakumaran, P. T. Kalaichelvan and V. G. Gnanasoundari, Inorg. Chim. Acta, 2017, 455, 283–297 CrossRef CAS.
- S. A. Elsayed, A. M. Noufal and A. M. El-Hendawy, J. Mol. Struct., 2017, 1144, 120–128 CrossRef CAS.
- T. Ahmad, K. V. Ramanujachary, S. E. Lofland and A. K. Ganguli, J. Mater. Chem., 2004, 14, 3406–3410 RSC.
- A. Bartyzel, J. Coord. Chem., 2013, 66, 4292–4303 CrossRef CAS.
- M.-J. Niu, D.-W. Sun, H.-H. Li, Z.-Q. Cao, S.-N. Wang and J.-M. Dou, J. Coord. Chem., 2014, 67, 81–95 CrossRef CAS.
- I. Dalle-Donne, D. Giustarini, R. Colombo, A. Milzani and R. Rossi, Free Radical Biol. Med., 2005, 38, 1501–1510 CrossRef CAS PubMed.
- N. R. Pramanik, S. Ghosh, T. K. Raychaudhuri, S. Chaudhuri, M. G. B. Drew and S. S. Mandal, J. Coord. Chem., 2007, 60, 2177–2190 CrossRef CAS.
- N. R. Pramanik, S. Ghosh, T. K. Raychaudhuri, S. Ray, R. J. Butcher and S. S. Mandal, Polyhedron, 2004, 23, 1595–1603 CrossRef CAS.
- S. K. Dutta, D. B. McConville, W. J. Youngs and M. Chaudhury, Inorg. Chem., 1997, 36, 2517–2522 CrossRef CAS.
- G. F. Sousa, C. C. Gatto, J. Ellena and J. D. Ardisson, J. Chem. Crystallogr., 2011, 41, 838–842 CrossRef CAS.
- R. Mukhopadhyay, S. Bhattacharjee, C. K. Pal, S. Karmakar and R. Bhattacharyya, J. Chem. Soc., Dalton Trans., 1997, 2267–2272 RSC.
- M. A. Ali, A. H. Mirza, M. H. S. A. Hamid, F. H. Bujang and P. V. Bernhardt, Polyhedron, 2004, 23, 2405–2412 CrossRef CAS.
- E. J. Gao, L. Wang, M. C. Zhu, L. Liu and W. Z. Zhang, Eur. J. Med. Chem., 2010, 45, 311–316 CrossRef CAS PubMed.
- Q. Mi, Y. Ma, X. Gao, R. Liu, P. Liu, Y. Mi, X. Fu and Q. Gao, J. Biomol. Struct. Dyn., 2016, 34, 2339–2350 CrossRef CAS PubMed.
- A. I. Matesanz, J. M. Pérez, P. Navarro, J. M. Moreno, E. Colacio and P. Souza, J. Inorg. Biochem., 1999, 76, 29–37 CrossRef CAS PubMed.
- S. Moradell, J. Lorenzo, A. Rovira, S. van Zutphen, F. X. Avilés, V. Moreno, R. de Llorens, M. A. Martinez, J. Reedijk and A. Llobet, J. Inorg. Biochem., 2004, 98, 1933–1946 CrossRef CAS PubMed.
- B. Valeur, Molecular Fluorescence Principles and Applications, Wiley-VCH Verlag GmbH, 2001, ISBNs: 3-527-29919-X (Hardcover); 3-527-60024-8 (Electronic) Search PubMed.
- L. Chen and X. Chen, J. Mol. Graphics Modell., 2012, 33, 35–43 CrossRef CAS PubMed.
- K. Kaneko, V. T. Chuang, A. Minomo, K. Yamasaki, N. V. Bhagavan, T. Maruyama and M. Otagiri, IUBMB Life, 2011, 63, 277–285 CrossRef CAS PubMed.
- F. Kratz, J. Controlled Release, 2008, 132, 171–183 CrossRef CAS PubMed.
- G. Fanali, A. di Masi, V. Trezza, M. Marino, M. Fasano and P. Ascenzi, Mol. Aspects Med., 2012, 33, 209–290 CrossRef CAS PubMed.
- J. Ghuman, P. A. Zunszain, I. Petitpas, A. A. Bhattacharya, M. Otagiri and S. Curry, J. Mol. Biol., 2005, 353, 38–52 CrossRef CAS PubMed.
- U. Kragh-Hansen, Biochim. Biophys. Acta, 2013, 1830, 5535–5544 CrossRef CAS PubMed.
- S. Sugio, A. Kashima, S. Mochizuki, M. Noda and K. Kobayashi, Protein Eng., 1999, 12, 439–446 CrossRef CAS.
- B. Rastegari, H. R. Karbalaei-Heidari, R. Yousefi, S. Zeinali and M. Nabavizadeh, Bioorg. Med. Chem., 2016, 24, 1504–1512 CrossRef CAS PubMed.
- L. Tabrizi, P. McArdle, A. Erxleben and H. Chiniforoshan, Eur. J. Med. Chem., 2015, 103, 516–529 CrossRef CAS PubMed.
- L. Yan, X. Wang, Y. Wang, Y. Zhang, Y. Li and Z. Guo, J. Inorg. Biochem., 2012, 106, 46–51 CrossRef CAS PubMed.
- T. S. Morais, F. C. Santos, L. Corte-Real and M. H. Garcia, J. Inorg. Biochem., 2013, 129, 94–101 CrossRef CAS PubMed.
- I. Matei and M. Hillebrand, J. Pharm. Biomed. Anal., 2010, 51, 768–773 CrossRef CAS PubMed.
- M. Tabatabaee, M. Ghassemzadeh, A. R. Dehghan, H. R. Khavasi and M. M. Heravi, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2006, 63, o42–o43 CrossRef.
- H. Liu, X. Shi, M. Xu, Z. Li, L. Huang, D. Bai and Z. Zeng, Eur. J. Med. Chem., 2011, 46, 1638–1647 CrossRef CAS PubMed.
- H. P. Klug and L. E. Alexander, X-Ray Diffraction Procedures: For Polycrystalline and Amorphous Materials, ed. Harold P. Klug and Leroy E. Alexander, Wiley-VCH, 2nd edn, p. 992. ISBN 0-471-49369-4, May 1974 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: CCDC 1851305–1851311 contains the supplementary crystallographic data for L and 1 to 6. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8ra07100d |
| This journal is © The Royal Society of Chemistry 2018 |