Open Access Article
Open Access ArticleDegradation of pendimethalin by the yeast YC2 and determination of its two main metabolites†
Yajie Han ab,
Zonggui Tangc,
Huifang Baod,
Dongmei Wue,
Xiaolin Dengf,
Gaowei Guof,
Bang-Ce Ye
ab,
Zonggui Tangc,
Huifang Baod,
Dongmei Wue,
Xiaolin Dengf,
Gaowei Guof,
Bang-Ce Ye *a and
Bin Dai*a
*a and
Bin Dai*a
aSchool of Chemistry and Chemical Engineering/The Key Lab. for Green Processing of Chemical Engineering of Xinjiang Bingtuan, Shihezi University, Shihezi 832003, People's Republic of China. E-mail: bcye@ecust.edu.cn; dbinly@126.com; Fax: +86993-2057270; Tel: +86993-2057270 Tel: +86993-2058176
bCollege of Life Science, Shihezi University, Shihezi 832003, People's Republic of China
cAnalysis and Testing Center, Xinjiang Agricultural Reclamation Academy of Sciences, Shihezi 832000, People's Republic of China
dInstitute of Applied Microbiology, Xinjiang Academy of Agricultural Sciences, Urumqi 830091, People's Republic of China
eBiotechnology Research Institue, Xinjiang Agricultural Reclamation Academy of Sciences, Shihezi, 832000, People's Republic of China
fDepartment of Chemistry, Teachers College, Shihezi University, Shihezi 832003, People's Republic of China
First published on 2nd January 2019
Abstract
In this study, we isolated a yeast strain, YC2, by enrichment culture from pendimethalin-contaminated soil. The analysis of its phenotypic features and 26S rRNA sequence confirmed that the strain YC2 is Clavispora lusitaniae. According to the kinetics of pendimethalin degradation, YC2 could degrade 74% of 200 mg L−1 pendimethalin in CMB liquid culture over 8 days. LC-MS/MS was used to identify the metabolites of pendimethalin after degradation. This study confirmed that its metabolites consist of 1,2-dimethyl-3,5-dinitro-4-N(buta-1,3-dien-2-yl)-dinitrobenzenamine-N-oxide and 1,2-dimethyl-3,5-dinitro-4-N(prop-1-en-2-yl)-dinitrobenzenamine-N-oxide, which were broken down by a series of enzymatic reactions to produce CO2 and H2O. Thus, the study herein sheds light on the role of yeast in the degradation of pendimethalin.
1. Introduction
Due to the wide use of pesticides in agriculture, soils and rivers have become significantly polluted.1 Consequently, these toxic pesticides pose a hazard to human health. According to the United States Environmental Protection Agency (EPA) and the World Health Organization (WHO), more than 1000 chemical compounds are used as pesticides. Pendimethalin-[N-(1-ethyl propyl)-3,4-dimethyl-2,6-dinitrobenzenenamine] is a dinitroaniline herbicide,2 which can control weeds during the growth of cotton, rice, soybean and tobacco.3 Pendimethalin is a micro-toxic substance; however, it may trigger thyroid follicular cell adenomas in rats, and therefore, has been recognized as a probable carcinogen to humans.4,5The chemical structure of pendimethalin is complex, and it can be strongly adsorbed by the soil but difficult to desorb. Additionally, the half-life of pendimethalin in soil is 90 days.6 The degradation of pendimethalin occurs mainly via biodegradation and photodegradation. However, its microbial remediation is an effective approach in the natural environment.7,8 Degradation reactions by microorganisms include oxidation, reduction, hydrolysis, and dealkylation. Furthermore, bacteria and fungi are the main organisms for pesticide degradation. Fungi fine-tune the molecular structure of pesticides to make them non-toxic, which are further degraded under the action of soil bacteria.9 There are some reports on the degradation of pendimethalin by microbes of Azotobacter chroococcum,7 Bacillus sp. Y3,10 Bacillus circulans,11 and fungi Aspergillus flavus.12 The latest study suggested that some fungi can degrade certain types of persistent or toxic environmental pollutants. Also, microbial communities can degrade pesticides from the environment.13,14 In previous reports, the microbial catabolism is initiated by different mechanisms, including nitroreduction, N-dealkylation, oxidation of benzene rings, and cyclization of side chains.15
In this study, we isolated a yeast strain, YC2, which is able to degrade pendimethalin. Strain YC2 initiated pendimethalin catabolism via the oxidation of the amine groups, generating two new metabolites, which were identified. Therefore, the catabolic pathway presented in this study is different from that in previous studies.
2. Materials and methods
2.1 Chemicals and reagents
Pendimethalin EC 33% was purchased from Jiangsu Fengshan Group, Corporation (in Jiangsu Province, China). The pendimethalin standard was purchased from Aladdin Bio-Chem Chemical Co. (Shanghai, China). All other chemicals were of analytical grade.The YPD culture medium contained 20 g L−1 of peptone, 20 g L−1 of glucose and 10 g L−1 of yeast extract. Czapek Modified A (CMA) was used to find isolate yeasts, and the medium contained 2.0 g L−1 NaNO3; 1.0 g L−1 K2HPO4; 0.5 g L−1 KCl; 0.5 g L−1 MgSO4·7H2O; and 0.01 g L−1 FeSO4·7H2O. To study the kinetics of pendimethalin degradation, Czapek Medium B (CMB) was used. The only difference is that Czapek medium A contains no sucrose and Czapek medium B contains 8 g sucrose per litre.
2.2 Soil
Yeast was collected from cotton cultivated soils in various areas in Shihezi (Xinjiang Province, China). These loamy soils were treated with pendimethalin annually for 15 years, and thus used for the isolation of the pendimethalin-degrading microbe. The soil samples were collected from the surface layer (2–20 cm). Selected samples were dried and sieved to 2 mm before the experiment.2.3 Enrichment and isolation of pendimethalin-degrading yeasts
The pendimethalin-degrading yeasts were isolated using the enrichment culture technique. To isolate the yeasts, 10 g agricultural soil was added to a beaker containing 100 mL of CMA culture media supplemented with pendimethalin (100 mg L−1). The mixture in the beaker was incubated on a thermostatic shaker (120 rpm) for 6 days at 30 °C in the dark. Then 1 mL of suspension was removed and transferred to a flask containing fresh medium, and the solution was supplemented with pendimethalin (200 mg L−1) and incubated for another 6 days under the same conditions. Finally, the concentration of pendimethalin was adjusted in the range of 100 to 3000 mg L−1 at 6 days intervals over 2 months. The last enrichment medium was diluted in a gradient, and the dilutions were transferred to CMA agar-solidified medium with pendimethalin (100 mg L−1), and then incubated at 30 °C.16–19 Using the streak plate technique, thirty strains were selected and stored at −80 °C with 30% glycerol.2.4 Degradation of pendimethalin by the isolated strains
To prepare the seed culture, the thirty strains were pre-incubated in YPD culture medium at 30 °C and shaken on a thermostatic shaker (120 rpm) for 18 h. Then 300 μL seed culture was added to 10 mL of CMB supplemented with 200 mg L−1 pendimethalin (in a 50 mL flask). The CMB medium was put in the shaker (120 rpm) at 30 °C. Then the cultures were centrifuged (9000 rpm, 10 min) and the supernatant was extracted with petroleum ether (10 mL × 3). All the fractions were dried over anhydrous sodium sulphate and then distilled. Chromatographic grade methanol was utilized to dissolve the residue from the extraction. The concentration of pendimethalin was determined by HPLC using a C8 column (5 μm, 3.9 × 150 mm). The mobile phase was a mixture of chromatographic pure methanol and deionized water (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, v/v) with a flow rate of 1 mL min−1, and the peaks were measured at 238 nm.
15, v/v) with a flow rate of 1 mL min−1, and the peaks were measured at 238 nm.
2.5 Identification of strain YC2
In the process of pendimethalin degradation, the best degrader of pendimethalin was selected and identified by 26S rRNA gene sequence analysis with the primers NL1 (5′-GCA TAT CAA TAA GCG GAG GAA AAG-3′) and NL4 (5′-GGT CCG TGT TTC AAG ACG G-3′). The NCBI BLASTN comparison software was used to reveal the taxonomic statue of the isolate.20 Furthermore, its microbiological properties were tested.2.6 Kinetics of pendimethalin degradation
The strain YC2 was cultured in YPD medium and incubated at 30 °C for 18 h in a thermostatic chamber. The culture was centrifuged at 5000 rpm for 10 min at 4 °C, and the cell pellets were re-suspended with sterile distilled and then adjusted to 1.5 OD600. Afterwards, the cell suspension was used as the inoculum strain. The pendimethalin from CMA and CMB (200 mg L−1) was used as the production medium with a 2% size inoculums strain, and the samples were incubated in an incubator (120 rpm) at 30 °C in the dark. The biodegradation of pendimethalin was evaluated at 24 h intervals.21–23 The pH value of the cultures was measured using a pH meter. The concentration of pendimethalin was determined by HPLC.2.7 Isolation and identification of metabolites
HPLC-MS/MS was used to identify the components of the metabolites. Chromatographic separation of the metabolites was conducted on an UltiMate 3000 UHPLC system (Dionex, Thermo Fisher Scientific, USA) equipped with a Phenomenex synergy C18 column (5.0 μm, 250 × 4.6 mm) through gradient elution with the mobile phase consisting of 0.1% formic acid-water solution (A), and acetonitrile (B) at a flow rate of 1.0 mL min−1. The peak was measured at 233 nm. The mass spectral analysis was performed using a Quadruple-Exactive Orbitrap mass spectrometer (Micromass, Thermo Fisher Scientific, USA), with a heat electrospray ionization probe (HESI) source. The used software for data collection was Xcalibur 2.2. Nitrogen was used as the sheath and auxiliary gas. The parameters of the instrument were as follows: sheath gas flow 50 arb, auxiliary gas flow 10 arb, source voltage 3.2 kV, capillary temperature 325 °C, and heat temperature 350 °C. The mass spectrometer was operated in Full MS/dd-MS2 experiment using the positive and negative modes, respectively. The full scan mode (from m/z 100–1500) was also used. The AGC target was 2 × 105 and C-Trap max injection time was 80 ms.3. Results and discussion
3.1 Isolation and identification of pendimethalin-degrading strain YC2
Using an enrichment procedure, thirty strains that could degrade pendimethalin were screened; however, only one yeast strain, designated as YC2, showed the highest ability to degrade pendimethalin, which could degrade above 60% of 200 mg L−1 pendimethalin within 5 days. Thus, strain YC2 was selected for further study.The strain YC2 colony grown on a YPD agar plate was nearly round, milky white and creamy, with a smooth surface, neat edges, viscous texture, buds and reproduction structures, and pseudohyphae. In addition, its microbiological, physiological and biochemical characters are shown in Tables 1, 2 and Fig. 1. Blast analysis of the 26S rRNA gene sequence of YC2 revealed a high sequence identity of up to 100% with Clavispora lusitaniae (Table 3). Thus, based on its phenotypic and phylogenetic properties, strain YC2 was identified as Clavispora lusitaniae.
| Sugar experiment | Sucrose | Raffinose | D-Xylose | Lactose | Honey disaccharide | Glucose |
|---|---|---|---|---|---|---|
| Fermentation | + | + | + | + | + | + |
| Assimilation | + | + | + | + | + | + |
| Starch hydrolysis test | Nitrogen assimilation experiment | Litmus milk test | |
|---|---|---|---|
| KNO3 | (NH4)2SO4 | ||
| − | − | + | − |
| Strain | Similar strain | Accession number | Ident. (%) |
|---|---|---|---|
| YC2 | Clavispora lusitaniae strain PMM10-1024036L | KP131848.1 | 100 |
| Clavispora lusitaniae strain ATCC 34449 | KU729100.1 | 100 | |
| Clavispora lusitaniae | KY102567.1 | 100 | |
| Clavispora lusitaniae | MG599148.1 | 100 |
3.2 Kinetics of pendimethalin degradation
The degradation and utilization of pendimethalin by Clavispora lusitaniae YC2 was identified on CMB culture, which was supplemented with 200 mg L−1 pendimethalin. The kinetics of pendimethalin degradation in the culture medium can be seen in Fig. 2. The result showed that strain YC2 could degrade 74% of 200 mg L−1 pendimethalin within 8 days of incubation, and Clavispora lusitaniae YC2 could efficiently degrade pendimethalin. Yeast can grow in the range of pH 3.0–7.5, and the optimum pH is 4.5 to 5.0. We found that strain YC2 reduced the pH from the initial 7.51 to 5.5 during the degradation of pendimethalin, which is beneficial to the yeast growth and degradation activity.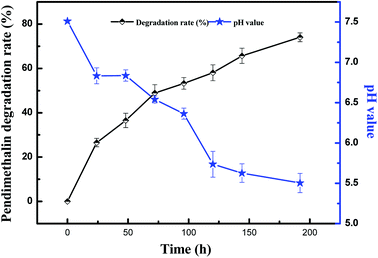 | ||
| Fig. 2 Kinetics of the degradation of pendimethalin and variations in pH in the YC2 liquid culture. The results are representative of three experiments. | ||
3.3 Identification of metabolites in liquid medium
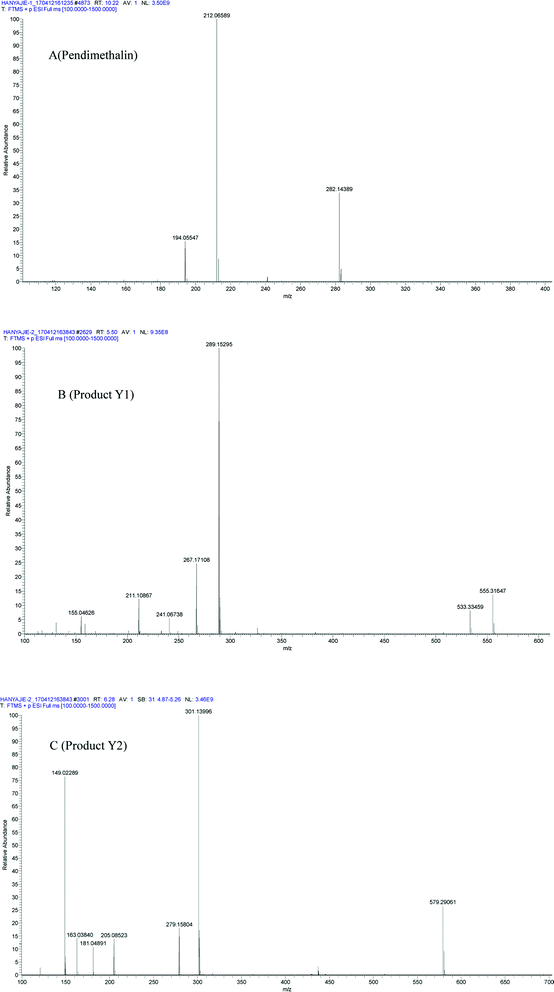
In liquid medium, according to HPLC and LC-MS/MS, there were two metabolites present during the degradation of pendimethalin. The ESI full scan is shown in Fig. 3, in which there are two peaks observed together with the parent peak (pendimethalin). The pendimethalin could be classified by comparing the reservation time of the standard (10.2 min). The other two peaks observed at 5.4 min and 6.2 min were denoted as Y1 and Y2, respectively.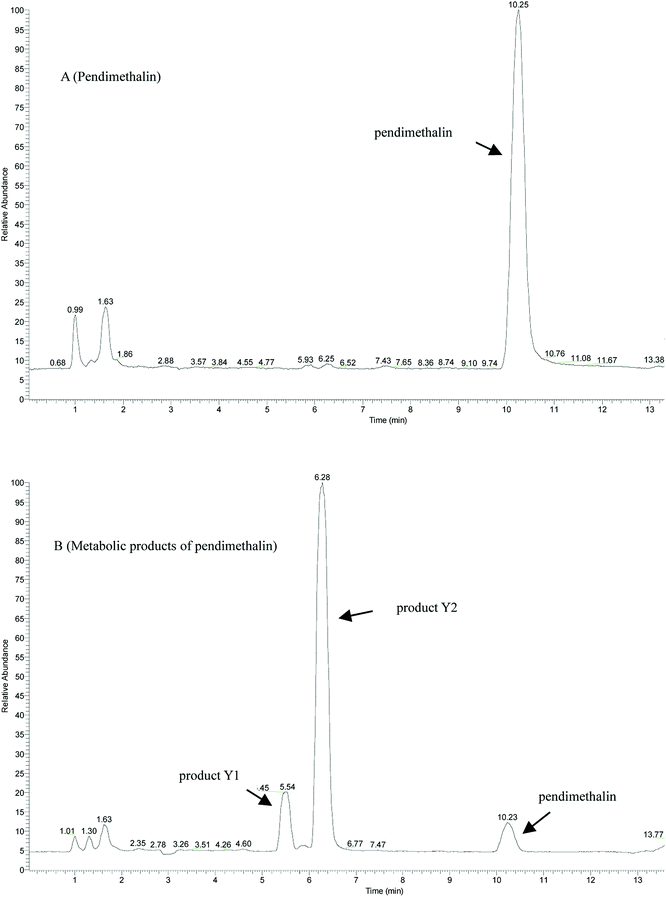 | ||
| Fig. 3 ESI full scan of the metabolites degraded by YC2: (A) pendimethalin standard and (B) metabolites of pendimethalin. | ||
Product Y1 was identified according to its mass spectrum data. As shown in Fig. 4B, its molecular ion appears at m/z 267 ([M + H]+), m/z 289 [M + Na]+, m/z 533 [2M + H]+, and m/z 555 [2M + Na]+, fitting the molecular formula C13H12N3O5. Based on the above results, metabolite Y1 is 1,2-dimethyl-3,5-dinitro-4-N(buta-1,3-dien-2-yl)-dinitrobenzenamine-N-oxide. Similarly, the mass spectrum of metabolite Y2 is shown in Fig. 4C, with its molecular ion at m/z 279 [M + H]+, m/z 301 [M + Na]+, and m/z 579 [2M + Na]+, fitting the molecular formula of C11H12N3O5. Thus, product Y2 was identified as 1,2-dimethyl-3,5-dinitro-4-N(prop-1-en-2-yl)-dinitrobenzenamine-N-oxide.
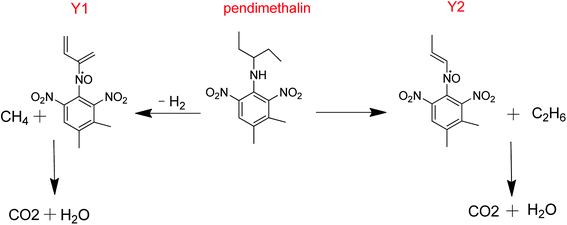
3.4 Metabolic pathway
The presented metabolic pathway of pendimethalin in liquid medium is illustrated in Fig. 5. Since the liquid medium was an aerobic environment, the oxidation of the amine groups generated the product Y1 with the loss of CH4 and H2, and similarly the formation of metabolite Y2 with the loss of C2H6. Finally, these products were broken down by a subsequent series of enzymatic reactions to form carbon dioxide and water.Previous reports have shown that some microorganisms can degrade pendimethalin, although the degradation pathways of pendimethalin are diverse. Two soil fungi, Fusarium oxysporum and Paecilomyces varioti degraded pendimethalin into two metabolites, that is N-(1-ethylpropyl)-3,4-dimethyl-2-nitrobenzene-l,6-diamine and 3,4-dimethyl-2,6-dinitroaniline.24 Rhizoctonia bataticola resolved pendimethalin into 3,4-dimethyl-2,6-dinitroaniline. Azotobacter chroococcum decomposed pendimethalin into six metabolites: 3,4-dimethyl-2,6-dinitroaniline, 6-aminopendimethalin, N-(2,6-dinitro-3,4-dimethyl) phenyl acetamide, 4,5-dimethyl-2-nitroaniline, (6-methyl-2,4-dinitro-3-(pentan-3-ylamino)phenyl)methanol, and 2-methyl-4-nitro-5-N-(1-cyclopropyl)-6-nitrosobenzyl alcohol.25 Pendimethalin was also degraded to 6-aminopendimethalin by nitroreduction, and other two downstream metabolites, 5-amino-2-methyl-3-nitroso-4-(pentan-3-ylamino)benzoic acid and 8-amino-2-ethyl-5-(hydroxymethyl)-1,2-dihydroquinoxaline-6-carboxylic acid were identified.26 Pendimethalin was also degraded to 1,3-dinitro-2-(pentan-3-ylamino)butane-1,4-diol by oxidative ring cleavage.7 Four metabolites were identified as N-(1-ethylpropyl)-3,4-dicarboxy-2,6-dinitrobenzenamine-N-oxide, N-(1-ethylpropyl)-3,4-dimethoxy-2,6-dinitrobenzenamine and benzimidazole-7-carboxyaldehyde in a bioslurry phase reactor.27
Therefore, our results are different from previous studies.5,20,28 The pathway in this study comes from the bioslurry reactor reported by Walker and Bond.27 Compared with other studies, the yeast in our study came from the natural soil, and we demonstrated that YC2 can be applied for the degradation of pendimethalin, which is new in the field of biodegradation mechanisms.
4. Conclusions
Strain YC2 was isolated from pendimethalin-contaminated soil and was characterized as Clavispora lusitaniae. Strain YC2 could degrade 74% of 200 mg L−1 pendimethalin within 8 days under incubation.Two metabolites, 2-dimethyl-3,5-dinitro-4-N(buta-1,3-dien-2-yl)-dinitrobenzenamine-N-oxide and 1,2-dimethyl-3,5-dinitro-4-N(prop-1-en-2-yl)-dinitrobenzenamine-N-oxide, were identified as the products of strain YC2-mediated degradation by LC-MS/MS. Herein, we summarized the metabolic pathway of pendimethalin in liquid medium, and offered different results compared with previous studies. These findings may be a new degradation mechanism that differs from that in previous research.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study is supported by the National Natural Science Foundation of China (grant 21575089), Scientific Research Foundation for Changjiang Scholars of Shihezi University (CJXZ201501) and the Key Laboratory for Green Processing of Chemical Engineering of Xinjiang Bingtuan (No. KF201603).References
- M. Younes and H. Galal-Gorchev, Pesticides in drinking water—a case study, Food Chem. Toxicol., 2000, 38(suppl. 1), S87–S90 CrossRef CAS PubMed.
- J. Lu, et al., Degradation of Pesticides in Nursery Recycling Pond Waters, J. Agric. Food Chem., 2006, 54(7), 2658–2663 CrossRef CAS PubMed.
- Y. D. Lee, et al., Loss of pendimethalin in runoff and leaching from turfgrass land under simulated rainfall, J. Agric. Food Chem., 2000, 48(11), 5376–5382 CrossRef CAS PubMed.
- L. Hou, et al., Pendimethalin exposure and cancer risk among pesticide applicators: a report from the U.S.-based agricultural health study, Ann. Epidemiol., 2004, 14(8), 608 CrossRef.
- B. Singh and K. Singh, Microbial degradation of herbicides, Crit. Rev. Microbiol., 2016, 42(2), 245 CAS.
- S. Venkata Mohan and M. Rama Krishna, et al., Solid phase bioremediation of pendimethalin in contaminated soil and evaluation of leaching potential, Bioresour. Technol., 2007, 98(15), 2905–2910 CrossRef CAS PubMed.
- R. K. Kole, et al., Bacterial degradation of the herbicide pendimethalin and activity evaluation of its metabolites, Bull. Environ. Contam. Toxicol., 1994, 52(5), 779 CrossRef CAS PubMed.
- C. Zhang, et al., Slurry-Phase Biological Treatment of 2,4-Dinitrotoluene and 2,6-Dinitrotoluene: Role of Bioaugmentation and Effects of High Dinitrotoluene Concentrations, Environ. Sci. Technol., 2000, 34(13), 2810–2816 CrossRef CAS.
- M. C. Diez, Biological aspects involved in the degradation of organic pollutants, J. Soil Sci. Plant Nutr., 2010, 10(3), 244–267 Search PubMed.
- H. Ni, et al., Biodegradation of Pendimethalin by Paracoccus sp. P13, Curr. Microbiol., 2018, 75(8), 1077–1083 CrossRef CAS PubMed.
- V. B. Megadi, et al., Biodegradation of pendimethalin by Bacillus circulans, Indian J. Biotechnol., 2010, 9(2), 173–177 CAS.
- A. S. Barua, et al., Degradation of pendimethalin by soil fungi, Pest Manage. Sci., 2010, 29(4), 419–425 CrossRef.
- A. A. Juwarkar, et al., A comprehensive overview of elements in bioremediation, Rev. Environ. Sci. Bio/Technol., 2010, 9(3), 215–288 CrossRef CAS.
- A. P. Pinto, et al., Degradation of terbuthylazine, difenoconazole and pendimethalin pesticides by selected fungi cultures, Sci. Total Environ., 2012, 435–436(7), 402–410 CrossRef CAS PubMed.
- R. K. Kole, et al., Bacterial degradation of the herbicide pendimethalin and activity evaluation of its metabolites, Bull. Environ. Contam. Toxicol., 1994, 52(5), 779–786 CrossRef CAS PubMed.
- N. K. Sahoo, et al., Biodegradation of 4-chlorophenol by Arthrobacter chlorophenolicus A6: effect of culture conditions and degradation kinetics, Biodegradation, 2011, 22(2), 275–286 CrossRef CAS PubMed.
- C. Li, et al., Biodegradation of methidathion by Serratia sp. in pure cultures using an orthogonal experiment design, and its application in detoxification of the insecticide on crops, Ann. Microbiol., 2013, 63(2), 451–459 CrossRef CAS.
- G. Chennappa, et al., Pesticide tolerant Azotobacter isolates from paddy growing areas of northern Karnataka, India, World J. Microbiol. Biotechnol., 2014, 30(1), 1–7 CrossRef CAS PubMed.
- A. P. Pinto, et al., Exploring the potential of novel biomixtures and Lentinula edodes fungus for the degradation of selected pesticides. Evaluation for use in biobed systems, Sci. Total Environ., 2016, 541, 1372–1381 CrossRef CAS PubMed.
- C. P. Kurtzman and C. J. Robnett, Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene, J. Clin. Microbiol., 1997, 35(5), 1216 CAS.
- Y. K. Kim, et al., Kinetics of endosulfan degradation by Phanerochaete chrysosporium, Biotechnol. Lett., 2001, 23(2), 163–166 CrossRef CAS.
- M. Goswami, et al., Kinetics of Chlorophenol Degradation by Benzoate-Induced Culture of Rhodococcus erythropolis M1, World J. Microbiol. Biotechnol., 2002, 18(8), 779–783 CrossRef CAS.
- S. S. Dash and S. N. Gummadi, Degradation Kinetics of Caffeine and Related Methylxanthines by Induced Cells of Pseudomonas sp, Curr. Microbiol., 2007, 55(1), 56–60 CrossRef CAS PubMed.
- S. B. Singh and G. Kulshrestha, Microbial degradation of pendimethalin, J. Environ. Sci. Health, Part B, 1991, 26(3), 309–321 CrossRef CAS PubMed.
- R. K. Kole, et al., Bacterial degradation of the herbicide pendimethalin and activity evaluation of its metabolites, Bull. Environ. Contam. Toxicol., 1994, 52(5), 779–786 CrossRef CAS PubMed.
- H. Ni, et al., Biodegradation of pendimethalin by Bacillus subtilis Y3, J. Environ. Sci., 2016, 41(3), 121–127 CrossRef PubMed.
- M. Ramakrishna, et al., Identification of metabolites during biodegradation of pendimethalin in bioslurry reactor, J. Hazard. Mater., 2008, 151(2–3), 658–661 CrossRef CAS PubMed.
- G. Kulshrestha and S. B. Singh, Influence of soil moisture and microbial activity on pendimethalin degradation, Bull. Environ. Contam. Toxicol., 1992, 48(2), 269–274 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra07872f |
| This journal is © The Royal Society of Chemistry 2019 |