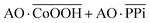The Boolean logic tree of molecular self-assembly system based on cobalt oxyhydroxide nanoflakes for three-state logic computation, sensing and imaging of pyrophosphate in living cells and in vivo†
Xin Xing
Zhang‡
,
Qiu Yan
Zhu‡
,
Jiao Yang
Lu
,
Fu Rui
Zhang
,
Wei Tao
Huang
 *,
Xue Zhi
Ding
and
Li Qiu
Xia
*,
Xue Zhi
Ding
and
Li Qiu
Xia
State Key Laboratory of Developmental Biology of Freshwater Fish, College of Life Science, Hunan Normal University, Changsha 410081, P. R. China. E-mail: vthuang@hunnu.edu.cn; Fax: (+86)731-8887-2905; Tel: (+86)731-8887-2905
First published on 24th October 2018
Abstract
Sensing of pyrophosphate (PPi) is helpful to better understand many life processes and diagnose various early-stage diseases. However, many traditional reported methods based on artificial receptors for sensing of PPi exhibit some disadvantages including difficulties in designing appropriate binding sites and complicated multi-step assembly/functionalization. Thus, it is significantly important and a big challenge to know how to use a simple molecular self-assembly or an interaction system to solve the above-mentioned limits to achieve the quantitative analysis of specific substances in the system. Based on the natural connection and similarity (such as stimulus responsiveness) between sensing and logic computing, in this study, the Boolean logic tree of molecular self-assembly system based on the cobalt oxyhydroxide (CoOOH) nanoplatform is constructed and applied to organize and connect “plug and play” molecular events (fluorescent dye, acridine orange and anion, PPi). By using molecules as inputs and the corresponding fluorescence signal as the output, the CoOOH-based molecular self-assembly system can be programmed for three-input fluorescent Boolean logic computation, fluorescent three-state logic computation, detection of PPi (linear range from 50 to 6400 nM with a detection limit of 20 nM) and even for imaging in living cancer cells and in vivo (in systems such as Zebrafish and Carassius auratus). Our approach adds a new dimension for expanding molecular logic computing and sensing systems, which will not only provide more opportunities for developing novel logic computing paradigms, but also be helpful in promoting the development and applications of intelligent molecular computing and sensing systems.
1. Introduction
Pyrophosphate anions (PPi) as important biological anions are involved in various cascading processes in the human body such as ATP hydrolysis, cellular metabolism, and DNA/RNA polymerization.1 Abnormal amounts of intracellular PPi are also a potential biomarker in the early diagnosis of cancer,2 arthritis,3 and urolithiasis.4 Therefore, sensing PPi has been the focus of research in recent years due to its chemical and biological significance. Recently, some innovative studies5,6 have proved that there is a natural connection and similarity (such as stimulus responsiveness) between sensing and logic computing. From a computing point of view, various PPi-involved biochemical reactions (such as ATP hydrolysis, cellular metabolism, DNA/RNA polymerization) keep living things alive by implementing molecular-level information processing, which can be considered as the combination of the increase or decrease of specific molecules as inputs producing some corresponding substances or energy changes as outputs. Thus, an increasing number of molecules or materials involved in molecular interactions or molecular sensing (such as molecular recognition or nano-assembly) were utilized as building blocks for the development of molecular information processing systems, such as Boolean logic computations,5,6 fuzzy logic computations,7,8 and even neural network computations.9,10 In fact, the Boolean logic computation in turn can also be applied for molecular sensing. Recently, we have demonstrated that the Boolean logic tree can be utilized to organize and connect “plug and play” chemical events with DNA, nanomaterials, and biomolecules for developing the chemical evolution sensing system and for applications in biosensing, molecular logic computation, and keypad lock security operation.7,11 The Boolean logic tree7 not only facilitates the organization and connection of lower-level molecular events, but also gives qualitative and/or quantitative information of specific analytes in the complex molecular system.11As for traditional sensing researches, fluorescent and colorimetric sensing of PPi12 using small molecular sensors13 has been developed based on hydrogen bonding, coordination chemistry, and aggregation-induced emission or quenching. Unfortunately, there are several limitations including difficulties in designing appropriate binding sites for PPi,14 complicated and time-consuming synthesis, and poor water solubility of artificial receptors. Moreover, there has been a growing interest in directly using or combining nanomaterials (gold nanoparticles, quantum dots, graphene oxide) with artificial receptors (such as metal-ion complexes) for sensing PPi.15 However, the existing technologies for PPi detection based on nano-assembly have several shortcomings including lack of sensitivity,16 complicated multi-step assembly/functionalization,17 and high toxicity,18 Thus, it is significantly important and a big challenge to know how to use a simple molecular self-assembly or interactions to construct a special Boolean logic system for solving the above-mentioned limits and achieving quantitative analysis of specific substances in the system.
Cobalt oxyhydroxide nanoflakes (CoOOH) with unique fluorescence quenching ability can interact with fluorescent reporters and molecular recognition elements for applications in biosensing (such as ascorbic acid19 in living cells and in vivo,20,21 T4 polynucleotide kinase activity,22 and DNA23) via Förster resonance energy transfer (FRET). Thus, in this study, the Boolean logic tree of molecular self-assembly system based on a CoOOH nanoplatform is constructed and applied to organize and connect “plug and play” molecular events using common fluorescent dye acridine orange (AO) and anion PPi. By using molecules as the inputs and the corresponding fluorescence signal as the output, the molecular self-assembly system can be programmed for three-input fluorescent Boolean logic computation, controllable and selectable fluorescent three-state logic computation, and detection and imaging of PPi in living cells and in vivo. This study adds a new dimension for expanding molecular logic computing and sensing systems, which will not only provide more opportunities for developing novel logic computing paradigms, but also promote the development and applications of intelligent molecular computing and sensing systems.
2. Experimental
2.1 Materials and reagents
CoCl2·6H2O was obtained from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). Adenosine triphosphate (ATP) was purchased from Macklin Reagents Co., Ltd (China). AO, PPi, NaClO, NaOH, glucose (Glu), fructose (Fru), adenine (A), thymine (T), adenosine diphosphate (ADP), adenosine monophosphate (AMP), glycine (Gly) and alanine (Ala) were purchased from Aladdin Reagents Co., Ltd (Shanghai, China). Aqueous solutions of P2O74−, PO43−, HPO42−, H2PO4−, HCO3−, CO32−, SO42−, S2−, NO3−, Cl−, and Ac− were prepared from Na4P2O7, Na3PO4·12H2O, Na2HPO4·2H2O, NaH2PO4·2H2O, NaHCO3, Na2CO3, Na2SO4, Na2S·9H2O, NaNO3, NaCl and NaAc, respectively. Aqueous solutions of Na+, Mg2+, Ca2+, Cu2+, K+ and Zn2+ were prepared from NaCl, MgCl2, CaCl2·2H2O, CuCl2·2H2O, KCl and ZnCl2, respectively. All the above inorganic salts were of analytical reagent grade and purchased from Aladdin Reagents Co., Ltd (Shanghai, China). All aqueous solutions were prepared with double-distilled water produced by a Milli-Q system (Millipore, Bedford, MA, USA, 18.2 MΩ cm).2.2 Instruments
Fluorescence measurements were performed using a SpectraMax M5 microplate spectrophotometer system (Molecular Devices, USA). Fluorescence emission spectra were measured at an excitation wavelength of 490 nm. UV-Vis absorption spectra were recorded on a Hach spectrophotometer DR6000 (USA). Transmission electron microscopy (TEM) images were obtained on a JEM-1200EX Transmission Electron Microscope (Japan) operated at 120 kV. Scanning electron microscopy (SEM) images were collected using an SU8010 Scanning Electron Microscope (Hitachi, Japan). Elemental composition was identified by X-ray photoelectron spectroscopy (XPS, Thermo Scientific escalab 250XI, USA). Fourier transform infrared (FT-IR) spectra of CoOOH nanoflakes were recorded by a Nicolet 6700 FT-IR spectrometer (Thermo Scientific, USA). A Zeiss Axio Observer A1 inverted fluorescence microscope (Carl Zeiss AG, Germany) was used to collect cell fluorescence images. ζ-Potentials were measured on Zetasizer Nano ZS (Malvern Instruments Ltd, UK).2.3 Preparation of CoOOH nanoflakes and AO–CoOOH complex
We synthesized CoOOH nanoflakes according to a previously reported method.10 Initially, 125 μl sample of sodium hydroxide (NaOH, 1 M) was added to 500 μl of CoCl2 (10 mM) solution and ultrasonicated for 1 min. Then, the solution was mixed with 25 μl of sodium hypochlorite (NaClO, 0.9 M), and the mixture was sonicated for 10 min. Subsequently, the mixture was centrifugated and washed with deionized water for three times. Lastly, the product was re-dispersed in 1 mL of water with the final concentration of 0.25 mg mL−1. Then, the CoOOH solution was added to the AO solution (the sodium acetate buffer solution, 10 mM, pH 4.0) and the mixture was incubated for 15 min. After that, the AO–CoOOH complex was prepared.2.4 In vitro detection of PPi
AO solution was diluted by the sodium acetate buffer solution (10 mM, pH 4.0). CoOOH nanoflakes (12 μg mL−1) were first incubated with the AO solution (10 μM) for 15 min at room temperature. Then, different concentrations (50, 200, 400, 800, 1600, 3200, 6400, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800, 25
800, 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 nM) of PPi were added into the AO–CoOOH mixed solution (10 μM
600 nM) of PPi were added into the AO–CoOOH mixed solution (10 μM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 μg mL−1). After 60 min of reaction, the fluorescence spectra were recorded at the excitation wavelength of 490 nm and the emission spectra were obtained from 510 to 750 nm. The selectivity for PPi was confirmed by adding potential interfering compounds instead of PPi in a similar way. For human serum analysis, the human serum (obtained from the hospital of Hunan Normal University) was diluted hundred-fold with deionized water and spiked with different concentrations of PPi. All experiments were performed at room temperature.
12 μg mL−1). After 60 min of reaction, the fluorescence spectra were recorded at the excitation wavelength of 490 nm and the emission spectra were obtained from 510 to 750 nm. The selectivity for PPi was confirmed by adding potential interfering compounds instead of PPi in a similar way. For human serum analysis, the human serum (obtained from the hospital of Hunan Normal University) was diluted hundred-fold with deionized water and spiked with different concentrations of PPi. All experiments were performed at room temperature.
2.5 Fluorescence microscopy imaging of PPi in living cancer cells and in Zebrafish
To explore the capability of the AO–CoOOH complex for monitoring intracellular PPi levels in cancer cells, murine colorectal carcinoma cell line CT26 was chosen as the model system. CT26 cells (CRL-2639, ATCC) were maintained in RPMI 1640 medium containing 10% fetal bovine serum (FBS, Hyclone) at 37 °C in a humidified environment with 5% CO2. CT26 cells were seeded into a 24-well glass-bottom plate (glass surface diameter: 14 mm) and cultured for 18 h. Cells were washed two times with 500 μL of PBS and then incubated with the AO–CoOOH complex (80 μL, 10 μM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 μg mL−1) in 500 μL of PBS at 37 °C in 5% CO2. After 3 h of incubation, the cells were washed two times with 500 μL of PBS; different concentrations of PPi (0.025, 0.1, and 2 mM) were added and incubated for 1 h and dispersed in 500 μL of the medium. Then, the cells were visualized using an inverted fluorescence microscope (Zeiss, Axio Observer A1) with a 40× LD Plan-Neofluar objective (NA = 0.6) at the excitation of 488 nm.
12 μg mL−1) in 500 μL of PBS at 37 °C in 5% CO2. After 3 h of incubation, the cells were washed two times with 500 μL of PBS; different concentrations of PPi (0.025, 0.1, and 2 mM) were added and incubated for 1 h and dispersed in 500 μL of the medium. Then, the cells were visualized using an inverted fluorescence microscope (Zeiss, Axio Observer A1) with a 40× LD Plan-Neofluar objective (NA = 0.6) at the excitation of 488 nm.
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Hunan Normal University and approved by the Animal Ethics Committee of Hunan Normal University. Zebrafish were maintained in an E3 embryo medium (15 mM NaCl, 0.5 mM KCl, 1 mM MgSO4, 1 mM CaCl2, 0.15 mM KH2PO4, 0.05 mM Na2HPO4, 0.7 mM NaHCO3, 5–10% methylene blue, pH 7.5). In fluorescence imaging experiments, three-day-old zebrafish were incubated with the AO–CoOOH complex (50 μL, 10 μM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 μg mL−1) in 1 mL of the E3 embryo medium for 1 h at 28 °C. After washing three times with PBS (pH 7.4) to remove the remaining probes, different concentrations of PPi (0.023, 0.093, 0.374 mM) were added and incubated for 1 h and dispersed in 1 mL of the medium. Then, the Zebrafish were visualized using an inverted fluorescence microscope (Zeiss, Axio Observer A1) with 10× A-Plan objective (NA = 0.25) at the excitation of 488 nm.
12 μg mL−1) in 1 mL of the E3 embryo medium for 1 h at 28 °C. After washing three times with PBS (pH 7.4) to remove the remaining probes, different concentrations of PPi (0.023, 0.093, 0.374 mM) were added and incubated for 1 h and dispersed in 1 mL of the medium. Then, the Zebrafish were visualized using an inverted fluorescence microscope (Zeiss, Axio Observer A1) with 10× A-Plan objective (NA = 0.25) at the excitation of 488 nm.
2.6 In vivo fluorescence imaging of PPi in freshwater fish Carassius auratus
The IVIS Spectrum imaging system (Caliper, MA) was used to determine the fluorescence intensities of the freshwater fish Carassius auratus (fluorescence excitation 500 nm, fluorescence emission 540 nm). Image scaling was normalized by converting total radiance efficiency. Fluorescence intensity was represented by a multicolor scale ranging from blue (least intense) to red (most intense). Signal intensity images were superimposed over gray scale reference photographs for anatomical representations. Also, 200 μL of the AO–CoOOH complex (0.67 mM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 μg mL−1) was injected intraperitoneally into Carassius auratus. After 1 h post injection, the fish were anesthetized by immersing in tricaine (50 mg L−1) and then imaged with the IVIS Spectrum system. After imaging, 200 μl of PPi solutions of different concentrations (5 and 100 mM) were injected intraperitoneally into the same position of the fish. After 1 h post injection, the fluorescent images were obtained with the IVIS Spectrum system.
80 μg mL−1) was injected intraperitoneally into Carassius auratus. After 1 h post injection, the fish were anesthetized by immersing in tricaine (50 mg L−1) and then imaged with the IVIS Spectrum system. After imaging, 200 μl of PPi solutions of different concentrations (5 and 100 mM) were injected intraperitoneally into the same position of the fish. After 1 h post injection, the fluorescent images were obtained with the IVIS Spectrum system.
3. Results and discussion
3.1 Characterization of CoOOH nanoflakes
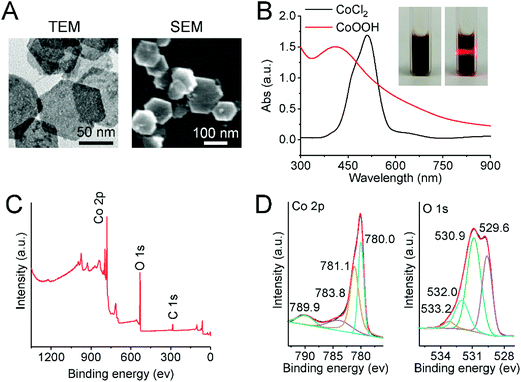
CoOOH nanoflakes were characterized using TEM, SEM, UV-Vis spectroscopy, XPS, FT-IR spectroscopy and ζ-potential. As shown in Fig. 1A, the TEM and SEM images of CoOOH nanoflakes clearly revealed that the morphology of CoOOH nanoflakes was hexagonal, which was consistent with previously reported results.11 The brown-black colored aqueous solution of CoOOH nanoflakes exhibited the Tyndall effect (Fig. 1B, inset) and had a strong absorption peak at 410 nm, whereas the characteristic absorption peak of purple CoCl2 was at 510 nm (Fig. 1B). Compositional analysis with X-ray photoelectron spectroscopy (XPS) revealed the signals of Co and O (Fig. 1C). The binding energies (BEs) of Co species were calibrated with reference to the aliphatic carbon C ls peak after setting its BE to 285.08 eV. The Co 2p peak was mainly deconvoluted into four peaks with BEs of 789.9, 783.8, 781.1, and 780.0 eV, whereas the O 1s peak was mainly deconvoluted into four peaks with BEs of 533.2, 532.0, 530.9, 529.6 eV (Fig. 1D). This confirmed the formation of CoOOH.24 The FT-IR spectrum of CoOOH nanoflakes is given in Fig. S1A.† The broad bands at 584 cm−1 and 1620 cm−1 were assigned to Co–O and Co–O2 bond stretching, respectively, whereas the peak at 3420 cm−1 was characteristic of the O–H bond stretching.25 The ζ-potential of CoOOH nanoflakes was measured to be about 29.4 mV (Fig. S1B†), indicating a positive charge on CoOOH nanoflake surfaces. The above data fully proved that the CoOOH nanoflakes were successfully prepared.3.2 Boolean logic tree analysis and logic computation of molecular self-assembly system based on the cobalt oxyhydroxide (CoOOH) nanoplatform
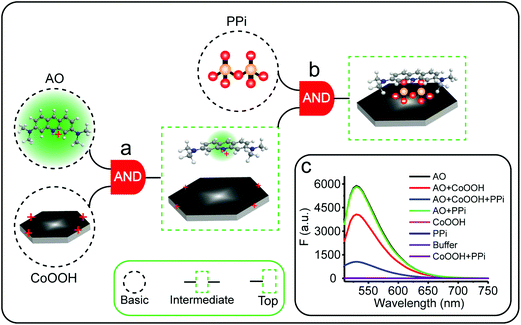
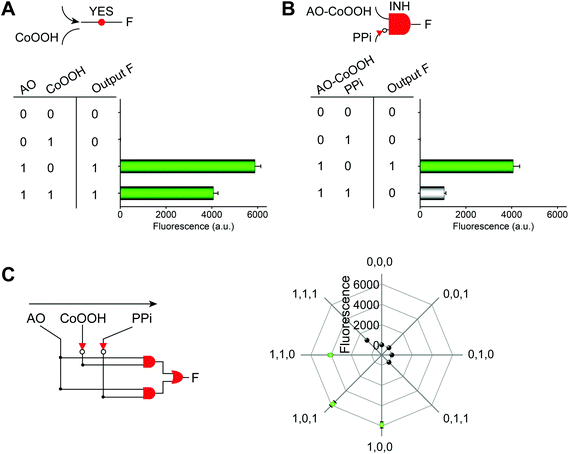
The Boolean logic tree consists of events (including basic, intermediate, and top events; Scheme 1, green box) and gates (Scheme 1, red box).7,11 The events are linked using “gate” symbols. As shown in Scheme 1, the AO dye (basic event) was self-assembled with CoOOH nanoflakes (basic event) to form the AND-gate AO–CoOOH complex (AO·CoOOH, intermediate event, the dot “·” is used to represent AND Boolean algebra), resulting in the fluorescence quenching of AO because of effective FRET between the fluorescent dye and CoOOH nanoflakes (Scheme 1a, c and Fig. S2†). AO·CoOOH (intermediate event) further self-assembled with PPi (basic event) with negative charges (ζ-potential −26.4 ± 4.7 mV), which served as a “bridge” connecting cationic AO (ζ-potential 12.7 ± 0.6 mV) and cationic CoOOH (ζ-potential 29.6 ± 0.7 mV, Table S1†) through electrostatic interactions and exported the AO–CoOOH–PPi ternary complex ((AO·CoOOH)·PPi, top event, ζ-potential 0.013 ± 0.4 mV), resulting in further fluorescence quenching of AO (Scheme 1b and c). Thus, we can represent the whole molecular self-assembly system as (AO·CoOOH)·PPi.The fluorescence changes in the different combinations of these aforementioned molecular events can be used for implementing logic computation. Logical outputs are defined as fluorescence (F) < 2000 (a.u.) for logical 0 and ≥2000 (a.u.) for logical 1. Self-assembly of AO and CoOOH functions as a YES fluorescence gate. The fluorescence output is high (1) only when AO is present (Scheme 1c and Fig. 2A). By combining the two input signals AO·CoOOH complex and PPi in accordance with the truth table, the fluorescence output is produced exclusively in the presence of AO·CoOOH and not PPi, corresponding to the INHIBIT (INH) fluorescence gate function (Scheme 1c and Fig. 2B). These molecular events (AO, CoOOH, PPi) not only can be connected to form a complex logic evolution system, but also use the fluorescence signal as the output for computing 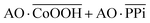 (Scheme 1c and Fig. 2C).
(Scheme 1c and Fig. 2C).
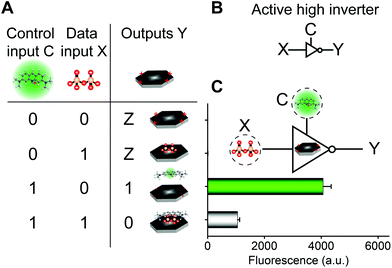
The aforementioned molecular self-assembly system based on the CoOOH nanoplatform was also used to implement three-state logic computation with one fluorescence output and two different kinds of inputs (a data input X and a control input C) for active-high inverter logic functions (Fig. 3). Three-state logic (or three-state buffer) allows an output port to assume a high impediment or nothing (Z) state in addition to the 0 and 1 logic levels, effectively controlling when an input signal passes through the device and when it does not.11,26 Logical outputs were examined by fluorescence analysis and defined as 20 < F < 2000 for logical 0, F ≥ 2000 for logical 1, and F < 20 for Z state. If the control input C = AO was absent (0) in the CoOOH nanoplatform, irrespective of the absence (0) or the presence (1) of the data input X = PPi, the CoOOH nanoplatform could not produce any fluorescence signal (Fig. 3C), that is, the output Y was nothing (Z). Once the control input C = AO was present (1) in the CoOOH nanoplatform and when the data input X = PPi was absent (0), the CoOOH self-assembled with AO; thus, the output Y was a high fluorescence signal (1). When the data input X = PPi was present (1), AO·CoOOH could further self-assemble with PPi with negative charges, which served as a “bridge” connecting cationic AO and cationic CoOOH through electrostatic interactions, resulting in clear fluorescence quenching of AO; thus, the output Y was a low fluorescence signal (0). Thus, the constructed three-state molecular logic computation implementing active-high inverter logic functions can allow the output ports to assume nothing (Z) state in addition to the 0 and 1 logic levels, effectively controlling when an input signal passes through the circuits.
3.3 Characterization of molecular self-assembly system
UV-Vis absorption spectra and FT-IR spectra were used for monitoring and characterizing the logic evolution process of the molecular self-assembly system at each step. Fig. S3† shows the absorption spectra of AO, CoOOH, PPi, and their mixtures. Interestingly, the spectrum of AO exhibited two peaks at 268 and 491 nm.27 The spectrum of CoOOH showed two peaks at 263 and 410 nm. The spectrum of PPi was flat and featureless. After the self-assembly of AO·CoOOH, the absorption spectrum showed a slight blueshift from 491 to 489 nm (as indicated by Fig. S3,† black arrow), indicating the adsorption of AO on the surface of CoOOH nanoflakes, resulting in the fluorescence quenching of AO. Meanwhile, the absorption spectra of the mixture solutions of CoOOH–PPi and AO–PPi remained almost unchanged. After the self-assembly of (AO·CoOOH)·PPi, the absorption spectrum of the ternary complex exhibited strong blueshifts from 491 to 467 nm (24 nm) and from 268 to 263 nm (5 nm, as indicated by Fig. S3,† red arrows). The band of (AO·CoOOH)·PPi showed a 24 nm blue-shift probably due to H-aggregation of AO molecules co-induced by CoOOH and PPi, which may be one main reason for further fluorescence quenching.27,28In addition, the evidence for interactions among AO, CoOOH, and PPi was provided by the FT-IR spectra shown in Fig. S4.† The spectrum of AO showed a rich collection of transmission bands around 1636 (Ar: aromatic backbone stretching vibration), 1594 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching vibration), 1251 (C–N stretching vibration), and 822 cm−1 (C–H deformation).27 The spectrum of PPi showed a very sharp and intense band around ∼735 cm−1 due to P2O74−.29 The peaks at 1123 and 1029 cm−1 corresponded to P–O asymmetric stretching vibration. The P–O symmetric stretching vibration was observed at 925 cm−1, and the O–P–O bending vibration was observed at 575 and 524 cm−1.30 By comparing the FT-IR spectra of AO, CoOOH, PPi and (AO·CoOOH)·PPi ternary complex, it was clear that the spectrum of the (AO·CoOOH)·PPi ternary complex showed a rich collection of transmission bands corresponding to the aromatic backbone stretching vibration (1647 cm−1), C
N stretching vibration), 1251 (C–N stretching vibration), and 822 cm−1 (C–H deformation).27 The spectrum of PPi showed a very sharp and intense band around ∼735 cm−1 due to P2O74−.29 The peaks at 1123 and 1029 cm−1 corresponded to P–O asymmetric stretching vibration. The P–O symmetric stretching vibration was observed at 925 cm−1, and the O–P–O bending vibration was observed at 575 and 524 cm−1.30 By comparing the FT-IR spectra of AO, CoOOH, PPi and (AO·CoOOH)·PPi ternary complex, it was clear that the spectrum of the (AO·CoOOH)·PPi ternary complex showed a rich collection of transmission bands corresponding to the aromatic backbone stretching vibration (1647 cm−1), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching vibration (around 1596 cm−1), and C–N stretching vibration (1260 cm−1) of AO as well as P–O asymmetric stretching vibration (1102 and 1026 cm−1) and P–O symmetric stretching vibration (923 cm−1) of PPi. The band in the region of 480–680 cm−1 could be assigned to Co–O and Co–N vibrations, proving the formation of cobalt–oxygen and cobalt–nitrogen bonds.31 Thus, the UV-Vis absorption and FT-IR results suggested that CoOOH and PPi may co-induce H-aggregation of AO molecules, resulting in the formation of the (AO·CoOOH)·PPi ternary complex probably by the coordination of Co2+ to the nitrogen atoms of acridine orange and the hydroxyl oxygen atoms of PPi.31
N stretching vibration (around 1596 cm−1), and C–N stretching vibration (1260 cm−1) of AO as well as P–O asymmetric stretching vibration (1102 and 1026 cm−1) and P–O symmetric stretching vibration (923 cm−1) of PPi. The band in the region of 480–680 cm−1 could be assigned to Co–O and Co–N vibrations, proving the formation of cobalt–oxygen and cobalt–nitrogen bonds.31 Thus, the UV-Vis absorption and FT-IR results suggested that CoOOH and PPi may co-induce H-aggregation of AO molecules, resulting in the formation of the (AO·CoOOH)·PPi ternary complex probably by the coordination of Co2+ to the nitrogen atoms of acridine orange and the hydroxyl oxygen atoms of PPi.31
3.4 Boolean logic tree of molecular self-assembly system based on the CoOOH nanoplatform for sensing and imaging of PPi in living cells and in vivo
Boolean logic tree7 not only facilitates the organization and connection of lower-level molecular events, but also gives qualitative and/or quantitative information of specific analytes in the complex molecular system.11 To investigate the PPi sensing performance of the Boolean logic tree of molecular self-assembly system based on cobalt oxyhydroxide nanoflakes, the experimental conditions were optimized. We investigated fluorescence responses of different ratios of AND-gate AO·CoOOH to PPi (Fig. S5A†). The fluorescence changes of AO·CoOOH to PPi (23.8 μM) reached maximal values when the concentration of CoOOH was 12 μg mL−1 (AO, 10 μM). Therefore, 12 μg mL−1 was used as the optimized concentration for CoOOH nanoflakes. We investigated the effect of pH on the fluorescence response of AO·CoOOH (10 μM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 μg mL−1) to PPi. The pH of the buffer in the range from 3.0 to 9.0 had diverse effects on the quenching efficiency (Fig. S5B†). The results certified that the optimal sensing performance was obtained at pH 4.0. We also monitored the fluorescence intensity change as a function of reaction time to study the interaction kinetics of AO and CoOOH nanoflakes. As shown in Fig. S5C,† with the increase in the reaction time, the fluorescence intensity decreased gradually and then reached equilibrium after 5 min. Then, the optimal reaction time between AND-gate AO·CoOOH and PPi was investigated. As shown in Fig. S5D,† the fluorescence intensity of the AO·CoOOH complex in the presence of PPi decreased gradually with the increase in the reaction time and then almost reached equilibrium after 60 min. Consequently, 60 min was chosen in the subsequent research.
12 μg mL−1) to PPi. The pH of the buffer in the range from 3.0 to 9.0 had diverse effects on the quenching efficiency (Fig. S5B†). The results certified that the optimal sensing performance was obtained at pH 4.0. We also monitored the fluorescence intensity change as a function of reaction time to study the interaction kinetics of AO and CoOOH nanoflakes. As shown in Fig. S5C,† with the increase in the reaction time, the fluorescence intensity decreased gradually and then reached equilibrium after 5 min. Then, the optimal reaction time between AND-gate AO·CoOOH and PPi was investigated. As shown in Fig. S5D,† the fluorescence intensity of the AO·CoOOH complex in the presence of PPi decreased gradually with the increase in the reaction time and then almost reached equilibrium after 60 min. Consequently, 60 min was chosen in the subsequent research.
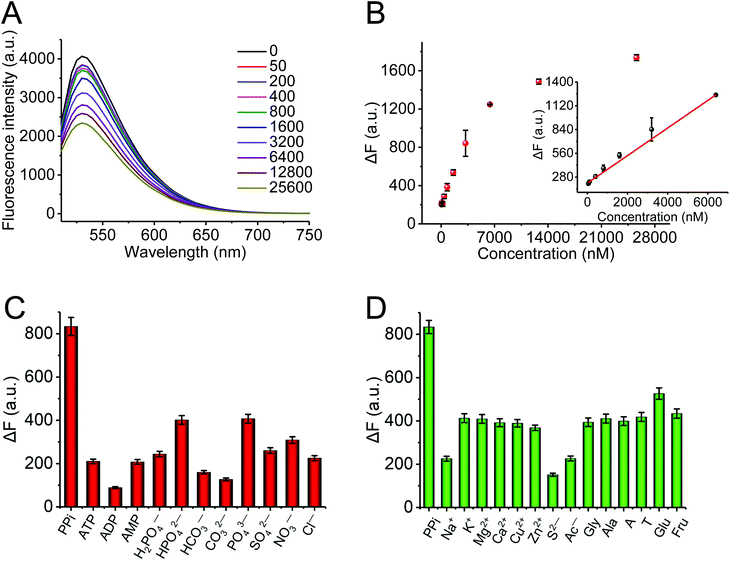
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 nM. Fig. 4B describes the plot of fluorescence intensity against the concentration of PPi. There was a good linear relationship between the fluorescence intensity and the concentration of PPi in the concentration range from 50 to 6400 nM (inset). The regression equation was ΔF = 0.16 [PPi] + 208.47 with a correlation coefficient of R2 = 0.999. The detection limit of PPi was 20 nM (3σ/slope). Fig. S6† further exhibits the relative fluorescence intensity of the AO·CoOOH complex in a 96-well plate in different conditions observed using the IVIS Spectrum imaging system; it clearly indicates the negative correlation between the fluorescence intensity of the AO·CoOOH complex and the PPi concentration. Compared with other reported optical PPi assays,16–18,32–36 our method has a much lower detection limit (Table S2†). The result revealed that the AO·CoOOH complex could afford a high sensitivity detection of PPi. Moreover, we further tested the fluorescence response of AO·CoOOH to potential interfering compounds including other anions, phosphate groups, biological molecules, and metal ions. As shown in Fig. 4C and D, in comparison with the interfering compounds, only PPi resulted in clear fluorescence change. The observations demonstrated that the AO·CoOOH complex presented excellent selectivity for the detection of PPi with high specificity.
600 nM. Fig. 4B describes the plot of fluorescence intensity against the concentration of PPi. There was a good linear relationship between the fluorescence intensity and the concentration of PPi in the concentration range from 50 to 6400 nM (inset). The regression equation was ΔF = 0.16 [PPi] + 208.47 with a correlation coefficient of R2 = 0.999. The detection limit of PPi was 20 nM (3σ/slope). Fig. S6† further exhibits the relative fluorescence intensity of the AO·CoOOH complex in a 96-well plate in different conditions observed using the IVIS Spectrum imaging system; it clearly indicates the negative correlation between the fluorescence intensity of the AO·CoOOH complex and the PPi concentration. Compared with other reported optical PPi assays,16–18,32–36 our method has a much lower detection limit (Table S2†). The result revealed that the AO·CoOOH complex could afford a high sensitivity detection of PPi. Moreover, we further tested the fluorescence response of AO·CoOOH to potential interfering compounds including other anions, phosphate groups, biological molecules, and metal ions. As shown in Fig. 4C and D, in comparison with the interfering compounds, only PPi resulted in clear fluorescence change. The observations demonstrated that the AO·CoOOH complex presented excellent selectivity for the detection of PPi with high specificity.
To evaluate the applicability of the AND-gate AO·CoOOH complex to real samples, we applied it to detect PPi in human serum. The human serum was diluted one hundred-fold with deionized water and spiked with different concentrations of PPi. Table 1 shows that the recoveries of PPi ranged from 102.34% to 114.06%, and the relative standard deviation varied from 0.49% to 1.20%. The results indicated that the AO·CoOOH complex has great potential for applications in a real biological system. Our molecular self-assembly system that was used for molecular logic gate and sensing operations relied on fluorescence quenching induced by CoOOH or/and PPi. Many factors (colloidal stability or interfering compounds) might lead to fluorescence signal reduction, thus resulting in the failure of the system. According to our observations and demonstrated results, the colloid solution was stable enough without precipitation during logic gate and sensing operations, and the AO·CoOOH system exhibited excellent sensitivity and selectivity towards PPi through fluorescence quenching, which proved that our quenching-based system possesses good accuracy and robustness.
| Sample | Added (μM) | Found (μM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Diluted human serum | 3.20 | 3.65 | 114.06 | 0.49 |
| 6.40 | 6.55 | 102.34 | 1.20 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
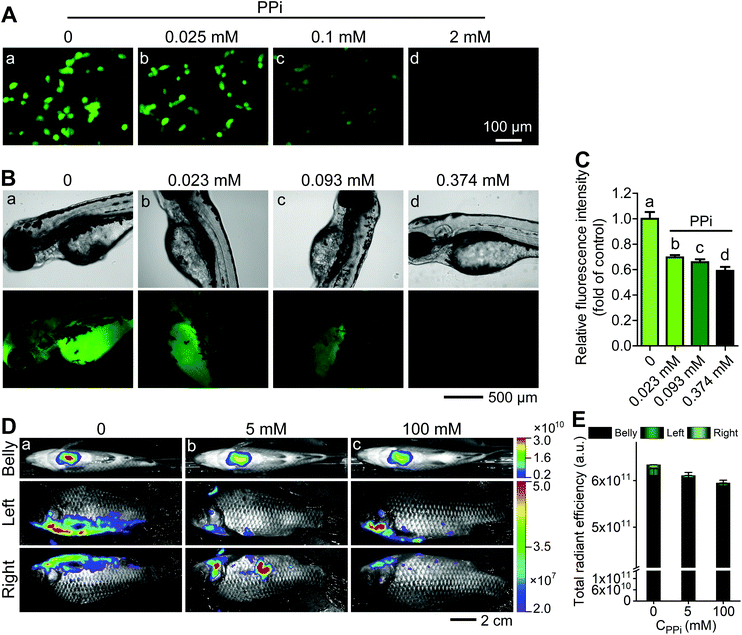
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 μg mL−1) for 2 h and 1 h, both CT26 cells and Zebrafish exhibited strong fluorescence signals (Fig. 5Aa and Ba). Then, different concentrations of PPi (0.025, 0.1, 2 mM for CT26 cells and 0.023, 0.093, 0.374 mM for Zebrafish) were subsequently added and incubated for another 1 h; it could be observed that the fluorescence was gradually quenched with the increase in the concentration of PPi (Fig. 5Ab–d, Bb–d, and C). Furthermore, we intraperitoneally injected the AO·CoOOH complex into Carassius auratus. After 1 h post injection, the fish exhibited strong fluorescence and the fluorescence intensity could be represented by multi-color distribution (Fig. 5Da). After 1 h post intraperitoneal injection of PPi, the fluorescence intensity of the fish decreased proportionately with increasing concentration of PPi (Fig. 5Db, c, and E). The pyrophosphate concentration in living things varies from the micromolar level to the millimolar level. For example, the pyrophosphate concentration is about 0.5 to 1.5 mM in Escherichia coli,37 over 10 mM in a yeast strain, and in the liver, the values range from a few micromolar to 1.4 mM.38 The optimized pH was 4.0 for in vitro sensing of PPi, and at the pH values above 6, molecular interactions among AO, CoOOH, and PPi may become weak probably due to deprotonation, resulting in reduction in the fluorescence quenching efficiency and sensitivity to PPi (Fig. S5B†); however, a sufficiently high concentration of PPi (such as sub-millimolar or millimolar level) at the physiological pH range could still cause remarkable fluorescence quenching of the AO·CoOOH system. The results indicated that the AO·CoOOH complex can be utilized to sense and image PPi in living cells and in Zebrafish and Carassius auratus in vivo.
12 μg mL−1) for 2 h and 1 h, both CT26 cells and Zebrafish exhibited strong fluorescence signals (Fig. 5Aa and Ba). Then, different concentrations of PPi (0.025, 0.1, 2 mM for CT26 cells and 0.023, 0.093, 0.374 mM for Zebrafish) were subsequently added and incubated for another 1 h; it could be observed that the fluorescence was gradually quenched with the increase in the concentration of PPi (Fig. 5Ab–d, Bb–d, and C). Furthermore, we intraperitoneally injected the AO·CoOOH complex into Carassius auratus. After 1 h post injection, the fish exhibited strong fluorescence and the fluorescence intensity could be represented by multi-color distribution (Fig. 5Da). After 1 h post intraperitoneal injection of PPi, the fluorescence intensity of the fish decreased proportionately with increasing concentration of PPi (Fig. 5Db, c, and E). The pyrophosphate concentration in living things varies from the micromolar level to the millimolar level. For example, the pyrophosphate concentration is about 0.5 to 1.5 mM in Escherichia coli,37 over 10 mM in a yeast strain, and in the liver, the values range from a few micromolar to 1.4 mM.38 The optimized pH was 4.0 for in vitro sensing of PPi, and at the pH values above 6, molecular interactions among AO, CoOOH, and PPi may become weak probably due to deprotonation, resulting in reduction in the fluorescence quenching efficiency and sensitivity to PPi (Fig. S5B†); however, a sufficiently high concentration of PPi (such as sub-millimolar or millimolar level) at the physiological pH range could still cause remarkable fluorescence quenching of the AO·CoOOH system. The results indicated that the AO·CoOOH complex can be utilized to sense and image PPi in living cells and in Zebrafish and Carassius auratus in vivo.
4. Conclusion
In summary, we demonstrated that the Boolean logic tree of molecular self-assembly system based on the CoOOH nanoplatform could be used for organizing and connecting “plug and play” molecular events (fluorescent dye, AO and anion, PPi). By using molecules as the inputs and the corresponding fluorescence signal as the output, the molecular Boolean logic tree system was programmed for three-input fluorescent Boolean logic computation and fluorescent three-state logic computation. Moreover, the stimulus responsiveness of the molecular Boolean logic tree system was also used for the fluorescent detection of PPi (linear range from 50 to 6400 nM with a detection limit of 20 nM) and even for imaging in living cancer cells and in vivo (in systems such as Zebrafish and Carassius auratus). Our study adds a new dimension for expanding molecular logic computing and sensing systems, which will not only provide more opportunities for developing novel logic computing paradigms, but also offer a potential sensing platform for understanding the role of important biomolecules (such as PPi) in physiological and pathological investigations. In the future, inspired by other complex biochemical cascading reactions39 (especially, enzyme-based catalytic processes), our proposed Boolean logic tree can be expanded to organize more complex systems and applied for the development of intelligent molecular computing and sensing systems (such as controllable and selectable sensing of multi-events).Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21505042), Scientific and Technological Plan Project of Changsha of China (No. KQ1707010), Hunan Province of China (No. 2016JJ3084), the Research Foundation of Education Bureau of Hunan Province (No. 15K084), the Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province (No. 20134486), Youth Foundation of Hunan Normal University (No. 31403), Doctoral Scientific Research Foundation of Hunan Normal University (No. 150612), Hunan Provincial Key Laboratory Open Foundation for Microbial Molecular Biology (No. 2014-03), and College Student Innovative Experiment Project of Hunan Province (No. 2017106).Notes and references
- J. K. Heinonen, Biological role of inorganic pyrophosphate, Springer Science & Business Media, 2001 Search PubMed.
- B. Muthuraj, S. Mukherjee, S. R. Chowdhury, C. R. Patra and P. K. Iyer, Biosens. Bioelectron., 2017, 89, 636–644 CrossRef CAS PubMed.
- A. K. Rosenthal, E. W. Campion and L. M. Ryan, N. Engl. J. Med., 2016, 374, 2575–2584 CrossRef CAS PubMed.
- S. H. Moochhala, J. A. Sayer, G. Carr and N. L. Simmons, Exp. Physiol., 2008, 93, 43–49 CrossRef CAS PubMed.
- A. P. de Silva and S. Uchiyama, Nat. Nanotechnol., 2007, 2, 399–410 CrossRef CAS PubMed.
- I. Willner, B. Shlyahovsky, M. Zayats and B. Willner, Chem. Soc. Rev., 2008, 37, 1153–1165 RSC.
- W. T. Huang, H. Q. Luo and N. B. Li, Anal. Chem., 2014, 86, 4494–4500 CrossRef CAS PubMed.
- W. T. Huang, J. R. Zhang, W. Y. Xie, Y. Shi, H. Q. Luo and N. B. Li, Biosens. Bioelectron., 2014, 57, 117–124 CrossRef CAS PubMed.
- K. M. Cherry and L. Qian, Nature, 2018, 559, 370–376 CrossRef CAS PubMed.
- L. Qian, E. Winfree and J. Bruck, Nature, 2011, 475, 368–372 CrossRef CAS PubMed.
- J. Y. Lu, X. X. Zhang, W. T. Huang, Q. Y. Zhu, X. Ding, L. Xia, H. Q. Luo and N. B. Li, Anal. Chem., 2017, 89, 9734–9741 CrossRef CAS PubMed.
- S. Lee, K. K. Y. Yuen, K. A. Jolliffe and J. Yoon, Chem. Soc. Rev., 2015, 44, 1749–1762 RSC.
- J. Wongkongkatep, A. Ojida and I. Hamachi, Top. Curr. Chem., 2017, 375, 30 CrossRef PubMed.
- K. A. Jolliffe, Acc. Chem. Res., 2017, 50, 2254–2263 CrossRef CAS PubMed.
- J. Wang, H. Chu, W. Chen and R. Sun, Chin. J. Inorg. Chem., 2016, 36, 2545–2558 CAS.
- Q. Wang, S. Zhang, H. Ge, G. Tian, N. Cao and Y. Li, Sens. Actuators, B, 2015, 207, 25–33 CrossRef CAS.
- L. Wang, Q. Song, Q. Liu, D. He and J. Ouyang, Adv. Funct. Mater., 2015, 25, 7017–7027 CrossRef CAS.
- T. Noipa, K. Ngamdee, T. Tuntulani and W. Ngeontae, Spectrochim. Acta, Part A, 2014, 118, 17–23 CrossRef CAS PubMed.
- Y. Cen, J. Tang, X. J. Kong, S. Wu, J. Yuan, R. Q. Yu and X. Chu, Nanoscale, 2015, 7, 13951–13957 RSC.
- N. Li, Y. H. Li, Y. Y. Han, W. Pan, T. T. Zhang and B. Tang, Anal. Chem., 2014, 86, 3924–3930 CrossRef CAS PubMed.
- H. M. Meng, X. B. Zhang, C. Yang, H. L. Kuai, G. J. Mao, L. Gong, W. H. Zhang, S. L. Feng and J. B. Chang, Anal. Chem., 2016, 88, 6057–6063 CrossRef CAS PubMed.
- Y. Cen, Y. Yang, R. Q. Yu, T. T. Chen and X. Chu, Nanoscale, 2016, 8, 8202–8209 RSC.
- Y. Q. Chang, Z. Zhang, H. Q. Liu, N. Wang and J. L. Tang, Analyst, 2016, 141, 4719–4724 RSC.
- J. Yang, H. Liu, W. N. Martens and R. L. Frost, J. Phys. Chem. C, 2010, 114, 111–119 CrossRef CAS.
- N. Li, Y. D. Zhu, T. Liu, S. G. Liu, S. M. Lin, Y. Shi, H. Q. Luo and N. B. Li, New J. Chem., 2017, 41, 1993–1996 RSC.
- N. Pandey and B. Choudhary, Analog Integr. Circ. Sig. Process., 2015, 84, 333–340 CrossRef.
- W. T. Huang, Y. Shi, W. Y. Xie, H. Q. Luo and N. B. Li, Chem. Commun., 2011, 47, 7800–7802 RSC.
- A. K. Shaw and S. K. Pal, J. Phys. Chem. B, 2007, 111, 4189–4199 CrossRef CAS PubMed.
- E. Salimi and J. Javadpour, J. Nanomater., 2012, 2012, 1–5 CrossRef.
- S. K. Gupta, M. Mohapatra, S. V. Godbole and V. Natarajan, RSC Adv., 2013, 3, 20046–20053 RSC.
- A. Pui, C. Policar and J. P. Mahy, Inorg. Chim. Acta, 2007, 360, 2139–2144 CrossRef CAS.
- Z. Hai, Y. Bao, Q. Miao, X. Yi and G. Liang, Anal. Chem., 2015, 87, 2678–2684 CrossRef CAS PubMed.
- S. Kim, M. S. Eom, S. K. Kim, S. H. Seo and M. S. Han, Chem. Commun., 2013, 49, 152–154 RSC.
- J. Deng, P. Yu, L. Yang and L. Mao, Anal. Chem., 2013, 85, 2516–2522 CrossRef CAS PubMed.
- L. J. Chai, J. Zhou, H. Feng, J. J. Lin and Z. S. Qian, Sens. Actuators, B, 2015, 220, 138–145 CrossRef CAS.
- L.-L. Tong, Z.-Z. Chen, Z.-Y. Jiang, M.-M. Sun, L. Li, J. Liu and B. Tang, Biosens. Bioelectron., 2015, 72, 51–55 CrossRef CAS PubMed.
- I. Mijakovic, S. Poncet, A. Galinier, V. Monedero, S. Fieulaine, J. Janin, S. Nessler, J. A. Marquez, K. Scheffzek, S. Hasenbein, W. Hengstenberg and J. Deutscher, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 13442–13447 CrossRef CAS PubMed.
- R. K. Airas and F. Cramer, Eur. J. Biochem., 1986, 160, 291–296 CrossRef CAS PubMed.
- A. Courbet, P. Amar, F. Fages, E. Renard and F. Molina, Mol. Syst. Biol., 2018, 14, e7845 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8an01565a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |