Biocatalytically induced surface modification of the tobacco mosaic virus and the bacteriophage M13†
Valentina
Vignali‡
a,
Barbara
S. Miranda‡
ab,
Irene
Lodoso-Torrecilla
a,
Cathelijn A. J.
van Nisselroy
a,
Bas-Jan
Hoogenberg
a,
Sybren
Dantuma
a,
Frank
Hollmann
 c,
Jan Willem
de Vries
d,
Eliza M.
Warszawik
d,
Rainer
Fischer
e,
Ulrich
Commandeur
e and
Patrick
van Rijn
c,
Jan Willem
de Vries
d,
Eliza M.
Warszawik
d,
Rainer
Fischer
e,
Ulrich
Commandeur
e and
Patrick
van Rijn
 *ad
*ad
aUniversity of Groningen, University Medical Center Groningen, Department of Biomedical Engineering-FB40, W. J. Kolff Institute-FB41. A. Deusinglaan 1, 9713AV Groningen, The Netherlands. E-mail: p.van.rijn@umcg.nl
bNational Institute of Science and Technology in Bioanalytics, Institute of Chemistry, University of Campinas, P. O. Box 6154, Campinas, Brazil
cDepartment of Biotechnology, Delft University of Technology, Van der Maasweg 9, 2629HZ Delft, The Netherlands
dDepartment of Polymer Chemistry, Zernike Institute for Advanced Materials, University of Groningen, Nijenborgh 4, 9747AG Groningen, The Netherlands
eInstitute of Biology VII, Molecular Biotechnology, RWTH Aachen University, Worringerweg 1, 52074 Aachen, Germany
First published on 28th November 2018
Abstract
Engineered viruses are finding an increasing number of applications in basic, translational research and materials science. Genetic and chemical engineering of capsids represents a key point for tailoring the properties of viral particles, but the synthetic efforts and limits accompanying these processes still hinder their usability. Here, a single-step highly selective biocatalytic functionalization approach is described, providing a general platform for virus–acrylate hybrid particles. The tobacco mosaic virus (TMV) and the bacteriophage M13 have been successfully modified via laccase induced free radical formation on the tyrosine residues through single electron oxidation as the initiating step and the free radicals subsequently react with acrylate-based monomers. This new approach can be extended to other biomolecular assemblies with surface exposed tyrosine residues, when the introduction of new functionalities is desired.
Engineered bionanostructures have great potential in drug/gene delivery systems, nanoelectronics, liquid crystals, and biosensors. Virus particles (VPs) are promising bionanomaterials in the aforementioned applications and have so far been used as nanocarrier platforms, nanoreactors, probes for imaging and sensing platforms.1 The applications of VPs can be modulated through the addition of new functionalities to their surface via genetic or synthetic modifications, which can also be sequentially performed.2 Although, these techniques offer great possibilities, they still present some limitations. Genetic modifications are restricted to the introduction of natural or unnatural peptides that ensure capsid formation preferably with a high production yield, these two requirements combined is a challenging process.3 Decorating viruses with novel functional groups such as fluorophores, inorganic structures, hydrophobic or charged moieties and stealth polymers has already been achieved.4 Several synthetic routes have been explored and even though surface modification can be achieved, a proper balance in terms of high specificity and viral structure maintenance is needed.5 Promising strategies as alternatives to the use of conventional peptide-coupling based chemistry consist of the functionalization of the tyrosine residues through diazonium coupling,6 which allows for subsequent modification such as by copper-catalyzed azide–alkyne cycloaddition (CuAAC).7 However, new approaches with less synthetic efforts, high chemical diversity, a limited number of synthetic steps, and associated purifications are desired in order to facilitate future application of viral particles.
Among the natural amino acid residues, tyrosine can be selectively activated by laccase via a single electron oxidation step which produces a free radical species.8 It was envisioned that the free radical could be used as a new grafting-from approach to modify the viral surface using laccase combined with a large library of acrylates, which offers tremendous chemical variability. Laccases are commercially available and require mild reaction conditions. Furthermore, as coenzyme-independent oxidoreductases they are very simple to use, requiring molecular oxygen as a cosubstrate and producing water as the sole by-product.9 The laccase-induced tyrosine oxidation generates a radical intermediate on the viral surface, which can subsequently react with acrylate-based monomer units. Depending on the monomer, novel properties can be introduced (Scheme 1).
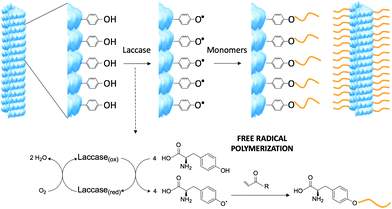 | ||
| Scheme 1 Modification of the tobacco mosaic virus via laccase induced free-radical oxidation. Laccase oxidizes the exposed tyrosine residues on the viral surface and the phenolic radicals newly formed triggers the functionalization of acrylate monomers. TMV structure created from the RCSB PDB (http://www.rcsb.org) of PDB ID 2OM3.13 | ||
The tobacco mosaic virus (TMV) and the bacteriophage M13 (M13) have been modified via the aforementioned biocatalytic approach. These two viruses have already proven their applicability in nanomedicine and materials science,10 but improvements in attaining a higher chemical diversity without the requirement of a complicated synthesis are needed in order to promote their large-scale application.
TMV is made up of 2130 identical coat protein (CP) subunits self-assembled in a helical fashion around a single strand RNA, forming tubes 18 nm in diameter and 300 nm in length.11 Previously, it was shown that TMV CP presents only one addressable tyrosine at position 139.6,7,12 The filamentous bacteriophage M13 instead is made up of 2700 copies of a major coat protein, p8, and few copies of minor coat proteins that enclose a circular single-stranded DNA molecule. The protein p8 contains two tyrosine residues at positions 21 and 24, both accessible as cosubstrates for enzymatic reactions.13 To demonstrate the ease of introducing chemical variability, molecular modifications using monomers that are hydrophilic, hydrophobic, charged, responsive, and fluorescent were performed. The reaction is straightforward and can be combined with different types of commercially available acrylates and other monomers, therefore this grafting from approach is extremely versatile with the possibility to introduce diverse chemical species.
Before applying this reaction to the viruses, the enzyme performance was tested, on the basis of previous work,14 at different pH, temperatures, and time of oxygen removal. For these preliminary tests, phenol was used as a model oxidation-initiator of the free radical polymerization, to mimic the tyrosine residue of the viral surface (ESI,† S1). The optimum conditions to obtain the highest amount of grafted monomers were determined to be phosphate buffer 0.1 mol L−1 at pH 6–7.5 and at 65–75 °C. For TMV modification, these conditions have been slightly adjusted to meet the integrity maintenance requirements compatibly. Although TMV is a robust bionanoparticle, it can still denature if maintained at high temperature for prolonged time and disassemble into smaller disk-like aggregates above pH 6.5.15 To minimize denaturation and the changes in aggregate morphology, a reaction mixture of 0.1 mol L−1 phosphate buffer pH 6 at 60 °C for 90 minutes was used. For M13, the reaction temperature has been lowered to 35 °C.
Different acrylate monomers were used: [2-(methacryloyloxy)ethyl]trimethylammonium chloride (METAC), N-isopropylacrylamide (NIPAAm), N-vinylcaprolactam (VCL), styrene (Styr) and fluorescein o-acrylate. These monomers were chosen due to their potential to add interesting properties to the viruses and also to evaluate the versatility of this new method to add diverse chemical functionalities. Additionally, fluorescein o-acrylate was used as a co-monomer at 1% or 10% together with the respective monomers for providing further control during the characterization steps and to produce fluorescent virus-hybrid particles. It is worth mentioning that styrene was dissolved in ethanol before being added to the reaction mixture, which had a final ethanol content of 10% (v/v). The addition of ethanol ensures sufficient solubility of styrene without denaturing the TMV while maintaining appropriate laccase activity, which is known to tolerate organic solvents up to 30% (v/v).14,16 After the reaction, the mixture was purified by dialysis against MilliQ water and characterized by transmission electron microscopy (TEM) (Fig. 1 and ESI,† SI2), matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-ToF) (Fig. 2A and ESI,† SI3), reversed-phase liquid chromatography (RPLC) (Fig. 2B), sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (ESI,† SI4), and fluorescence spectrometry (ESI,† SI5).
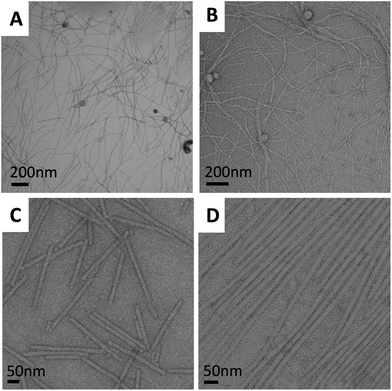 | ||
| Fig. 1 Transmission electron micrographs of the virus particles. (A) M13 unmodified (B) M13–NIPAM hybrid (C) TMV unmodified and (D) TMV–NIPAM hybrid. | ||
The TEM images of the TMV and M13 modified particles reveal that the viral structure is maintained, indicating their stability towards the reaction conditions (Fig. 1). The unmodified TMV and M13 normally appear as single linear filaments while the hybrid particles occur in an assembled state, which is probably caused due to a drying effect related to the altered surface properties. In general, longer structures were observed for the TMV. Hybrid particles assembled in a head-to-tail manner even when the TMV concentration was reduced in the reaction mixture. This occurs most likely due to the interaction between the hydrophobic ends, which was also found in previous studies with polyaniline deposition onto the surface of TMV.4e
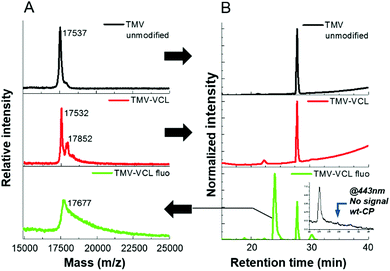
The covalent nature of the monomer grafting to the viral surface has been verified using MALDI-ToF, reversed-phase liquid chromatography and SDS-PAGE. The MALDI-ToF spectrum of the unmodified TMV (Fig. 2A) shows a single narrow signal corresponding to a mass of 17.5 kDa, which can be attributed to the TMV CP. In the spectra of the viral particles after biocatalytic modification (Fig. 2A and Fig. S3, ESI†), the peak at 17.5 kDa is still present, but additionally another broad signal representing a heavier species is detected. This indicates that the products of the reactions are particles made up of a mixture of modified and unmodified CPs. The presence of the latter is probably due to the fact that the laccase needs time and spatial freedom to properly activate the tyrosine moieties and during the activation and modification, other tyrosine moieties located nearby are most likely sterically hindered. The distribution representing the modified CPs indicates that the obtained conjugates are not monodisperse, which is to be expected from a free radical approach as it generally provides highly dispersed polymers and is susceptible to side reactions and termination. Moreover, since oxygen acts as an inhibitor of the free radical polymerization, but is required as a cosubstrate for the biocatalytic process, the control on the polydispersity of the growing chains of monomer is challenging. Alternatively, hydrogen peroxide could be used as a cosubstrate and it could lead to improved polymerization due to the absence of oxygen, but it would also initiate polymerization and make the reaction mixture more complex. By analyzing the signals, it has been determined that with this biocatalytic approach it was possible to graft oligomeric structures composed of more than 15 monomer units of NIPAAm on a single protein, and over 10 units of VCL and METAC. Through the integration of the signals provided by MALDI-ToF, the degree of modification for each type of acrylate was assessed (Table S1, ESI†). Subsequently, the unmodified CPs and the CP–VCL conjugates of the TMV capsids were separated via reversed-phase chromatography. Using a detection wavelength of 254 nm, the elution of only one protein fraction was detected (Fig. 2B TMV–VCL, 28 min) which was identical to the unmodified CPs (Fig. 2A and B TMV unmodified). The detection of the CP–VCL conjugates was most likely hindered due to the modification of the tyrosine moiety from a phenol-like structure to an alkylated phenol. The substitution pattern of phenol-based structures greatly affects the wavelength of maximum absorption as well as the molar absorption coefficient.17 By introducing a fluorescent co-monomer it was nevertheless possible to unambiguously differentiate between CP conjugates (fraction eluted at 24 min) and unmodified CPs using a detection wavelength of 254 nm (Fig. 2B, TMV–VCL fluo). An unidentified minor signal at 30 min was also detected. A second detection wavelength of 443 nm was used to identify the fluorescent co-monomer. Only the fraction eluted at 24 min displayed a signal, confirming the presence of CP conjugates (Fig. 2B, inset). The MALDI-ToF analysis of the isolated fraction at 24 min shows a peak at 17.677 kDa with a mass corresponding to the functionalized CP, whilst the characteristic peak of the unmodified TMV CP is absent (Fig. 2A). This indicates that the modification is of covalent, rather than of non-covalent, nature. As can be seen from the SDS-PAGE results of the viral particles (Fig. S4, ESI†), the unmodified TMV contains a majority of sub-units with a molecular weight (Mw) of approximately 17 kDa. A small band around 35 kDa is also present, probably corresponding to the CP dimers whose interactions are resistant to the denaturing conditions of the SDS-page. On the other hand, all TMV-modified hybrids presented a protein band more distributed towards a higher molecular weight than 17 kDa. Additionally, there is a large build-up around two times the molecular weight of the CP. This suggests that the presence of new functionalities can stabilize the interaction between the dimers, as previously observed for the unmodified TMV. The overall results provide supporting evidence that CP conjugates of a higher molecular weight than the unmodified CP are formed, as was observed by MALDI-ToF, further emphasizing the covalent nature of the modification through the free radical biocatalytic grafting reaction.
To investigate the potential of introducing new properties onto the viral surface through this functionalization, the fluorescence signal of the fluorescein o-acrylate virus-hybrids has also been measured (Fig. S5, ESI†). Both the TMV and M13 hybrids emitted a well detectable signal not only when functionalized solely with fluorescein o-acrylate but also when the fluorophore was added as 10% co-monomer in the reaction with another monomer, NIPAAm.
In summary, a new biocatalytic route has been demonstrated to modify TMV and M13 with chemically diverse oligomeric structures with a wide variety of properties by an easy and straightforward approach to tailor the overall properties of such particles. Future endeavours to obtain better control over the polydispersity and polymer distribution may be achieved by omitting oxygen from the reaction. Additionally, the translation towards a controlled radical polymerization mechanism could be investigated to reduce the inhibitory effect of oxygen on the polymerization and to increase control over the polymer length, which might also be achieved by separating the activation and polymerization steps.14 This new synthetic approach enables virus particles to become more generally applicable due to less synthetic efforts. Additionally, the introduction of polymers might also greatly affect the assembly dynamics of the virus similar to that found for other systems as the microenvironment greatly affects the assembly, and may lead to new assembled virus particles.18 Although TMV and M13 were used in this study, the approach is not limited to these viruses and any tyrosine-bearing protein structure can be potentially modified in this fashion. When protein-based particles are used for therapeutic applications, they trigger an immune response, which leads to particle degradation. This disadvantage offers an additional reason for pursuing easy protein modification with polymer forming approaches, in fact the grafting of “stealth” monomers to proteins through this biocatalytic approach could allow prolonged non-detection times by the immune system improving the pharmacokinetic profile.19 Moreover, the exposure of tyrosine in vivo may lead to the nitration of this residue which is known to trigger an immune response.20 Therefore, in addition to providing a general synthetic tool for expanding the use of bionanostructures in various applications, targeting and modifying the tyrosine residue as presented here is regarded as a dual approach to potentially lower immune recognition.
The authors would like to thank Joop de Vries, Dr Marc C. A. Stuart, Dr Philipp T. Kühn and Mark Loznik for technical assistance. Prof. Andreas Herrmann is kindly acknowledged for providing access to equipment. Barbara S. Miranda would like to thank the National Institute for Science and Technology in Bioanalytics and Science Without Borders Program for funding. The project leading to this application has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 713482.
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) H. Masarapu, B. K. Patel, P. L. Chariou, H. Hu, N. M. Gulati, B. L. Carpenter, R. A. Ghiladi, S. Shukla and N. F. Steinmetz, Biomacromolecules, 2017, 18, 4141–4153 CrossRef CAS PubMed; (b) M. Comellas-Aragonès, H. Engelkamp, V. I. Claessen, N. A. J. M. Sommerdijk, A. E. Rowan, P. C. M. Christianen, J. C. Maan, B. J. M. Verduin, J. J. L. M. Cornelissen and R. J. M. Nolte, Nat. Nanotechnol., 2007, 2, 635–639 CrossRef; (c) W. G. Kim, H. Song, C. Kim, J. S. Moon, K. Kim, S. W. Lee and J. W. Oh, Biosens. Bioelectron., 2016, 85, 853–859 CrossRef CAS; (d) F. Zang, S. Chu, K. Gerasopoulos, J. N. Culver and R. Ghodssi, Nanotechnology, 2017, 28, 265301 CrossRef; (e) A. M. Wen and N. F. Steinmetz, Chem. Soc. Rev., 2016, 45, 4074–4126 RSC.
- S. Lin, H. Yan, L. Li, M. Yang, B. Peng, S. Chen, W. Li and P. R. Chen, Angew. Chem., Int. Ed., 2013, 52, 13970–13974 CrossRef CAS.
- (a) C. M. Guenther, B. E. Kuypers, M. T. Lam, T. M. Robinson, J. Zhao and J. Suh, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2014, 6, 548–558 CAS; (b) I. F. Bin Mohamed Suffian, M. Garcia-Maya, P. Brown, T. Bui, Y. Nishimura, A. R. B. M. J. Palermo, C. Ogino, A. Kondo and K. T. Al-Jamal, Sci. Rep., 2017, 7, 43160 CrossRef.
- (a) Y. Wang, J. Liu, L. Cao, W. Wang, Y. Sun, Z. Yin and Z. Lou, ChemBioChem, 2018, 19, 1465–1470 CrossRef CAS; (b) A. G. Malyutin, R. Easterday, Y. Lozovyy, A. Spilotros, H. Cheng, O. R. Sanchez-Felix, B. D. Stein, D. G. Morgan, D. I. Svergun and B. Dragnea, et al. , Chem. Mater., 2015, 17, 327–335 CrossRef; (c) H. Bludau, A. E. Czapar, A. S. Pitek, S. Shukla, R. Jordan and N. F. Steinmetz, Eur. Polym. J., 2017, 88, 679–688 CrossRef CAS PubMed; (d) J. Rong, F. Oberbeck, X. Wang, X. Li, J. Oxsher, Z. Niu and Q. Wang, J. Mater. Chem., 2009, 19, 2841–2845 RSC; (e) Z. Niu, M. Bruckman, V. S. Kotakadi, J. He, T. Emrick, T. P. Russell, L. Yang and Q. Wang, Chem. Commun., 2006, 3019–3021 RSC.
- M. T. Smith, A. K. Hawes and B. C. Bundy, Curr. Opin. Biotechnol., 2013, 24, 620–626 CrossRef CAS.
- T. L. Schlick, Z. Ding, E. W. Kovacs and M. B. Francis, J. Am. Chem. Soc., 2005, 127, 3718–3723 CrossRef CAS.
- M. A. Bruckman, G. Kaur, L. A. Lee, F. Xie, J. Sepulveda, R. Breitenkamp, X. Zhang, M. Joralemon, T. P. Russell and T. Emrick, et al. , ChemBioChem, 2008, 9, 519–523 CrossRef CAS.
- (a) M. L. Mattinen, K. Kruus, J. Buchert, J. H. Nielsen, H. J. Andersen and C. L. Steffensen, FEBS J., 2005, 272, 3640–3650 CrossRef CAS PubMed; (b) K. Minamihata, M. Goto and N. Kamiya, Bioconjugate Chem., 2011, 22, 74–81 CrossRef CAS PubMed; (c) M. L. Mattinen, M. Hellman, P. Permi, K. Autio, N. Kalkkinen and J. Buchert, J. Agric. Food Chem., 2006, 54, 8883–8890 CrossRef CAS.
- (a) S. Riva, Trends Biotechnol., 2006, 24, 219–226 CrossRef CAS; (b) F. Hollmann and I. W. C. E. Arends, Polymers, 2012, 4, 759–793 CrossRef.
- (a) A. E. Czapar, Y. R. Zheng, I. A. Riddel, S. Shukla, S. G. Awuah, A. J. Lippard and N. F. Steinmetz, ACS Nano, 2016, 10, 4119–4126 CrossRef CAS; (b) M. Bäcker, C. Koch, S. Eiben, F. Geiger, F. Eber, H. Gliemann, A. Poghossian, C. Wege and M. J. Schöning, Sens. Actuators, B, 2017, 238, 716–722 CrossRef; (c) J.-S. Moon, W.-G. Kim, C. Kim, G.-T. Park, J. Heo, S. Y. Yoo and J.-W. Oh, Mini-Rev. Org. Chem., 2015, 12, 271–281 CrossRef CAS; (d) K. S. Sunderland, M. Yang and C. Mao, Angew. Chem., Int. Ed., 2017, 56, 1964–1992 CrossRef CAS.
- P. J. G. Butler, Philos. Trans. R. Soc., B, 1999, 354, 537–550 CrossRef CAS.
- M. A. Bruckman and N. F. Steinmetz, Methods Mol. Biol., 2014, 1108, 173–185 CrossRef CAS.
- M. J. Glucksman, S. Bhattacharjee and L. Makowski, J. Mol. Biol., 1992, 226, 455–470 CrossRef CAS.
- F. Hollmann, Y. Gumulya, C. Tölle, A. Liese and O. Thum, Macromolecules, 2008, 41, 8520–8524 CrossRef CAS.
- (a) M. A. Lauffer and W. C. Price, J. Biol. Chem., 1940, 133, 1–15 CAS; (b) A. Klug, Philos. Trans. R. Soc., B, 1999, 354, 531–535 CrossRef CAS; (c) J. M. Alonso, T. Ondarçuhu and A. M. Bittner, Nanotechnology, 2013, 24, 105305 CrossRef.
- J. M. Alonso, M. L. Górzny and A. M. Bittner, Trends Biotechnol., 2013, 31, 530–538 CrossRef CAS.
- J. M. Antosiewicz and D. Shugar, Biophys. Rev., 2016, 8, 151–161 CrossRef CAS PubMed.
- (a) J. Wang, K. Liu, R. Xing and X. Yan, Chem. Soc. Rev., 2016, 45, 5589 RSC; (b) R. Xing, C. Yuan, S. Li, J. Song, J. Li and X. Yan, Angew. Chem., Int. Ed., 2018, 57, 1537 CrossRef CAS.
- (a) K. L. Lee, S. Shukla, M. Wu, N. R. Ayat, C. E. El Sanadi, A. M. Wen, J. F. Edelbrock, J. K. Pokorski, U. Commandeur and G. R. Dubyak, et al. , Acta Biomater., 2015, 19, 166–179 CrossRef CAS; (b) I. Ozer and A. Chilkoti, Bioconjugate Chem., 2017, 28, 713–723 CrossRef CAS.
- L. H. Jones, A. Narayanan and E. C. Hett, Mol. BioSyst., 2014, 10, 952–969 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8cc08042a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |