Sustainable production of medium chain fatty acids (MCFA) with an enriched mixed bacterial culture: microbial characterization using molecular methods
M. Venkateswar
Reddy
 *a,
S. Venkata
Mohan
b and
Young-Cheol
Chang
*a,
S. Venkata
Mohan
b and
Young-Cheol
Chang
 *a
*a
aDepartment of Applied Sciences, College of Environmental Technology, Muroran Institute of Technology, 27-1 Mizumoto, Muroran, 050-8585, Japan. E-mail: mvr_234@yahoo.co.in; ychang@mmm.muroran-it.ac.jp; Fax: +81-143-46-5757; Tel: +81-80-9001-5196 Tel: +81-143-46-5757
bBioengineering and Environmental Sciences (BEES), CSIR-Indian Institute of Chemical Technology (CSIR-IICT), Hyderabad 500007, India
First published on 6th November 2017
Abstract
Chain elongation is the process by which bacteria convert ethanol and short chain fatty acids (SCFA) into medium chain fatty acids (MCFA). In the present study, a bacterial mixed culture was enriched and used together with synthetic waste (SW) to produce commercially valuable SCFA and MCFA for 91 days by anaerobic fermentation. For the first time, the effects of substrate, electron donor, and methane inhibition on MCFA production were evaluated. The produced SCFA, MCFA and biogas were analyzed using chromatography techniques. Butyrate (C4) and caproate (C6) were the dominant products in SCFA and MCFA, respectively. Bacteria in conditions of methane inhibition produced the highest concentration of butyrate (8.2 g l−1), caproate (8.6 g l−1), heptanoate (0.7 g l−1), and caprylate (0.23 g l−1). The dominance of Clostridia, Rumen bacterium, and the Actinomyces group in the enriched culture were identified by denaturing gradient gel electrophoresis (DGGE) analysis. Studies are being conducted under pipeline with enriched cultures to produce SCFA and MCFA using the real waste streams.
1. Introduction
Anaerobic fermentation processes based on biomass and mixed cultures are being developed to produce chemicals and fuels.1–3 In one of these processes called chain elongation, bacteria convert ethanol and short chain fatty acids (SCFA) into medium chain fatty acids (MCFA). MCFA are saturated monocarboxylic acids with six to eight carbon atoms.1–4 Chain elongation is an interesting process because the MCFA products can be used as antimicrobials,5 corrosion inhibitors,6 and precursors in biodiesel7 and bioplastics8 production. So far, it is unknown which products are produced most favourably. The precursors of MCFA, such as SCFA and ethanol, can be produced from lignocellulosic materials9,10 which cannot be consumed by humans. This way, the food-fuel argument can be evaded for the chain elongation process. Chain elongation can be integrated into existing biorefineries which produce ethanol from sugar and have organic residues available for SCFA production. The chain elongation process adds value to ethanol, and the energy needed to separate the MCFA from the broth is likely lower than that required for ethanol distillation due to the low solubility of the MCFA.4 Chain elongation has been recently reported in microbial electrosynthesis systems (MES) by researchers. Bajracharya et al. (2017) aimed to develop a stable and robust CO2-reducing biocathode from a mixed culture, evading methane production.11 Biomass growth and steady acclimation to CO2 electro-reduction gifted a production of acetate and butyrate by a chain elongation process. Schievano et al. (2016) and Roy et al. (2016) reported the production of various chemicals through chain elongation processes in electro-fermentation and bioelectrochemical systems, respectively.12,13Chain elongation can be performed under non-sterile conditions with a mixed microbial culture. Consequently, a sterilization step for the feedstock is not necessary. Chain elongation can use organic residues as a source for SCFA production. Acidification of organic waste leads to the generation of SCFA, specifically if methanogens are inhibited. Acetate uptake by methanogens can be achieved by the lowering of pH,9 and other methods such as heat shocks are also used to diminish the influence of methanogens. Attempts have been made to produce MCFA with organic residues. Kenealy et al. (1995) reported the production of caproate (C6) from cellulose and ethanol with a defined co-culture.14 Grootscholten et al. (2013) used mixed cultures to produce MCFA from municipal waste and ethanol in a single stage reactor.1 Nonetheless, the hydrolysis rate of the municipal waste with ethanol supplementation was lower than that without ethanol supplementation. Higher concentrations of undissociated MCFA and ethanol might be the reason for the lower hydrolysis rate. To decrease the effect of undissociated MCFA, these components could be removed from the reactor by in-line extraction.4 However, even if in-line extraction were effective in removing the toxicity of undissociated MCFA, ethanol toxicity might still affect the hydrolysis rate. The stability of the chain elongation process using a mixed culture is affected by competitive processes like excessive oxidation of ethanol to acetate, acetotrophic methanogenesis, and MCFA oxidation to acetate.2,15 These competitive processes should be controlled to establish effective MCFA production. The objective of this work, therefore, was to enrich mixed bacterial cultures for production of MCFA through anaerobic fermentation. The evaluation of MCFA production was conducted under various conditions i.e., with methane inhibition, without methane inhibition, without electron donors, and without substrate. The dominant MCFA producers present in the enriched mixed bacterial cultures were identified using denaturing gradient gel electrophoresis (DGGE). Meanwhile, since acetate is the chief intermediate product of anaerobic digestion, we used acetate as a substrate in the synthetic waste in our viability study.
2. Materials and methods
2.1 Biocatalyst
A mixed anaerobic culture obtained from Gifu University, Japan, was used for enrichment. Enrichment was done by growing the mixed culture in synthetic medium. Serum bottles of 120 ml with butyl rubber stoppers and aluminium caps were used; each bottle contained synthetic medium and the mixed anaerobic culture. The synthetic medium contained 1.0 g (NH4)2SO4, 1.0 g K2HPO4, 0.2 g NaH2PO4, 0.2 g MgSO4·7H2O, 0.05 g NaCl, 0.05 g CaCl2, 8.3 mg FeCl3·6H2O, 1.4 mg MnCl2·4H2O, 1.17 mg Na2MoO4·2H2O, and 1 mg ZnCl2 per liter of deionized water. A solution of trace elements was added to the SW at a concentration of 1 ml l−1. The trace element solution contained 0.786 g CuSO4·5H2O, 5.0 g FeSO4·7H2O, 12.609 g NaMoO4·2H2O, 4.05 g NiCl2·6H2O, 4.398 g ZnSO4·7H2O, 2.453 g CoCl2·6H2O, 0.75 g KI, 3.0 g H3BO3, 5.0 g MnCl2·4H2O and 5.0 g EDTA per liter of distilled water. The synthetic medium contained commercial SCFA such as acetate (8 g l−1), propionate (1 g l−1), and butyrate (1 g l−1) as carbon sources. Ethanol (10 g l−1) was also added as the electron donor to stimulate the chain elongation process for the production of MCFA. Before inoculation, the pH was adjusted to 7 with 5 M NaOH solution. After inoculation with bacteria, the bottles were closed and capped. The headspace was flushed with nitrogen for 5 minutes. The bottles were incubated at 37 °C in a rotating shaker at 120 rpm for 12 days. Liquid samples were anaerobically taken, centrifuged in a reaction tube and used for SCFA and MCFA analysis. Enrichment was done in three repeated cycles; each cycle was carried out for 12 days. During enrichment cycles, bacteria effectively utilized SCFA and ethanol.2.2 Synthetic waste (SW)
The SW was prepared as suggested by the Biological Resource Center, National Institute of Technology and Evaluation (NBRC), Japan with some modifications. It contained 8 g sodium acetate, 1 g yeast extract, 27 g ethanol, 2 g (NH4)2SO4, 2 g K2HPO4, 0.4 g NaH2PO4, 0.10 g NaCl, 0.2 g MgSO4·7H2O, 0.05 g CaCl2, 8.3 mg FeCl3·6H2O, 1.4 mg MnCl2·4H2O, 1.2 mg Na2MoO4·2H2O, and 1 mg ZnCl2 per liter of deionized water. The pH of the SW was adjusted to 7 and it was autoclaved before being added to the serum bottles. Ethanol was added after sterilization.2.3 SCFA and MCFA production
Experiments were performed in triplicate in this study. Serum bottles of 120 ml with butyl rubber stoppers and aluminium caps were used for all of the experiments. Each bottle contained 40 ml of SW, and the pH was adjusted to 7 with 5 M NaOH solution before inoculation. For SCFA and MCFA production, 2 ml of the pre-grown enriched culture was inoculated into the different serum bottles containing SW. MCFA production from acetate was stimulated by the addition of ethanol as an electron donor. After inoculation with bacteria, the bottles were closed and capped. The headspace was flushed with nitrogen for 5 minutes. The bottles were incubated at 37 °C in a rotating shaker at 120 rpm for 91 days. Liquid samples were anaerobically taken and collected in a reaction tube. The liquid samples were centrifuged (5 min, 8000 × g) and used for SCFA and MCFA analysis as mentioned in Sections 2.4.1 and 2.4.2. Biogas present in the head space was measured as mentioned in Section 2.4.3.2.4 Analysis
2.5 Microbial diversity analysis
Denaturing gradient gel electrophoresis (DGGE) analysis was performed for the identification of the dominant organisms present in the mixed culture, and to evaluate their role in the production of MCFA. Samples were collected from the serum bottles at different time intervals, i.e., day 0, day 30 and day 60. DNA extraction, amplification of the 16S rRNA genes, and DGGE analysis of total community DNA were determined as described by Reddy and Mohan.163. Results and discussion
3.1 Acetate concentration decrement
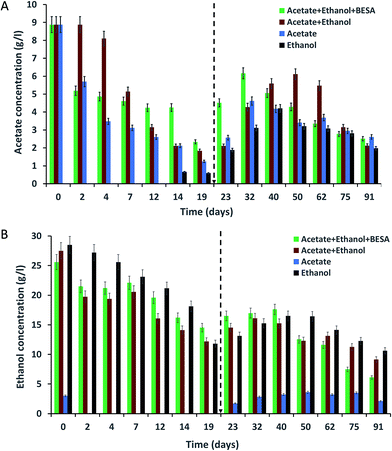
Enriched bacterial culture were inoculated into SW and incubated for 91 days at 37 °C supplemented with acetate or ethanol or a mixture of both acetate and ethanol. The acetate concentrations in the SW were analyzed at different time intervals using HPLC (Fig. 1A). For all of the experiments, the initial (day 0) acetate concentration was 8.8 g l−1 and it showed a continuous decrement up to the 19th day. Experiment-3 showed the highest removal of acetate on day 19 (87%) followed by experiment-2 (80%) and experiment-1 (74%). Even though acetate was properly utilized by the bacteria, significant amounts of MCFA were not produced until the 19th day. Hence, 10 ml of SW was added to the serum bottles on day 19 in order to maintain the acetate concentration for chain elongation which can lead to enhanced MCFA production.After adding 10 ml of SW, the acetate concentration increased in all of the experiments for a certain period and thereafter decreased. In experiment-1, the acetate concentration increased from 2.9 to 4.5 g l−1, then decreased until the end of the experiment. A higher increment (1.2–6.1 g l−1) in the acetate concentration was observed in experiment-2. After a certain time, the acetate concentration showed a gradual decrement due to the utilization of acetate for butyrate and MCFA production through the chain elongation process. This was supported by the increment in butyrate and MCFA production after the 19th day. The experiment conducted without acetate in the SW (experiment-4) did not showed acetate up to day 12, but a small amount of acetate was observed on the 14th day (0.66 g l−1). Even though we did not provided acetate, the enriched culture synthesized acetate from ethanol. Vasudevan et al. (2014) reported the complete utilization of acetate (2.3 g l−1) by an enriched mixed culture by the 25th day.17 Contrary to this, Steinbusch et al. (2011) reported that acetate (5 g l−1) was not completely utilized by an enriched mixed culture within 117 days.3 In the present study also, acetate was not completely utilized by the enriched mixed culture.
3.2 Ethanol concentration decrement
Changes in the ethanol concentration at different time intervals were analyzed for all of the experiments (Fig. 1B). The same amount of ethanol was used to supplement the SW in experiments-1, 2 and 4. After inoculating the SW with bacteria on day 0, the ethanol concentrations were varied (i.e., experiment-1: 25.6 g l−1; experiment-2: 27.5 g l−1; and experiment-4: 28.5 g l−1). A decrease in ethanol concentration was observed over time. Ethanol removal by the 19th day was high in experiment-4 (59%) followed by experiment-2 (56%) and experiment-1 (44%). Even though ethanol was being properly utilized, the bacteria produced lower amounts of SCFA and MCFA. After adding SW to the serum bottles on the 19th day, the concentration of ethanol, SCFA and MCFA was increased.Experiments conducted without ethanol (experiment-3) also showed 2.2 g l−1 of ethanol on day 0; this may be due to the carry-over of ethanol from the pre-culture medium. From the 2nd day, ethanol was not observed in the SW due to its utilization by the bacteria. After the 19th day, the ethanol concentration increased, reaching 3.62 g l−1 (50th day) and then decreased. Reports are available about the complete utilization of ethanol by mixed bacterial culture. Vasudevan et al. (2014) reported the complete utilization of ethanol (11.4 g l−1) on the 20th day by using a two year old enriched mixed culture.17 Steinbusch et al. (2011) reported that after 40 days, ethanol (4 g l−1) was completely consumed by an enriched mixed culture.3 However, in our study, we did not observe the complete utilization of ethanol by the enriched culture.
3.3 SCFA production
The concentrations of SCFA, like butyric (C4) and valeric (C5) acids, were identified at different time intervals for all of the experiments. Bacteria in experiment-1 showed high amounts of butyric acid due to the presence of BESA (Fig. 2A). BESA inhibits the methanogenic process and directs the metabolic pathway towards fatty acid production.3 C4 production started from the 7th day (0.62 g l−1), and the concentration increased up to the 19th day (3.03 g l−1). The highest production was observed on the 40th day (8.25 g l−1) followed by the 91st day (6.72 g l−1), and 75th day (6.46 g l−1). Experiment-2 also showed a good amount of C4 production. Production started from the 2nd day (0.79 g l−1), and after that it increased i.e., 1.76 g l−1 on the 7th day, and 1.73 g l−1 on the 19th day (Fig. 2A). Higher butyric acid production was observed on the 62nd day (6.48 g l−1) and 91st day (5.56 g l−1). Compared with experiment-2, experiment-1 showed 2.07 times higher C4 production on the 40th day. Experiments conducted with only acetate (experiment-3) and only ethanol (experiment-4) did not show C4 production up until the 19th day (data not shown). After adding 10 ml of SW on the 19th day, C4 production was initiated. Experiment-3 showed a low amount of C4 on the 19th day (0.47 g l−1). The production was high on the 91st day (3.33 g l−1). Experiment-4 showed a good concentration of C4 on the 32nd day (2.35 g l−1). Butyric acid is a key precursor for the production of MCFA from acetate and ethanol. Many authors have reported the production of butyric acid from acetate and ethanol by C. kluyveri. Barker et al. (1945) reported that C. kluyveri can metabolize acetate and ethanol under anaerobic conditions and produce hydrogen, butyric acid, and caproic acids.18 They noted that if acetate is present in excess, butyric acid is the main product, while if ethanol is present in excess, caproic acid is the main product.18 Butyric and caproic acids are formed simultaneously in their study rather than successively during part of the fermentation.18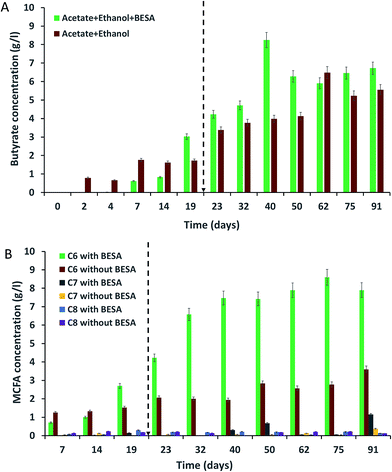 | ||
| Fig. 2 Production of (A) butyrate (C4); and (B) MCFA (caproate-C6, heptanoate-C7, caprylate-C8) from synthetic waste using enriched mixed bacterial cultures. | ||
Among all of the experiments, experiment-2 showed the highest amounts of C5. The highest concentration of C5 was observed on the 62nd day (2.16 g l−1) followed by the 14th day (1.55 g l−1), 91st day (0.19 g l−1), and 75th day (0.18 g l−1). Bacteria in experiment-1 produced their highest concentration of C5 on the 62nd day (0.81 g l−1), followed by the 75th day (0.44 g l−1), 91st day (0.42 g l−1), and 32nd day (0.34 g l−1). Experiments-3 and 4 did not show C5 production until the 40th day; 0.21 and 0.31 g l−1 of C5 were observed on the 75th day for experiments-3 and 4, respectively. Researchers have previously reported the production of valeric acid (C5) during anaerobic fermentation. Ding et al. (2010) reported the production of 160 and 450 mg l−1 of C5 from sucrose and lactose as substrates, respectively, using mixed culture as inoculum.19 Weimer et al. (2015) reported 1.6 g l−1 of C5 production using co-cultures of Clostridium kluyveri 3231B with mixed ruminal bacteria fermented with mixtures of cellulosic biomass and ethanol.20 Steinbusch et al. (2011) reported the presence of C5 in the medium fermented with a mixed microbial culture,3 but they did not include quantitative results for C5. C5 was likely produced by reverse β-oxidation reactions involving ethanol as the electron donor and propionate as the electron acceptor.20 Compared with that of C4, C5 production was low in this study. Further research is necessary to find out the reason for this.
3.4 MCFA production by chain elongation
The enriched bacterial culture mainly produced three types of MCFA, i.e., caproate (C6), heptanoate (C7), and caprylate (C8). C6 was the dominant product in all of the experiments. Bacteria in experiment-1 produced high amounts of C6 due to the presence of BESA (Fig. 2B). BESA inhibits the methanogenic process and directs the metabolic pathway towards fatty acid production.3 Bacteria started to produce C6 from the 7th day (0.71 g l−1), and thereafter, production was enhanced with increasing time, i.e., 4.22 g l−1 on the 23rd day, 6.58 g l−1 on the 32nd day, and 7.47 g l−1 on the 40th day. Bacteria produced the highest concentration of C6 on the 75th day (8.6 g l−1) followed by the 91st day (7.9 g l−1), and 62nd day (7.89 g l−1). After experiment-1, bacteria in experiment-2 showed the next best levels of C6 production. Production was highest on the 91st day (3.6 g l−1), followed by the 50th day (2.83 g l−1), 75th day (2.78 g l−1), 62nd day (2.57 g l−1), and 23rd day (2.06 g l−1). BESA showed a significant influence on MCFA production, providing a 3.1-fold enhancement on the 75th day. However, BESA is an expensive chemical, so its usage for MCFA production is not feasible in large scale studies. Further research is required for the replacement of BESA with a low cost chemical, and to find alternative methods to inhibit methane production and subsequently enhance MCFA production. Experiments conducted with acetate (experiment-3) and ethanol (experiment-4) showed C6 production after the 32nd day (data not shown). Experiment-3 showed the highest C6 production on the 91st day (2.08 g l−1), followed by the 50th day (2.04 g l−1) and 40th day (1.89 g l−1). Experiment-4 showed C6 production on the 40th day (0.89 g l−1), 62nd day (0.71 g l−1), 75th day (0.81 g l−1), and 91st day (0.73 g l−1).Unlike the C6 production, bacteria produced lower amounts of C7 and C8 (Fig. 2B). Bacteria in experiment-1 produced 0.7 g l−1 of C7 on the 50th day, while experiment-2 gave 0.4 g l−1 on the 91st day, and experiment-3 gave 0.18 g l−1 on the 75th day. Experiments-1 and 3 produced 0.21 and 0.19 g l−1 of C8 respectively on the 40th day. Experiment-2 showed 0.23 g l−1 of C8 on the 14th day. Stable MCFA production was not observed in experiments-3 and 4 due to the lack of ethanol and acetate, respectively. Yin et al. (2017) conducted batch experiments to investigate the effect of the acetate/ethanol ratio and initial ethanol concentration on caproate production, and they reported that the highest caproate concentration of 8.42 g l−1 was achieved from high ethanol strength wastewater with an ethanol/acetate ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (550 mM total carbon).21 Leng et al. (2017) considered thermodynamic and physiological factors in the co-production of 1,3-propanediol and caproate from crude glycerol.22 Caproate formation was increased in their studies by intermittent ethanol supplementation, with the ideal caproate generation attained at the ethanol–acetate molar ratio of 3. Kucek et al. (2016) reported that in ethanol-fed systems, even-numbered carboxylates (butyrate, caproate) are the predominant products produced via the reverse β-oxidation pathway.23 Steinbusch et al. (2011) reported that C6 was mainly produced by the enriched mixed culture, and that the production of caproate (8.27 g l−1) and caprylate (0.32 g l−1) were observed from 50 mM ethanol and 50 mM acetate.3 Kenealy et al. (1995) used co-cultures of ruminal cellulolytic bacteria with C. kluyveri. They reported the production of 2.6 g l−1 of butyrate and 4.6 g l−1 of caproate from 4.4 g l−1 ethanol and 6.0 g l−1 cellulose.14 Ding et al. (2010) used undefined mixed cultures and reported the production of acetate (0.6–1.2 g l−1), butyrate (0.87–1.74 g l−1), and caproate (1.27–2.66 g l−1) from 20 g l−1 of glucose substrate.19 Grootscholten et al. (2013) reported the production of butyrate (1 g l−1), caproate (11.1 g l−1), and caprylate (0.6 g l−1) with a mixed culture by using 34 g l−1 acetate and 71.5 g l−1 ethanol.1 Ethanol and acetate were converted into MCFA by ethanol–acetate metabolism. The ethanol–acetate metabolism is quite unique for anaerobes and can be described by three coupled reactions.3,4 First, ethanol is converted to acetate by producing NADH and ATP via substrate-level phosphorylation. Second, the fatty acid (as the acetate) is elongated in a cyclic pathway to butyrate using CoA, NADH and FADH2 through the coupling of two acetyl-CoA moieties to butyryl-CoA in a cyclic loop. Third, further chain elongation to caproate occurs from butyrate and ethanol in a similar loop by coupling butyryl-CoA with acetyl-CoA. This mechanism results in the addition of two carbons to the original carboxylic acid in every loop.
1 (550 mM total carbon).21 Leng et al. (2017) considered thermodynamic and physiological factors in the co-production of 1,3-propanediol and caproate from crude glycerol.22 Caproate formation was increased in their studies by intermittent ethanol supplementation, with the ideal caproate generation attained at the ethanol–acetate molar ratio of 3. Kucek et al. (2016) reported that in ethanol-fed systems, even-numbered carboxylates (butyrate, caproate) are the predominant products produced via the reverse β-oxidation pathway.23 Steinbusch et al. (2011) reported that C6 was mainly produced by the enriched mixed culture, and that the production of caproate (8.27 g l−1) and caprylate (0.32 g l−1) were observed from 50 mM ethanol and 50 mM acetate.3 Kenealy et al. (1995) used co-cultures of ruminal cellulolytic bacteria with C. kluyveri. They reported the production of 2.6 g l−1 of butyrate and 4.6 g l−1 of caproate from 4.4 g l−1 ethanol and 6.0 g l−1 cellulose.14 Ding et al. (2010) used undefined mixed cultures and reported the production of acetate (0.6–1.2 g l−1), butyrate (0.87–1.74 g l−1), and caproate (1.27–2.66 g l−1) from 20 g l−1 of glucose substrate.19 Grootscholten et al. (2013) reported the production of butyrate (1 g l−1), caproate (11.1 g l−1), and caprylate (0.6 g l−1) with a mixed culture by using 34 g l−1 acetate and 71.5 g l−1 ethanol.1 Ethanol and acetate were converted into MCFA by ethanol–acetate metabolism. The ethanol–acetate metabolism is quite unique for anaerobes and can be described by three coupled reactions.3,4 First, ethanol is converted to acetate by producing NADH and ATP via substrate-level phosphorylation. Second, the fatty acid (as the acetate) is elongated in a cyclic pathway to butyrate using CoA, NADH and FADH2 through the coupling of two acetyl-CoA moieties to butyryl-CoA in a cyclic loop. Third, further chain elongation to caproate occurs from butyrate and ethanol in a similar loop by coupling butyryl-CoA with acetyl-CoA. This mechanism results in the addition of two carbons to the original carboxylic acid in every loop.
3.5 Biogas composition
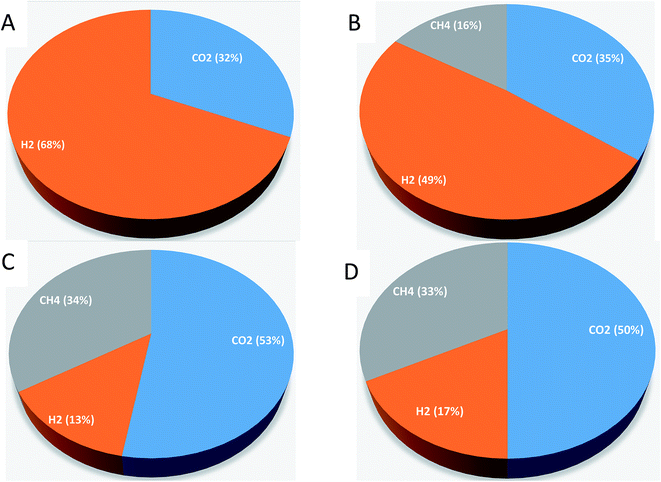
Gas samples were taken in order to measure the CO2, H2 and CH4 percentages in the produced biogas by analyzing the sample with GC-TCD. Samples were measured on 40th day. For experiment-1, the biogas contained 32 % CO2 and 68% H2 (Fig. 3). For experiment-2, CO2 (35%), H2 (49%), and CH4 (16%) were observed on the 40th day. For experiment-3, the biogas contained 53% CO2, 13% H2, and 34% CH4; for experiment-4, CO2 (50%), H2 (17%), and CH4 (33%) were observed on the 40th day. Compared with CO2 and H2, a lower percentage of CH4 was observed. Grootscholten et al. (2013) reported that the CH4 percentage in the gas phase of the reactors was below one percent.1 Zhu et al. (2015) also noted the low CH4 percentage in their studies.24 They reported the biogas composition of CO2 (41%), H2 (42%) and CH4 (0.3%) by using yellow water as the substrate (brown and sticky muddy liquid produced from solid-fermentation contains 96.5 g l−1 of lactic acid, 50.5 g l−1 of ethanol, and 15.6 g l−1 of glucose) and pit mud as the source of the inoculum. Agler et al. (2011) also reported a lesser amount of CH4 production in the mixed culture reactor using beer from the ethanol industry for chain elongation of SCFA.9 Contrary to our results, Steinbusch et al. (2011) reported CO2 measurements that were below the detection limit in their experiments.3 The produced H2 in our experiments might be used for MCFA production. Steinbusch et al. (2011) reported that it is feasible to produce MCFA from acetate using both ethanol and H2 as suitable electron donors.33.6 Change in pH
The initial pH of the SW was adjusted to 7 before it was added to the serum bottles. The pH showed a decreasing trend with time for all of the experiments, which might be due to the production of SCFA and MCFA by the enriched mixed culture (Fig. 4). The decreasing trend continued up to the 19th day, but after adding SW on the 19th day, the pH was slightly increased until the 23rd day and then it decreased again. Experiment-1 showed a pH of 6.9 on day 0, which decreased to 5.3 by the 19th day. The pH then decreased from 5.54 (on the 23rd day) to 5.48 (on the 91st day). For experiment-2, the pH decreased from 6.8 (on day 0) to 5.17 (on the 91st day). Experiment-3 showed a decrement of pH from 6.8 to 4.99. Experiment-4 showed a decrement of pH from 6.9 (on day 0) to 5.32 (on the 23rd day) and it decreased further to 4.87 by the 91st day. The observed lower C8 concentrations in this study might be due to acidic pH values. Steinbusch et al. (2011) reported that higher MCFA production was observed at pH 7 when acetate, ethanol and H2 were fermented at different pH conditions, i.e., pH 5.5 and 7.3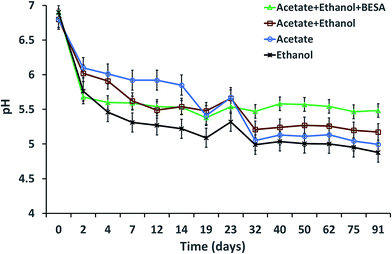 | ||
| Fig. 4 pH variation with respect to time during MCFA production under various experimental conditions. | ||
3.7 Microbial characterization
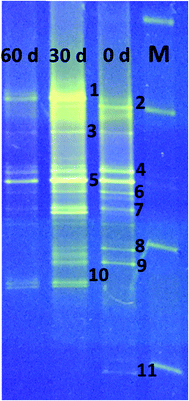
DGGE analysis was used to determine the structural composition of the bacteria present in the enriched mixed culture on days 0, 30, and 60 of the experiments. Among all the bands, 11 dominant bands were observed, which were phylogenetically related to phyla Firmicutes (54%), Bacteroidia (27%), Actinobacteria (9%), and rumen bacteria (9%). All the bacteria are related to classes Clostridia (36%), un-cultured bacteria (27%), Actinobacteria (9%), Anaerovorax (9%), Romboutsia (9%) and rumen bacteria (9%) (Table 1, Fig. 5). Clostridia and its members were found to be predominant in enriched mixed culture. Clostridia are Gram positive, rod shaped, obligate anaerobes capable of producing endospores. C. sporogenes Cu-R6 (band-2) and uncultured Clostridia (band-7) were observed to be dominant on day 0 and day 30. C. propionicum (band-10) were dominant on the 30th and 60th days, but not observed on day 0. Contrary to this C. coccoides (band-11) were observed on day 0, but not observed on the 30th and 60th days. Many studies have reported the capacity of Clostridia sp. to produce MCFA. Jeon et al. (2013) isolated Clostridium sp. BS-1 (KCCM 10991P) from the sludge sample of a wastewater treatment plant, and used it for the production of caproic acid with galactitol.25 They reported that 32 g l−1 of caproic acid was observed in the solvent due to effective extraction and the maintenance of a low (1–2 g l−1) concentration. Kurakawa et al. (2015) reported that the strictly anaerobic C. coccoides group constitutes 25–60% of the total among a variety of human intestinal bacteria, and is the most dominant bacterial group.26 Kuchta and Abeles (1985) reported that C. propionicum converts lactate to propionate, hydrates acrylate to lactate, and reduces acrylate to propionate.27 Steinbusch et al. (2011) reported that a relative of C. kluyveri is very likely responsible for the MCFA production in their experiment, as it dominated the microbial population in the MCFA producing bioreactor.3| Band no. | Closest relative | Similarity (%) | Accession no. | Phylogenetic affiliation |
|---|---|---|---|---|
| 1 | Rumen bacterium | 79 | HM597709 | — |
| Anaerobic bacterium S6 | 79 | EF029509 | ||
| 2 | Clostridium sporogenes Cu-R6 | 91 | KU587045 | Firmicutes |
| Clostridium Hs50 | 91 | AB673454 | ||
| 3 | Uncultured bacterium | 81 | AB552906 | Bacteroidia |
| Uncultured bacterium | 79 | AB240476 | ||
| 4 | Uncultured bacterium | 86 | KU241934 | Bacteroidia |
| Uncultured bacterium | 86 | KX589545 | ||
| 5 | Uncultured bacterium | 85 | KM656233 | Bacteroidia |
| Uncultured bacterium | 84 | KM295153 | ||
| 6 | Romboutsia DYFP104 | 87 | KT002352 | Firmicutes |
| Romboutsia BDYFP92 | 87 | KT002340 | ||
| 7 | Uncultured Clostridia | 94 | KP717478 | Firmicutes |
| Uncultured Clostridia | 94 | EU887998 | ||
| 8 | Uncultured Anaerovorax sp. | 100 | EU073780 | Firmicutes |
| Uncultured Clostridiales | 99 | KJ600140 | ||
| 9 | Actinomyces sp. NB15 | 98 | KT452786 | Actinobacteria |
| Actinomyces sp. MS2 | 98 | HF952919 | ||
| 10 | Clostridium propionicum JCM 1430 | 88 | NR_113408 | Firmicutes |
| Clostridium sp. Val-6 | 88 | EU937739 | ||
| 11 | Clostridium coccoides | 97 | EF025906 | Firmicutes |
| Clostridium sp. | 97 | Y10584 |
After Clostridia, un-cultured bacteria were found to be most prevalent in our study. Un-cultured bacteria (bands-3, 4, 5) were observed to be dominant on day 0, day 30 and day 60. No reports were available about the production of MCFA using un-cultured bacteria. Actinomyces, belonging to the class of Actinobacteria, were also found to be prominent. Actinomyces species are facultative anaerobes, although some are obligate anaerobes. Actinomyces species are Gram-positive, rod-shaped, and may form endospores. They are known for the important role they play in soil ecology: they produce several enzymes that help to degrade the organic plant material, lignin and chitin. Wallace et al. (1995) reported the synthesis of branched-chain and straight-chain fatty acid in three actinomycetes. Actinomyces (band-9) were observed to be dominant on days 0 and 30, but were not observed on the 60th day.28
Rumen bacterium (band-1) was observed on the 30th and 60th days. Many researchers have isolated MCFA producing bacteria from rumen. C. kluyveri isolated from bovine rumen is the most extensively studied bacteria with regards to chain elongation.29Eubacterium pyruvativorans was isolated from sheep rumen fluid and it produced valerate and caproate from propionate and butyrate respectively.30,31 Elsden et al. (1956) isolated Eubacterium limosum from the rumen of a sheep that was fed a molasses-based diet.32E. limosum produced acetate, butyrate, and caproate during growth in methanol with CO2, acetate and butyrate. Megasphaera elsdenii was isolated from the rumen of sheep and it ferments glucose, fructose, lactate and sucrose with the formation of acetate, propionate, butyrate, valerate, caproate, H2, and CO2.33 Weimer et al. (2015) added ethanol to switch grass and alfalfa stems to achieve a caproate production rate of 3.1 g l−1 d−1 with rumen fluid and an augmented C. kluyveri.20 Bright bands of Romboutsia (band-6) were observed on day 0 and day 30. Romboutsia is a Gram-positive, rod-shaped, non-motile, spore-forming obligatory anaerobic bacterium. Gerritsen et al. (2014) isolated the bacteria R. ilealis sp. CRIB from the gastrointestinal tract of a rat and reported that it produced acetic acid, lactic acid, and propionic acid from peptone-yeast extract-glucose medium.34 Bright bands of Anaerovorax sp. (band-8) were observed on day 0 and day 30; no band was observed on day 60. Matthies et al. (2000) reported that A. odorimutans, which is strictly anaerobic, Gram-positive, and non-spore-forming, ferments putrescine to acetate, butyrate, and H2.35
4. Conclusions
A bacterial mixed culture which can produce good amounts of short chain fatty acids (SCFA) and medium chain fatty acids (MCFA) was enriched and used to produce SCFA and MCFA from acetate and ethanol. The effects of substrate, electron donor, and methane inhibition on MCFA production were evaluated. Among the various conditions tested, bacteria grown with methane inhibition produced the highest concentrations of butyrate, caproate, and caprylate. CO2, and H2 were present at the highest percentages in the biogas. DGGE analysis revealed that bacteria related to Clostridia, Rumen bacterium, and the Actinomyces group were dominant in the enriched mixed culture.Conflicts of interest
The authors have declared no conflict of interest.Acknowledgements
This work was supported by the Japan Society for the Promotion of Science (15F15352). Dr M. V. Reddy gratefully acknowledges the JSPS for providing a Postdoctoral fellowship (ID No: P15352).References
- T. I. M. Grootscholten, K. D. F. Borgo, H. V. M. Hamelers and C. J. N. Buisman, Biomass Bioenergy, 2013, 48, 10–16 CrossRef CAS.
- T. I. M. Grootscholten, D. P. B. T. B. Strik, K. J. J. Steinbusch, C. J. N. Buisman and H. V. M. Hamelers, Appl. Energy, 2014, 116, 223–229 CrossRef CAS.
- K. J. J. Steinbusch, H. V. M. Hamelers, C. M. Plugge and C. J. N. Buisman, Energy Environ. Sci., 2011, 4, 216–224 CAS.
- M. T. Agler, C. M. Spirito, J. G. Usack, J. J. Werner and L. T. Angenent, Energy Environ. Sci., 2012, 5, 8189–8192 CAS.
- M. K. Woolford, J. Sci. Food Agric., 1975, 26, 219–228 CrossRef CAS PubMed.
- Y. I. Kuznetsov and K. A. Ibatullin, Prot. Met., 2002, 38, 439–443 CrossRef CAS.
- M. Renz, Eur. J. Org. Chem., 2005, 6, 979–988 CrossRef.
- B. Witholt and B. Kessler, Curr. Opin. Biotechnol., 1999, 10, 279–285 CrossRef CAS PubMed.
- M. T. Agler, B. A. Wrenn, S. H. Zinder and L. T. Angenent, Trends Biotechnol., 2011, 29, 70–78 CrossRef CAS PubMed.
- N. Sarkar, S. K. Ghosh, S. Bannerjee and K. Aikat, Energy, 2012, 37, 19–27 CAS.
- S. Bajracharya, R. Yuliasni, K. Vanbroekhoven, C. J. N. Buisman, D. P. B. T. B. Strik and D. Pant, Bioelectrochemistry, 2017, 113, 26–34 CrossRef CAS PubMed.
- A. Schievano, T. Pepesciarria, K. Vanbroekhoven, H. D. Wever, S. Puig, S. J. Andersen, K. Rabaey and D. Pant, Trends Biotechnol., 2016, 34, 866–878 CrossRef CAS PubMed.
- S. Roy, A. Schievano and D. Pant, Bioresour. Technol., 2016, 213, 129–139 CrossRef CAS PubMed.
- W. R. Kenealy, Y. Cao and P. J. Weimer, Appl. Microbiol. Biotechnol., 1995, 44, 507–513 CrossRef CAS PubMed.
- H. Seedorf, W. F. Fricke, B. Veith, H. Brüggemann, H. Liesegang, A. Strittmatter, M. Miethke, W. Buckel, J. Hinderberger, F. Li, C. Hagemeier, R. K. Thauer and G. Gottschalk, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 2128–2133 CrossRef CAS PubMed.
- M. V. Reddy and S. V. Mohan, Bioresour. Technol., 2012, 103, 313–321 CrossRef PubMed.
- D. Vasudevan, H. Richter and L. T. Angenent, Bioresour. Technol., 2014, 151, 378–382 CrossRef CAS PubMed.
- H. A. Barker, M. D. Kamen and B. T. Bornstein, Proc. Natl. Acad. Sci. U. S. A., 1945, 31(12), 373–381 CrossRef CAS.
- H. B. Ding, G. Y. A. Tan and J. Y. Wang, Bioresour. Technol., 2010, 101, 9550–9559 CrossRef CAS PubMed.
- P. J. Weimer, M. Nerdahl and D. J. Brandl, Bioresour. Technol., 2015, 175, 97–101 CrossRef CAS PubMed.
- Y. N. Yin, Y. F. Zhang, D. B. Karakashev, J. L. Wang and I. Angelidaki, Bioresour. Technol., 2017, 241, 638–644 CrossRef CAS PubMed.
- L. Leng, P. Yang, Y. Mao, Z. Wu, T. Zhang and P. H. Lee, Water Res., 2017, 114, 200–209 CrossRef CAS PubMed.
- L. A. Kucek, M. Nguyen and L. T. Angenent, Water Res., 2016, 93, 163–171 CrossRef CAS PubMed.
- X. Zhu, Y. Tao, C. Liang, X. Li, N. Wei, W. Zhang, Y. Zhou, Y. Yang and T. Bo, Sci. Rep., 2015, 5, 1–9 Search PubMed.
- B. S. Jeon, C. Moon, B. C. Kim, H. Kim, Y. Um and B. I. Sang, Enzyme Microb. Technol., 2013, 53, 143–151 CrossRef CAS PubMed.
- T. Kurakawa, K. Ogata, K. Matsuda, H. Tsuji, H. Kubota, T. Takada, Y. Kado, T. Asahara, T. Takahashi and K. Nomoto, PLoS One, 2015, 10, 1–19 Search PubMed.
- R. D. Kuchta and R. Abeles, J. Biol. Chem., 1985, 260, 13181–13189 CAS.
- K. K. Wallace, B. Zhao, H. A. I. McArthur and K. A. Reynolds, FEMS Microbiol. Lett., 1995, 131, 227–234 CrossRef CAS PubMed.
- P. J. Weimer and D. M. Stevenson, Appl. Microbiol. Biotechnol., 2012, 94, 461–466 CrossRef CAS PubMed.
- R. J. Wallace, N. McKain, N. R. McEwan, E. Miyagawa, L. C. Chaudhary, T. King, N. D. Walker, J. H. A. Apajalahti and C. J. Newbold, Int. J. Syst. Evol. Microbiol., 2003, 53, 965–970 CrossRef CAS PubMed.
- B. R. Genthner, C. L. Davis and M. P. Bryant, Appl. Environ. Microbiol., 1981, 42, 12–19 CAS.
- S. R. Elsden, B. E. Volcani, F. M. C. Gilchrist and D. Lewis, J. Bacteriol., 1956, 72, 681–689 CAS.
- K. Choi, B. S. Jeon, B. C. Kim, M. K. Y. Oh, Y. Um and B. I. Sang, Appl. Biochem. Biotechnol., 2013, 171, 1094–1107 CrossRef CAS PubMed.
- J. Gerritsen, S. Fuentes, W. Grievink, L. V. Niftrik, B. J. Tindall, H. M. Timmerman, G. T. Rijkers and H. Smidt, Int. J. Syst. Evol. Microbiol., 2014, 64, 1600–1616 CrossRef CAS PubMed.
- C. Matthies, S. Evers, W. Ludwig and B. Schink, Int. J. Syst. Evol. Microbiol., 2000, 4, 1591–1594 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2018 |