Ultrasmall hybrid protein–copper sulfide nanoparticles for targeted photoacoustic imaging of orthotopic hepatocellular carcinoma with a high signal-to-noise ratio†
Huixiang
Yan‡
 ab,
Jingqin
Chen‡
bc,
Ying
Li
d,
Yuanyuan
Bai
b,
Yunzhu
Wu
a,
Zonghai
Sheng
e,
Liang
Song
ab,
Jingqin
Chen‡
bc,
Ying
Li
d,
Yuanyuan
Bai
b,
Yunzhu
Wu
a,
Zonghai
Sheng
e,
Liang
Song
 b,
Chengbo
Liu
b,
Chengbo
Liu
 *b and
Hai
Zhang
*b and
Hai
Zhang
 *af
*af
aDepartment of Ultrasound, The Second Clinical College of Jinan University, Shenzhen People's Hospital, 518020 Shenzhen, China. E-mail: szzhhans.scc.jnu@foxmail.com; Tel: +86 (755) 2553-3497
bResearch Laboratory for Biomedical Optics and Molecular Imaging, Shenzhen Key Laboratory for Molecular Imaging, Institute of Biomedical and Health Engineering, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China. E-mail: cb.liu@siat.ac.cn; Tel: +86 (755) 8639-2240
cShenzhen College of Advanced Technology, University of Chinese Academy of Sciences, Shenzhen 518055, China
dDivision of Radiology, Department of Medicine, the University of Hong Kong Shenzhen Hospital, China
ePaul C. Lauterbur Research Center for Biomedical Imaging, Institute of Biomedical and Health Engineering, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China
fDepartment of Ultrasound, First Affiliated Hospital of Southern University of Science and Technology, The Shenzhen Medical Ultrasound Engineering Center, Shenzhen People's Hospital, 518020 Shenzhen, China
First published on 10th October 2018
Abstract
Although photoacoustic imaging combined with second near infrared (NIR II) molecular probes for tumor diagnosis has drawn tremendous attention during the past few decades, the targeted photoacoustic imaging of orthotopic hepatocellular carcinoma (HCC) still remains a challenge due to high liver vascularization and non-specificity of probes in liver tumors. Herein, we report on cyclic arginine–glycine–aspartic acid (cRGD) peptide conjugated ultrasmall CuS nanoparticles (CuS@BSA-RGD NPs) which encapsulate bovine serum albumin (BSA) and possess high optical absorption at 1064 nm. The encapsulation of BSA results in great biocompatibility of CuS@BSA-RGD NPs along with excellent photostability and physiological stability. The cRGD conjugation enables the improvement of tumor uptake of CuS@BSA-RGD NPs by virtue of its positive tumor cell targeting capability. The efficient accumulation of CuS@BSA-RGD NPs in the tumor over time after intravenous administration to orthotopic HCC bearing mice was achieved, which resulted in highly sensitive photoacoustic visualization of the tumor region. Toxicity studies indicate that CuS@BSA-RGD NPs exhibited negligible systemic toxicity in vivo. The results demonstrate that the CuS@BSA-RGD NPs might hold great promise for future imaging and diagnosis of cancer.
1. Introduction
In recent decades, photoacoustic imaging technology using noninvasive light and ultrasound to deliver potent optical contrast in deep tissue at high resolution1 has made significant progress in preclinical disease diagnosis.2–4 Photoacoustic imaging based on endogenous photo-absorbers (e.g. blood and melanin) has been employed in the noninvasive imaging of tumor angiogenesis5–8 and functional vascular structures,9,10 oxygen hypermetabolism8,11 and disordered hemodynamics,12 with both high contrast and excellent spatial resolution. However, for specific photoacoustic imaging of tumors, the high background signal of a photoacoustic signal observed from endogenous absorbers significantly reduced the imaging signal-to-noise ratio (SNR). Therefore, exogenous contrast agents have been developed to increase the SNR of photoacoustic imaging for tumor diagnosis, such as subcutaneous13–15 and orthotopic tumors.16–19 Due to the limitation of interference of the background signal and the demands for high penetration depth, a higher imaging SNR is critical for photoacoustic imaging in deep orthotopic tumors. To date, only a few studies of photoacoustic imaging in orthotopic hepatocellular carcinoma (HCC) have been reported, mainly due to the challenge of a large blood background signal in the liver resulting from high vascularization. Importantly, the liver is the major metabolic organ of the body, resulting in a significant accumulation of a large number of imaging probes without tumor specificity.20,21 Therefore, there is an increasing demand for developing exogenous contrast agents to enhance the SNR of photoacoustic imaging in orthotopic HCC.The ideal strategy to enhance the SNR of imaging is to increase the sensitivity of contrast agents and decrease the background signal. Currently, various NIR photoacoustic contrast agents have been developed, including carbon materials,22–24 precious metal materials,25,26 transition metal sulfides,27–29 and small molecule dyes.30–33 Although these materials showed good absorption properties at excitation wavelengths in the range of 650–950 nm (NIR I), the laser wavelengths in the NIR I region have shown high light absorption and photon scattering in tissues, resulting in an unsuitable large background signal (e.g. blood signal), and limited photoacoustic imaging in deep tissues.34 The second NIR region (NIR II, 1000–1700 nm) lasers which have lower optical absorption and photon scattering properties are the ideal candidates for the generation of low background signals to overcome the drawbacks mentioned above.35 Hence, the NIR II photoacoustic contrast agents have attracted increasing scientific interest in biomedical applications, specifically, for orthotopic HCC imaging.
With rapid advances in nanotechnology, various types of nanomaterials with high NIR II absorption, such as gold nanorods,36 copper sulphide (CuS) nanoparticles,37 silver nanoplates,38 phosphorus phthalocyanines,39 semiconductor materials (SPNs),40–43etc. have been reported as photoacoustic contrast agents for photoacoustic imaging with a high SNR. Among these nanomaterials, CuS based nanoparticles are promising materials due to their high NIR II absorption,37,44 low toxicity45 and efficient biodegradability46 during normal metabolic processes. Gao et al. reported a protein-modified CuS nanoparticle for photoacoustic imaging and nuclear magnetic bimodal imaging of mouse subcutaneous tumors.44 Besides, Ku et al. have prepared a CuS semiconductor nanoparticle for the photoacoustic imaging of orthotopic brain tumors in deep brain regions.37 However, to the best of our knowledge, no study has used photoacoustic imaging of targeted orthotopic HCC with NIR II wavelengths to achieve a high SNR.
Owing to the high biocompatibility and facile chemical modification, BSA, has been extensively used in drug delivery systems, metal nanoparticle fabrication and imaging contrast agent prepration.47–51 Since tumor vessels and most tumor cells overexpress αvβ3 and αvβ5 integrin receptors, cRGD which possesses excellent selectivity and high binding affinity to these receptors can be used as a tumor targeting ligand.52,53 In this work, we reported on a contrast reagent based on NIR II ultrasmall CuS nanoparticles with bovine serum albumin (BSA) encapsulation and cyclic arginine–glycine–aspartic acid (cRGD) peptide conjugation (CuS@BSA-RGD NPs) for active targeted photoacoustic imaging in orthotopic HCC. The CuS@BSA-RGD NPs showed negligible cytotoxicity, low hemolysis, efficient cellular uptake and high SNR photoacoustic imaging in vivo at 1064 nm. High quality contrast enhanced photoacoustic images delineating the tumor region were obtained after the intravenous injection of CuS@BSA-RGD NPs into orthotopic HCC-bearing mice. Our results indicate that CuS@BSA-RGD NPs have great potential for being used as a photoacoustic contrast agent for future biomedical imaging applications. To date, the results reported here describe the first attempt to use CuS@BSA-RGD NPs as a targeted NIR II probe for photoacoustic imaging of orthotopic HCC.
2. Materials and methods
2.1. Materials
Copper chloride (CuCl2), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), indocyanine green (ICG), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), and dimethyl sulfoxide (DMSO) were obtained from Sigma-Aldrich (St Louis, MO). Bovine serum albumin (BSA) was purchased from Biosharp Company. ICG-Sulfo-OSu was obtained from AAT Bioquest. Inc. Sodium sulfide nonahydrate (Na2S·9H2O) and sodium hydroxide (NaOH) were purchased from Aladdin. The cRGD peptide was purchased from GL Biochem (Shanghai) Ltd. Fetal bovine serum (FBS), Dulbecco's modified Eagle's medium (DMEM) and Trypsin-EDTA were obtained from Gibco BRL (Eggenstein, Germany). Ultra-purified water (18.25 MΩ cm, 25 °C) used throughout all experiments was derived from a Milli-QPlus System (Millipore Corporation, Bedford, USA). Other chemicals used this study were of analytical reagent grade and used as received.2.2. Synthesis of CuS@BSA-RGD NPs
0.25 g of BSA dissolved in 7.5 mL of ultrapure water was used as the core reagent. Next, 0.268 g of CuCl2 and 0.48 g of Na2S·9H2O were dissolved in 10 mL of ultrapure water to form the reaction solution (0.2 M). Then, 1 mL of the copper solution was added to the BSA solution. 0.5 mL of a 0.4 M NaOH solution was added to adjust the pH, followed by the addition of 1.5 mL of sodium sulfide solution. Then, the mixture was heated to 90 °C and stirred for about 30 min. The aqueous solution was then cooled down to room temperature. Then, 40 mg of EDC was added to the prepared CuS@BSA solution at room temperature with slight stirring. After 20 min, 32 mg of NHS was added to the mixture and allowed to react for another 2 h in the dark. 1 mL of cRGD (5 mg mL−1) was added to the solution. The resulting mixture was vigorously stirred for 12 h. The aqueous solution was filtered through a polyethersulfone (PES) syringe driven filter (0.22 μm) and centrifuged three times using a 30 K centrifugal filter unit at 8000 rpm for 15 min. The product was dialyzed against deionized water for 2 days using a dialysis bag with a molecular weight cut-off (MWCO) of 3500 Da. The aqueous solution was then stored at 4 °C.2.3. Characterization of CuS@BSA-RGD NPs
Transmission electron microscopy (TEM) imaging was performed using a FEI Tecnai Sphera (Tecnai G2 F20 S-TWIN, FEI, USA) instrument. Dynamic light scattering (DLS) and zeta potential measurements of the CuS@BSA-RGD NPs were carried out using a Zetasizer Nano-ZS (Malvern Instruments, UK). UV spectra were recorded on a UV-vis-NIR spectrophotometer (UV-2600, Shimadzu, Japan) with the corresponding samples placed in a 1 cm quartz cuvette. All photoacoustic measurements were carried out on a custom-built acoustic-resolution photoacoustic computed tomography (AR-PACT) system and acoustic-resolution photoacoustic microscopy (AR-PAM) system with a tunable pulsed optical parametric oscillator (OPO) laser (Vibrant 532 I HE, Opotek, USA). Detailed information of this custom-built system can be found in our previous literature reports.54,552.4. Photoacoustic performance
The CuS@BSA-RGD NP dispersions at various concentrations (31.25, 62.5, 125, 250 and 500 μg mL−1) were filled in the individual holes of a custom-made agar plate and imaged with excitation wavelengths of 780 nm and 1064 nm. The photoacoustic signal of each sample was adopted by averaging over the sample area. To study the photostability of CuS@BSA-RGD NPs, 3000 laser pulses were used to irradiate the CuS@BSA-RGD NPs and real-time recorded any resulting photoacoustic signals. To investigate the in vivo photoacoustic performance, CuS@BSA-RGD NPs were blended with Matrigel (BD Biosciences) at a series of concentrations. The final percentage in the mixtures of Matrigel was 50% (v/v) for all solutions. The Matrigel–nanoparticle mixture (50 μL) was then injected subcutaneously on the dorsal areas of male nude mice to form inclusions of CuS@BSA-RGD NPs. Matrigel alone was imaged as a control. In order to verify that the material penetration efficiency at an irradiation wavelength of 1064 nm was higher than that at a shorter wavelength, various wavelengths (700, 780, 850, 900, and 1064 nm) were selected and irradiated at the same energy on the same biological tissue samples side-by-side.2.5. Cellular uptake
Fluorescence images were recorded to confirm the different ways of internalization of nanoparticles into cells. CuS@BSA-RGD NPs and CuS@BSA NPs were labeled by ICG-Sulfo-OSu, an ICG derivative, respectively. Briefly, 0.5 mg of ICG-Sulfo-OSu powder was added to 1 mL of Cu-based solutions (≈1 mg mL−1) and stirred at room temperature for 12 h. Then, the unbound ICG-Sulfo-OSu was removed through dialyzation against deionized water for 2 days and the solid ICG-Sulfo-OSu was eliminated through centrifugation at 8000g for 15 min. The hybrid was re-dispersed using deionized water to form the ICG labeled products CuS@BSA-ICG-RGD NPs and CuS@BSA-ICG NPs, respectively. All procedures were conducted in the absence of direct light. The absorbance and fluorescence spectra of ICG labeled CuS@BSA-RGD NPs and CuS@BSA NPs were measured using a UV-vis-NIR and fluorescence spectrometer to confirm ICG conjugation. After culturing in a confocal dish for 24 h, Hep G2 cells were incubated with ICG labeled CuS@BSA-RGD NPs and CuS@BSA NPs (≈0.2 mg mL−1) for 3 h, respectively. The redundant nanocomposites outside the cells were washed off with PBS and ultrapure water three times. Subsequently, the cells were successively fixed with 4% paraformaldehyde solution and 4′,6-diamidino-2-phenylindole (DAPI, 10 μg mL−1) for 10 min and 3 min respectively. For DAPI and ICG excitation, fluorescence images of the stained cells were then recorded using a confocal laser scanning microscope (CLSM, Leica TCS SP5, Germany) at 405 nm and 633 nm, respectively.2.6. In vitro biocompatibility
The human liver cancer cell line (Hep G2) was obtained from the American Type Culture Collection (ATCC). The Hep G2 cells were cultured in DMEM medium supplemented with 10% (v/v) fetal bovine serum, 1% (v/v) penicillin or 1% (v/v) streptomycin and incubated at 37 °C under a 5% CO2 atmosphere. Hep G2 human liver tumor cells were used to evaluate the cytotoxicity of CuS@BSA-RGD NPs. Cells in the log phase of growth were seeded into 96-well plates at a density of 6 × 104 cells per well and incubated for 24 h for cell attachment. CuS@BSA-RGD NP solutions were then appropriately diluted with fresh culture medium and added to the wells (200 μL). After 24 h of continuous exposure to various concentrations of CuS@BSA-RGD NPs, the culture media were replaced by fresh culture media to remove any free CuS@BSA-RGD NPs in solution. The relative cell viability (RCV) was investigated using standard MTT assays and measured using a 96-well plate reader (BioTek SynergyTM 4) at 450 nm using the following equation:where At, Apc and Anc represent the absorbance of the tested group, and the positive and negative controls, respectively. In addition, whole blood (≈1 mL) from Balb/c nude mice was collected and centrifuged (1500 rpm, 3 min) to separate the red blood cells (RBCs). The RBCs were washed with PBS 3 times. 10% RBCs (v/v, in PBS) were incubated at a series of concentrations of CuS@BSA-RGD NPs (25, 50, 100, 150 and 200 μg mL−1) at 37 °C for 4 h. After centrifugation (8000 rpm, 1 min), the supernatant of the suspensions was collected and analyzed by using a UV-Vis-NIR spectrometer at 541 nm. The hemolysis percentage (HP) was calculated using the equation as follows:
where Dt, Dpc and Dnc are the absorbance of the tested sample, and the positive (deionized water) and negative (PBS) controls, respectively.
2.7. Animal model
Male Balb/c nude mice (20 ± 2 g, 6–8 weeks old) were purchased from the Beijing Vital River Laboratory Animal Technology Co., Ltd. All animal procedures were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals of Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, and approved by the Animal Ethics Committee of Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences. To establish a human liver cancer orthotopic Hep G2 tumor model, Hep G2 cells (2 × 105) were injected into the liver of male Balb/c nude mice. After ≈2 weeks of growth, the sonograms of the HCC were acquired by using an ultrasound device with a 40 MHz high-frequency transducer (Vevo 2100; VisualSonics, Toronto, Canada). Then, the mice were used for subsequent experiments. Finally, the cells were validated by Hematoxylin and Eosin (H&E) staining.2.8. In vivo photoacoustic imaging of orthotopic HCC
Mice bearing Hep G2 tumors were administered 100 μL of the CuS@BSA-RGD NPs at a concentration of 1 mg mL−1via tail vein intravenous injection. At predetermined time points, the tumor regions were imaged using a custom-built AR-PACT system with excitation wavelengths of 780 nm and 1064 nm. After imaging, the mice were sacrificed and their major organs were harvested, including the heart, liver, spleen, lungs and kidneys.2.9. In vivo toxicity
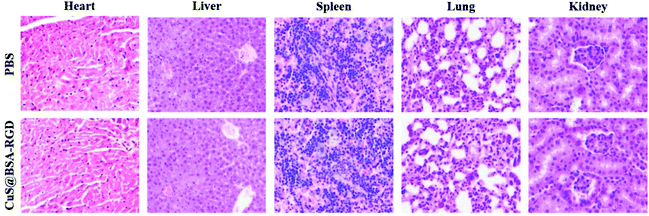
A dosage of 20 mg kg−1 of CuS@BSA-RGD NPs was administered via tail vein injection into healthy Balb/c mice (five mice per group), respectively. PBS was used as the control. Fourteen days after the injection of CuS@BSA-RGD NPs, the blood (about 1 mL per animal) of the mice was collected via orbit blood withdrawal for subsequent blood biochemistry assays and blood panel tests. The latter were performed at the Shenzhen Center for Disease and Prevention. Moreover, the major organs (including the heart, liver, kidneys, lungs and spleen) from CuS@BSA-RGD NP treated mice were harvested. Through a series of processes, including fixation in 10% neutral buffered formalin, embedding into paraffin and sectioning at 5 μm thickness, the tissues were stained with H&E and examined using a digital microscope.3. Results and discussion
3.1. Synthesis and characterization of CuS@BSA-RGD NPs
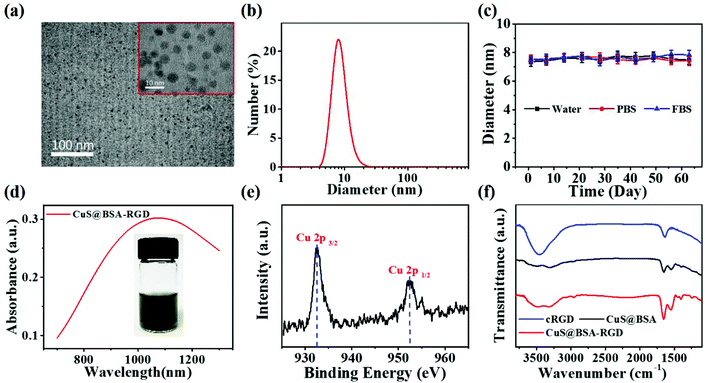
The CuS@BSA-RGD NPs were fabricated via three individual synthetic steps, followed by the conjugation with RGD (Scheme 1a), for high SNR photoacoustic imaging of HCC (Scheme 1b). HR-TEM images reveal that the CuS@BSA-RGD NPs have uniform size, and their morphology is spherical (Fig. 1a). The average diameter of CuS@BSA-RGD NPs was about 7 nm (Fig. 1b). The zeta potential of CuS@BSA-RGD NPs and CuS@BSA NPs was −26.8 mV and −28.7 mV (Fig. S1†). After two months of storage in water followed by PBS and FBS at 25 °C, no size change of CuS@BSA-RGD NPs was found, indicating that the CuS@BSA-RGD NPs possess excellent dispersibility and stability (Fig. 1c). Furthermore, we have demonstrated that BSA, a natural active protein, could effectively stabilize CuS nanoparticles through facile encapsulation.56–59 The optical absorption spectrum obtained showed that CuS@BSA-RGD NPs feature remarkable absorption properties at 1064 nm in the NIR-II window (Fig. 1d). To examine the chemical status of the Cu element and verify its composition in CuS@BSA-RGD NPs, we analyzed the X-ray photoelectron spectroscopy (XPS) of CuS@BSA-RGD (Fig. 1e) and CuS@BSA NPs (Fig. S2†). As shown in the Cu 2p spectrum, two peaks at 932.7 eV and 952.8 eV corresponding to Cu(II) 2p3/2 and Cu(II) 2p1/2 were essentially identical binding energies for Cu2+ in CuS, which were consistent with the reported literature.44,60 Additionally, the FT-IR spectrum of CuS@BSA (Fig. 1f) shows two peaks at ∼1644 cm−1 and ∼1532 cm−1, revealing the existence of the characteristic bands of BSA.61 Compared to CuS@BSA, a stronger broad peak between 3700 cm−1 and 3100 cm−1 was observed from the FT-IR spectrum of CuS@BSA-RGD, indicating the existence of a –NH–CO– bond between BSA and cRGD. The results demonstrated that BSA and cRGD were successfully covalently grafted onto BSA via an amidation reaction.62,63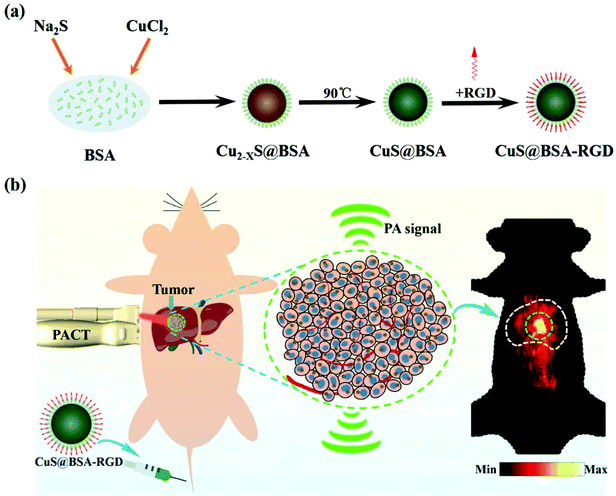 | ||
| Scheme 1 (a) The schematic synthesis of CuS@BSA-RGD nanoparticles and (b) the application of photoacoustic imaging in an orthotopic HCC model. | ||
3.2 Photoacoustic properties of CuS@BSA-RGD NPs
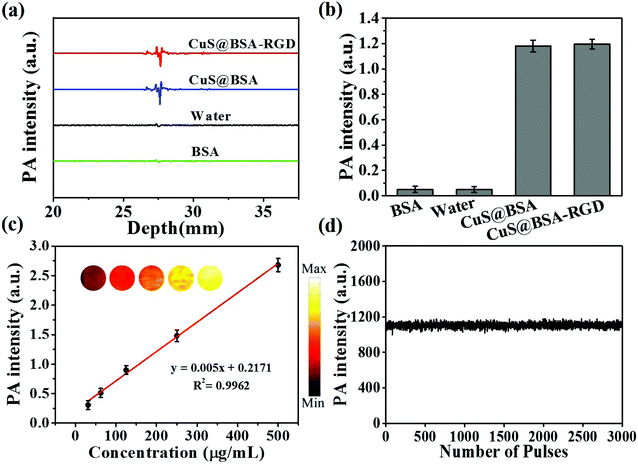
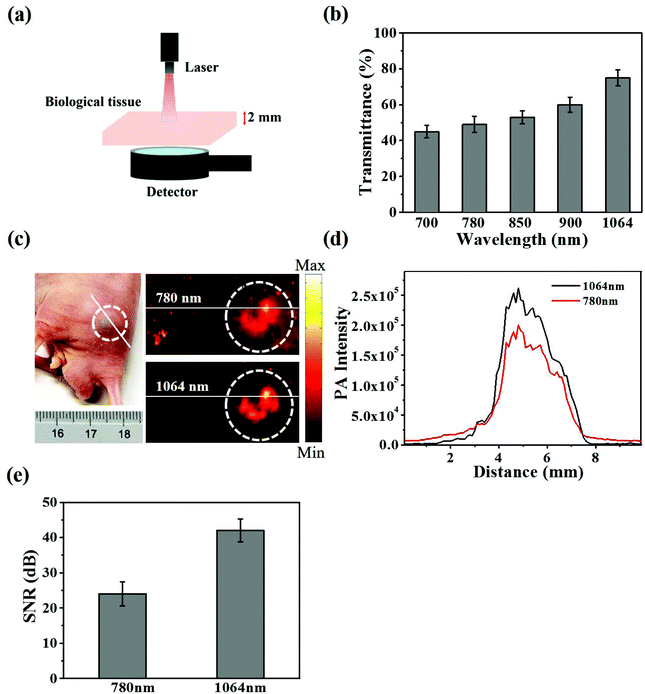
As shown in Fig. 2a and b, CuS@BSA-RGD NPs and CuS@BSA NPs showed a strong photoacoustic signal at 1064 nm pulse laser excitation. However, BSA and water showed negligible photoacoustic signals. Fig. 2c shows that the photoacoustic signal increased linearly (R2 = 0.9962), with an enhancement of CuS@BSA-RGD NPs at a concentration ranging from 31.25 to 500 μg mL−1. After irradiation with 3000 laser pulses and 10 mJ cm−2 laser fluence, the photoacoustic signal of CuS@BSA-RGD NPs showed no significant attenuation, indicating the excellent photostability of CuS@BSA-RGD NPs (Fig. 2d). These results demonstrate that CuS@BSA-RGD NPs have excellent photoacoustic properties at pulse laser excitation (1064 nm).In order to evaluate the laser penetration ability at various laser wavelengths in biological tissues, the laser energy after penetrating into 2 mm of chicken breast tissue was measured (Fig. 3a). As shown in Fig. 3b, the transmittance percentage of a 1064 nm laser was higher than that of other wavelengths (700, 780, 850 and 900 nm). In addition, Fig. 3c shows the photoacoustic maximum amplitude projection (MAP) images of the right back of nude mice post subcutaneous injection of CuS@BSA-RGD NPs and upon excitation at 780 nm and 1064 nm. The photoacoustic signal intensity in two solid red lines at a pulse laser wavelength of 1064 nm (Fig. 3c) was 1.37-fold higher than that of the photoacoustic signal intensity at a pulse laser wavelength of 780 nm (Fig. 3d). The SNR of the photoacoustic MAP images at 1064 nm laser excitation was calculated to be 1.75 fold higher than that at a laser excitation of 780 nm (Fig. 3e). The high SNR resulted from the intense photoacoustic signal of CuS@BSA-RGD NPs and lower background noise because of the low tissue absorption and scattering at 1064 nm laser excitation.
3.3. Cellular uptake and quantitative assay
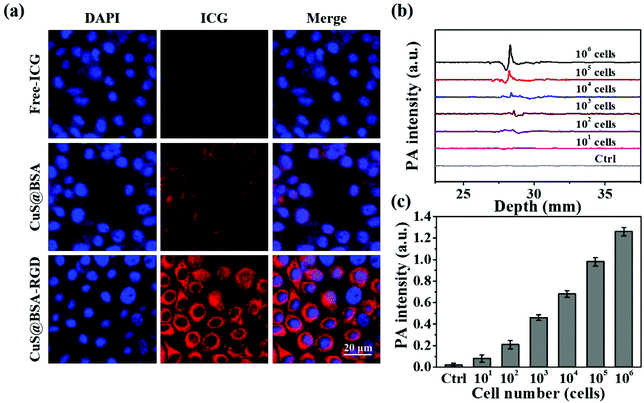
As shown in Fig. 4a, CuS@BSA-RGD NPs labeled with ICG incubated with Hep G2 cells for 3 h exhibit the strongest ICG fluorescence signal in the cytoplasm compared with free ICG and CuS@BSA NPs. The enhanced cellular uptake of CuS@BSA-RGD NPs might be attributed to the RGD receptor integrin αvβ3 on Hep G2 cell membranes mediated by the cell endocytosis pathway.64 Moreover, Hep G2 cells with the inclusion of CuS@BSA-RGD NPs were collected and their photoacoustic signal was detected using the AR-PAM system. As shown in Fig. 4b and c, the control Hep G2 cells without CuS@BSA-RGD NPs presented a negligible photoacoustic signal. After 3 h of incubation, a strong photoacoustic signal could be observed. The photoacoustic intensity increased linearly with the augment of cell numbers (Fig. 4c). The limit of detection is about 100 cells. These findings demonstrated that CuS@BSA-RGD NPs could be efficiently internalized by Hep G2 cells and generate intense photoacoustic signals, potentially allowing for the utilization of CuS@BSA-RGD NPs for highly sensitive photoacoustic imaging in orthotopic HCC.3.4. In vitro biocompatibility of CuS@BSA-RGD NPs
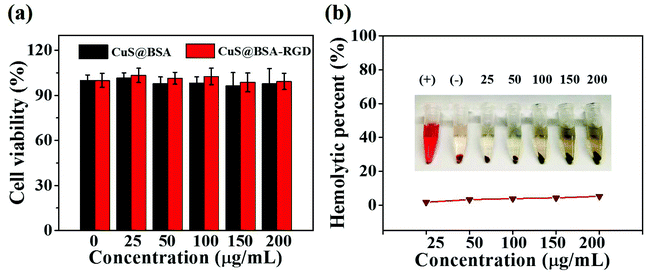
The cytotoxic properties of CuS@BSA NPs and CuS@BSA-RGD NPs were tested using a standard MTT assay. CuS@BSA NPs and CuS@BSA-RGD NPs at different concentrations incubated with Hep G2 cells for 24 h showed no obvious cytotoxic effects, indicating that the nanoparticles have low cytotoxicity (Fig. 5a). Moreover, the effects of CuS@BSA-RGD NPs on the hemolytic behavior of red blood cells were studied to evaluate the hemocompatibility. As shown in Fig. 5b, the hemolysis percentages of CuS@BSA-RGD NPs were lower than 5.5% at the tested concentrations, demonstrating a suitable blood compatibility of the CuS@BSA-RGD NPs. Furthermore, these results suggested that the CuS@BSA-RGD NPs also have appropriate in vitro biocompatibility.3.5. In vivo photoacoustic imaging of orthotopic HCC
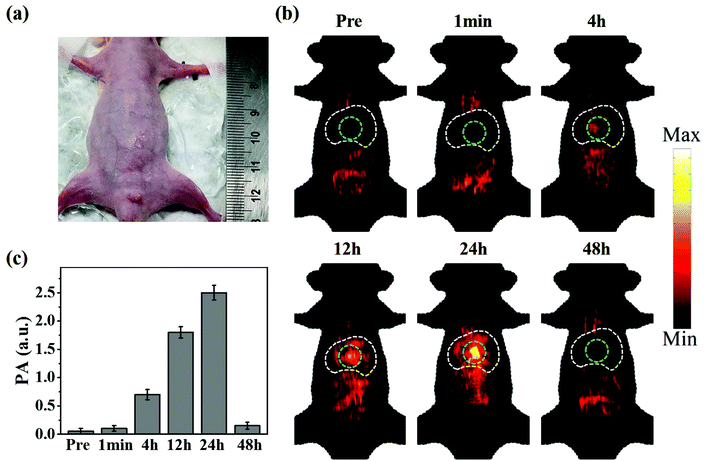
The successfully established orthotopic HCC model was verified using ultrasound images and liver tissue H&E staining images (Fig. S3 and S4†). Fig. 6a shows a photograph of the orthotopic HCC bearing mouse used in this study. CuS@BSA-RGD NPs were injected via the tail vein and the photoacoustic signal was monitored at different time points. As shown in Fig. 6b, weak photoacoustic signals in the thorax and abdominal region could be attributed to the intestine and blood vessels which were observed prior to CuS@BSA-RGD NP injection. Post injection and with time, stronger photoacoustic signals could be detected in the liver region that reached a peak value at 24 h post injection. The latter finding is most likely due to the liver metabolism and enrichment effect of nanoparticles. Upon target accumulation of CuS@BSA-RGD NPs, 24 h post injection, the tumors could be clearly detected via photoacoustic imaging. The PA signals were greatly reduced at 48 h post injection compared to at 24 h post injection. The statistical results of the photoacoustic signal in the tumors are shown in Fig. 6c. In contrast, CuS@BSA NPs showed no significant tumor accumulation effect (Fig. S5†). As shown in Fig. S6,† the liver bearing HCC was taken out for photoacoustic imaging compared to the normal liver at 24 h post injection. PA signals in the region of HCC are stronger than that in the region without a tumor or in the normal liver, which is in agreement with the result of in vivo photoacoustic imaging. Moreover, the amount of Cu in the normal liver and HCC was quantified by ICP-OES and the result is shown in Fig. S7.† In addition, the blood circulation time shows that the CuS@BSA-RGD NPs were able to circulate in blood for 48 h (Fig. S8†), which allows nanoparticles to accumulate in HCC via the EPR effect effectively. These results confirm the accumulation of the nanomaterials in HCC. The imaging depth of the tumor site was found to be ≈2 mm below the skin. The high SNR images of the tumors could mainly be attributed to the high photoacoustic signal of the CuS@BSA-RGD NPs and the low tissue light absorption and low background signal caused by the photon scattering of a 1064-nm pulse laser. These results demonstrated that CuS@BSA-RGD NPs represent a highly efficient NIR II photoacoustic contrast agent for the orthotopic HCC model.3.6. In vivo toxicity of CuS@BSA-RGD NPs
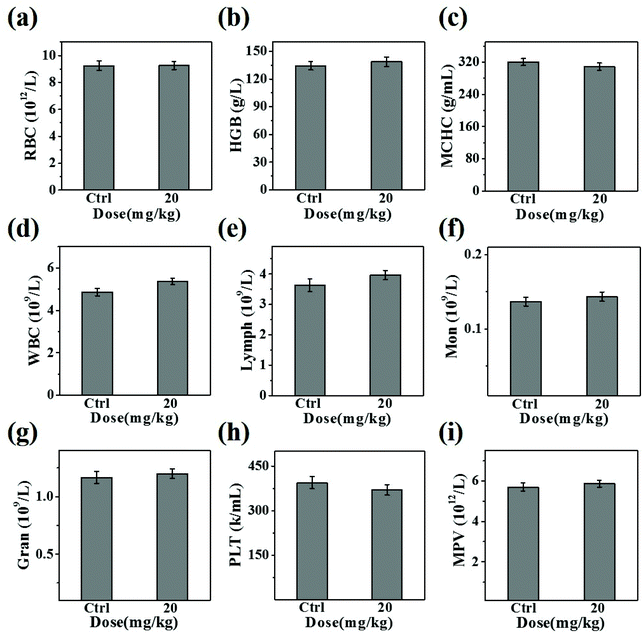
The in vivo toxicity of CuS@BSA-RGD NPs was investigated via blood and histology analysis in healthy mice. CuS@BSA-RGD NPs at a dosage of 20 mg kg−1 were administered to the mice via tail vein injection. After 14 days, the complete blood panel parameters of the mice in the CuS@BSA-RGD NP treated group were measured, and the results of the CuS@BSA-RGD NP treated group were found to be almost the same as those of the control group (Fig. 7a–i). Meanwhile, the H&E staining images of the main organs (heart, liver, spleen, lungs and kidneys) showed no obvious organ damage or inflammation (Fig. 8). Therefore, the excellent biocompatibility and biosafety of the CuS@BSA-RGD NPs may render this material a promising agent for wide in vivo biomedical applications.4. Conclusion
Collectively, ultrasmall CuS@BSA-RGD NPs were successfully synthesized with great stability and biocompatibility for the photoacoustic imaging of targeted orthotopic HCC. The CuS@BSA-RGD NPs exhibited strong absorbance in the NIR II window. Therefore, photoacoustic imaging in deep tissue areas with a lower background was achieved because the biological tissues show low optical absorption and photon scattering at the NIR II wavelengths. Moreover, the active tumor targeting capability of the NPs enables them to accumulate significantly in the tumor region. Thus, highly sensitive photoacoustic imaging of orthotopic HCC was achieved, and the orthotopic HCC region showed significantly enhanced photoacoustic signals (consequently much higher SNR) compared to that of the normal liver tissue. In addition, the CuS@BSA-RGD NPs were non-toxic both in vitro and in vivo. The first time validation of the CuS@BSA-RGD NPs for the active targeted photoacoustic imaging of the orthotopic HCC model revealed its great potential for highly sensitive and accurate HCC detection in future translational medicine.Author contributions
H. Z. and C. L. conceived the study. H. Y., J. C., Y. L., Y. B. and Y. W. performed the experiments. H. Y. and J. C. analysed the data. H. Y., J. C., Y. L., Y. B., Y. W., Z. S. and L. S. interpreted the results. H. Y. and J. C. prepared the manuscript with input from all co-authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge financial support through the following grants: Shenzhen Science and Technology Innovation grant JCYJ20160422153149834, JCYJ20170413153129570, JCYJ20160531175040976, JCYJ20150521144321005, JCYJ20160608214524052 and JCYJ20150731154850923; National Natural Science Foundation of China (NSFC) grants 81771841, 91739117, 81522024, 81427804, 81430038 and 61475182; National Key Basic Research (973) Program of China grants 2014CB744503 and 2015CB755500; Guangdong Natural Science Foundation grants 2014B050505013 and 2014A030312006; Guangzhou Science and Technology Innovation grant 201604020144.Notes and references
- L. V. Wang and S. Hu, Science, 2012, 335, 1458–1462 CrossRef CAS PubMed.
- L. Li, L. Zhu, C. Ma, L. Lin, J. Yao, L. Wang, K. Maslov, R. Zhang, W. Chen, J. Shi and L. V. Wang, Nat. Biomed. Eng., 2017, 1, 0071 CrossRef PubMed.
- K. S. Valluru, K. E. Wilson and J. K. Willmann, Radiology, 2016, 280, 332–349 CrossRef PubMed.
- M. Jeon, J. Kim and C. Kim, Med. Biol. Eng. Comput., 2016, 54, 283–294 CrossRef PubMed.
- M. Omar, M. Schwarz, D. Soliman, P. Symvoulidis and V. Ntziachristos, Neoplasia, 2015, 17, 208–214 CrossRef PubMed.
- T. Jin, H. Guo, H. Jiang, B. Ke and L. Xi, Opt. Lett., 2017, 42, 4434–4437 CrossRef PubMed.
- R. Lin, J. Chen, H. Wang, M. Yan, W. Zheng and L. Song, Quant. Imaging Med. Surg., 2015, 5, 23 Search PubMed.
- J. Yao, K. I. Maslov and L. V. Wang, Technol. Cancer Res. Treat., 2012, 11, 301–307 CrossRef PubMed.
- T. J. Allen, O. Ogunlade, E. Zhang and P. C. Beard, Biomed. Opt. Express, 2018, 9, 650 CrossRef PubMed.
- N. Liu, S. Yang and D. Xing, Opt. Lett., 2018, 43, 138–141 CrossRef PubMed.
- C. Yeh, S. Hu, J. Liang, L. Li, B. Soetikno, Z. H. Lu, R. E. Sohn, K. Maslov, J. M. Arbeit and L. V. Wang, Proc. SPIE, 2015, 9323, 93233H CrossRef.
- A. Berezhnoi, M. Schwarz, A. Buehler, S. V. Ovsepian, J. Aguirre and V. Ntziachristos, J. Biophotonics, 2018, e201700359 CrossRef PubMed.
- J. Chen, X. Li, X. Liu, H. Yan, Z. Xie, Z. Sheng, X. Gong, L. Wang, X. Liu, P. Zhang, H. Zheng, L. Song and C. Liu, Biomater. Sci., 2018, 6, 1503–1516 RSC.
- J. Chen, C. Liu, G. Zeng, Y. You, H. Wang, X. Gong, R. Zheng, J. Kim, C. Kim and L. Song, Nanoscale Res. Lett., 2016, 11, 85 CrossRef PubMed.
- L. Chen, X. Zhou, W. Nie, W. Feng, Q. Zhang, W. Wang, Y. Zhang, Z. Chen, P. Huang and C. He, ACS Appl. Mater. Interfaces, 2017, 9, 17786–17798 CrossRef CAS PubMed.
- J. Chen, C. Liu, D. Hu, F. Wang, H. Wu, X. Gong, X. Liu, L. Song, Z. Sheng and H. Zheng, Adv. Funct. Mater., 2016, 26, 8715–8725 CrossRef CAS.
- Q. Chen, C. Liang, X. Sun, J. Chen, Z. Yang, H. Zhao, L. Feng and Z. Liu, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5343–5348 CrossRef CAS PubMed.
- M. K. Gurka, D. Pender, P. Chuong, B. L. Fouts, A. Sobelov, M. W. McNally, M. Mezera, S. Y. Woo and L. R. McNally, J. Controlled Release, 2016, 231, 60–67 CrossRef CAS PubMed.
- S. Liang, C. Li, C. Zhang, Y. Chen, L. Xu, C. Bao, X. Wang, G. Liu, F. Zhang and D. Cui, Theranostics, 2015, 5, 970–984 CrossRef CAS PubMed.
- T. Sun, Y. S. Zhang, B. Pang, D. C. Hyun, M. Yang and Y. Xia, Angew. Chem., Int. Ed., 2014, 53, 12320–12364 CAS.
- D. Lee, S. Beack, J. Yoo, S.-K. Kim, C. Lee, W. Kwon, S. K. Hahn and C. Kim, Adv. Funct. Mater., 2018, 28, 1800941 CrossRef.
- L. Xie, G. Wang, H. Zhou, F. Zhang, Z. Guo, C. Liu, X. Zhang and L. Zhu, Biomaterials, 2016, 103, 219–228 CrossRef CAS PubMed.
- J. Liu, C. Wang, X. Wang, X. Wang, L. Cheng, Y. Li and Z. Liu, Adv. Funct. Mater., 2015, 25, 384–392 CrossRef CAS.
- A. Zerda, Z. Liu, S. Bodapati, R. Teed, S. Vaithilingam, B. T. Khuri-Yakub, X. Chen, H. Dai and S. S. Gambhir, Nano Lett., 2010, 10, 2168–2172 CrossRef PubMed.
- G. Li, Y. Chen, L. Zhang, M. Zhang, S. Li, L. Li, T. Wang and C. Wang, Nano-Micro Lett., 2018, 10, 7 CrossRef PubMed.
- Y. Liu, J. J. Yin and Z. Nie, Nano Res., 2014, 7, 1719–1730 CrossRef CAS.
- J. Wang, H. Liu, Y. Liu, C. Chu, Y. Yang, Y. Zeng, W. Zhang and G. Liu, Biomater. Sci., 2018, 6, 586–595 RSC.
- W. He, K. Ai, C. Jiang, Y. Li, X. Song and L. Lu, Biomaterials, 2017, 132, 37–47 CrossRef CAS PubMed.
- L. Xi, S. R. Grobmyer, G. Zhou, W. Qian, L. Yang and H. Jiang, J. Biophotonics, 2014, 7, 401–409 CrossRef CAS PubMed.
- K. E. Wilson, S. V. Bachawal, L. Abou-Elkacem, K. Jensen, S. Machtaler, L. Tian and J. K. Willmann, Theranostics, 2017, 7, 1463 CrossRef CAS PubMed.
- G. Ferrauto, F. Carniato, E. Di Gregorio, L. Tei, M. Botta and S. Aime, Nanoscale, 2017, 9, 99–103 RSC.
- S. Sreejith, J. Joseph, M. Lin, N. V. Menon, P. Borah, H. J. Ng, Y. X. Loong, Y. Kang, S. W.-K. Yu and Y. Zhao, ACS Nano, 2015, 9, 5695–5704 CrossRef CAS PubMed.
- T. Guan, W. Shang, H. Li, X. Yang, C. Fang, J. Tian and K. Wang, Bioconjugate Chem., 2017, 28, 1221–1228 CrossRef CAS PubMed.
- M. Smith, M. C. Mancini and S. Nie, Nat. Nanotechnol., 2009, 4, 710 CrossRef PubMed.
- J. Kim, S. Park, G. B. Park, W. Choi, U. Jeong and C. Kim, Proc. SPIE, 2018, 10494, 104944I Search PubMed.
- S. Hannah, D. VanderLaan, Y.-S. Chen and S. Y. Emelianov, Biomed. Opt. Express, 2014, 5, 3042–3052 CrossRef PubMed.
- G. Ku, M. Zhou, S. Song, Q. Huang, J. Hazle and C. Li, ACS Nano, 2012, 6, 7489 CrossRef CAS PubMed.
- K. A. Homan, M. Souza, R. Truby, G. P. Luke, C. Green, E. Vreeland and S. Emelianov, ACS Nano, 2012, 6, 641–650 CrossRef CAS PubMed.
- Y. Zhou, D. Wang, Y. Zhang, U. Chitgupi, J. Geng, Y. Wang, Y. Zhang, T. R. Cook, J. Xia and J. F. Lovell, Theranostics, 2016, 6, 688 CrossRef CAS PubMed.
- J. Wu, L. You, L. Lan, H. J. Lee, S. T. Chaudhry, R. Li, J. X. Cheng and J. Mei, Adv. Mater., 2017, 29, 1703403 CrossRef PubMed.
- Y. Jiang, P. K. Upputuri, C. Xie, Y. Lyu, L. Zhang, Q. Xiong, M. Pramanik and K. Pu, Nano Lett., 2017, 17, 4964–4969 CrossRef CAS PubMed.
- J. Zhang, C. Yang, R. Zhang, R. Chen, Z. Zhang, W. Zhang, S.-H. Peng, X. Chen, G. Liu, C. S. Hsu and C.-S. Lee, Adv. Funct. Mater., 2017, 27, 1605094 CrossRef PubMed.
- Z. Cao, L. Feng, G. Zhang, J. Wang, S. Shen, D. Li and X. Yang, Biomaterials, 2018, 155, 103–111 CrossRef CAS PubMed.
- D. Gao, Z. Sheng, Y. Liu, D. Hu, J. Zhang, X. Zhang, H. Zheng and Z. Yuan, Adv. Healthcare Mater., 2016, 6, 1601094 CrossRef PubMed.
- M. Zhou, J. Li, S. Liang, A. K. Sood, D. Liang and C. Li, ACS Nano, 2015, 9, 7085–7096 CrossRef CAS PubMed.
- L. Guo, I. Panderi, D. D. Yan, K. Szulak, Y. Li, Y.-T. Chen, H. Ma, D. B. Niesen, N. Seeram, A. Ahmed, B. Yan, D. Pantazatos and W. Lu, ACS Nano, 2013, 7, 8780 CrossRef CAS PubMed.
- T. Yang, Y. Wang, H. Ke, Q. Wang, X. Lv, H. Wu, Y. Tang, X. Yang, C. Chen, Y. Zhao and H. Chen, Adv. Mater., 2016, 28, 5923–5930 CrossRef CAS PubMed.
- W. Yang, W. Guo, W. Le, G. Lv, F. Zhang, L. Shi, X. Wang, J. Wang, S. Wang, J. Chang and B. Zhang, ACS Nano, 2016, 10, 10245–10257 CrossRef CAS PubMed.
- W. Yang, X. Wu, Y. Dou, J. Chang, C. Xiang, J. Yu, J. Wang, X. Wang and B. Zhang, Biomaterials, 2018, 161, 256–269 CrossRef CAS PubMed.
- W. Yang, W. Guo, J. Chang and B. Zhang, J. Mater. Chem. B, 2017, 5, 401–417 RSC.
- C. Ding, Y. Xu, Y. Zhao, H. Zhong and X. Luo, ACS Appl. Mater. Interfaces, 2018, 10, 8947–8954 CrossRef CAS PubMed.
- M. Schottelius, B. Laufer, H. Kessler and H.-J. Wester, Acc. Chem. Res., 2009, 42, 969–980 CrossRef CAS PubMed.
- C. Zhan, B. Gu, C. Xie, J. Li, Y. Liu and W. Lu, J. Controlled Release, 2010, 143, 136–142 CrossRef CAS PubMed.
- H. Wang, C. Liu, X. Gong, D. Hu, R. Lin, Z. Sheng, C. Zheng, M. Yan, J. Chen, L. Cai and L. Song, Nanoscale, 2014, 6, 14270–14279 RSC.
- M. Li, C. Liu, X. Gong, R. Zheng, Y. Bai, M. Xing, X. Du, X. Liu, J. Zeng, R. Lin, H. Zhou, S. Wang, G. Lu, W. Zhu, C. Fang and L. Song, Biomed. Opt. Express, 2018, 9, 1408–1422 CrossRef PubMed.
- Q. Chen and Z. Liu, Adv. Mater., 2016, 28, 10557–10566 CrossRef CAS PubMed.
- J. Xie, Y. Zheng and J. Y. Ying, J. Am. Chem. Soc., 2009, 131, 888–889 CrossRef CAS PubMed.
- H. Mao, W. Zhang, W. Zhou, B. Zou, B. Zheng, S. Zhao and F. Huo, ACS Appl. Mater. Interfaces, 2017, 9, 24649–24654 CrossRef CAS PubMed.
- B. Wang, R. Gui, H. Jin, W. He and Z. Wang, Talanta, 2018, 178, 1006–1010 CrossRef CAS PubMed.
- J. Zhang, J. Yu, Y. Zhang, Q. Li and J. R. Gong, Nano Lett., 2011, 11, 4774–4779 CrossRef CAS PubMed.
- C. Zhang, Y. Y. Fu, X. Zhang, C. Yu, Y. Zhao and S.-K. Sun, Dalton Trans., 2015, 44, 13112–13118 RSC.
- C. Liu, J. Chen, Y. Zhu, X. Gong, R. Zheng, N. Chen, D. Chen, H. Yan, P. Zhang, H. Zheng, Z. Sheng and L. Song, Nano-Micro Lett., 2018, 10, 1–12 CrossRef PubMed.
- H. S. Seo, Y. M. Ko, J. W. Shim, Y. K. Lim, J.-K. Kook, D.-L. Cho and B. H. Kim, Appl. Surf. Sci., 2010, 257, 596–602 CrossRef CAS.
- S. Li, J. Wei, L. Yuan, H. Sun, Y. Liu, Y. Zhang, J. Li and X. Liu, Cancer Biother. Radiopharm., 2011, 26, 529–538 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8bm00767e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |

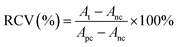
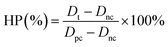
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: