Open Access Article
Open Access ArticleRetracted Article: Down-regulation of MRPS23 inhibits LPS-induced proliferation and invasion via regulation of the NF-κB signaling pathway in osteosarcoma cells†
Ai-Guo Liu‡
a,
Ke-Lin Xu‡ *b,
Wei-Lin Wanga,
Bing-Kang Zhoua and
Qing-Gong Guoa
*b,
Wei-Lin Wanga,
Bing-Kang Zhoua and
Qing-Gong Guoa
aDepartment of Orthopaedics, The First Affiliated Hospital of Henan University, Kaifeng 475000, China
bDepartment of Orthopaedics and Traumatology, Wuxi Hospital Affiliated to Nanjing University of Chinese Medicine, No. 8 Zhongnan West Road, Wuxi 214000, China. E-mail: xukelin_orth@163.com; Fax: +86-510-88859999; Tel: +86-510-88859999
First published on 4th April 2019
Abstract
Mitochondrial ribosomal protein S23 (MRPS23), encoded by a nuclear gene, is a participant in the translation of mitochondrial proteins. Recently, MRPS23 has been reported to be overexpressed in many types of cancers and have a close association with cancer progression. However, the specific roles of MRPS23 in osteosarcoma (OS) remain unknown. In this study, we investigated the expression pattern and biological functions of MRPS23 in OS cells. Our results demonstrated that MRPS23 was up-regulated in OS tissues and cell lines. Down-regulation of MRPS23 significantly inhibited OS cell proliferation and invasion induced by lipopolysaccharide (LPS) in vitro. Furthermore, the in vivo experiments showed that MRPS23 down-regulation markedly suppressed OS cell growth and metastasis induced by LPS. Mechanistically, down-regulation of MRPS23 inhibited the activity of NF-κB signaling pathway in OS cells. In conclusion, these findings indicated that MRPS23 may be a potential therapeutic target for OS treatment.
Introduction
Osteosarcoma (OS), a common type of bone sarcomas, frequently occurs in children, adolescents or young adults.1,2 This malignant neoplasm arises from primitive transformed cells of mesenchymal origin, exhibits osteoblastic differentiation and produces malignant osteoid.3–5 OS is characterized by aggressiveness, local recurrence and distant metastasis.6 In the past, surgery was the main therapeutic measure for OS patients and the 5 year survival rate was only 20% due to the micrometastasis overlooked at diagnosis.7 Currently, multimodal therapies have been applied for OS and the 5 year survival rate has been increased to 70% for patients with localized disease.8,9 Unfortunately, OS patients with metastasis still suffer from a poor prognosis and their 5 year survival rate remains only 30%.10,11 Therefore, exploring novel biomarkers and therapeutic approaches relevant to metastasis is urgently needed for a better clinical management of OS patients.Mitochondria are bioenergetic, biosynthetic and signaling organelles.12–14 They not only play a significant role in the adaptation to environmental changes but also function as the main energy and power centers in cells.15,16 Increasing evidence has demonstrated that mitochondria take part in many other essential functions such as cell death, apoptosis and autophagy apart from energy metabolism.17–19 In addition, mitochondria have been found serving as important mediators of tumorigenesis.20,21 Thus, many researchers have focused on a group of unique proteins called mitochondrial ribosomal proteins (MRPs). These proteins are completely encoded by nuclear genes and mainly assist with the process of mitochondrial protein translation in mitochondria.22 More importantly, many studies have reported other functions of MRPs in tumorigenesis.23–25 For example, MRPS23, a member of the MRP family, has been demonstrated to be aberrantly expressed in some types of cancers and closely associated with development of these cancers.26–28 However, the biological functions of MRPS23 in OS remain unclear.
In this study, we investigated the roles and mechanisms of MRPS23 in OS progression induced by lipopolysaccharide (LPS). The results showed that MRPS23 was up-regulated in OS tissues and cell lines. Furthermore, down-regulation of MRPS23 significantly inhibited LPS-induced OS cell proliferation and invasion in vitro and in vivo and these effects were mediated partly via the NF-κB pathway.
Results
Expression of MRPS23 is elevated in OS tissues and cell lines
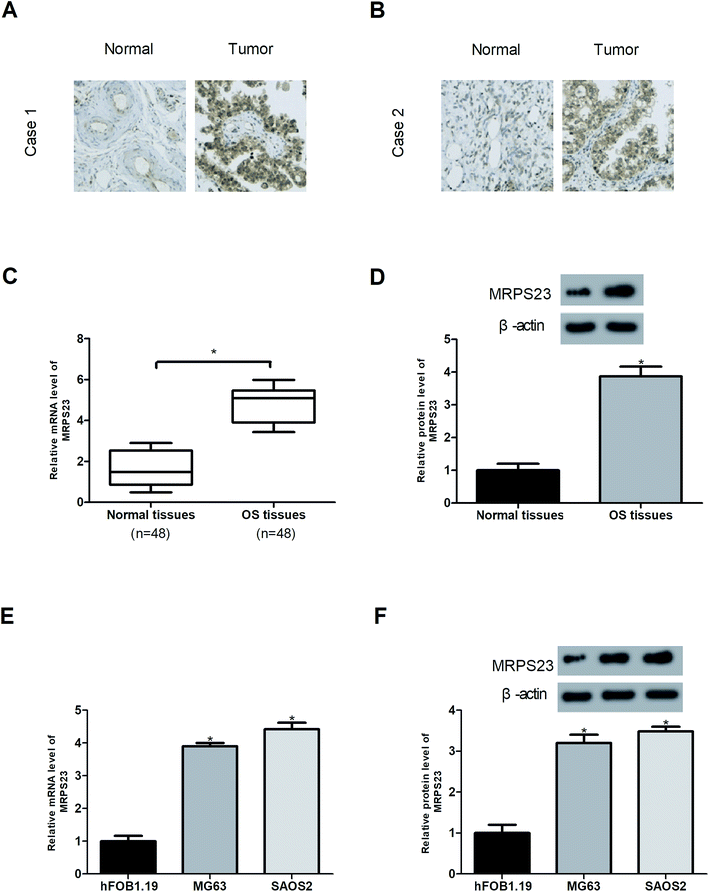
The expression of MRPS23 in OS tissues was examined by immunohistochemistry, RT-PCR and western blot analysis. The results showed that MRPS23 staining was strong in OS tissues but rarely observed in corresponding normal bone tissues (Fig. 1A and B). In addition, MRPS23 mRNA and protein expression levels were markedly higher in OS tissues than in the adjacent normal bone tissues (Fig. 1C and D). Furthermore, we investigated the expression of MRPS23 in OS cell lines. As shown in Fig. 1E and F, MRPS23 was significantly up-regulated at both mRNA and protein levels in OS cell lines (SAOS2 and MG63) in comparison with the normal osteoblast cell line hFOB1.19.Down-regulation of MRPS23 induces loss of mitochondrial membrane potential in OS cells and inhibits LPS-induced OS cell proliferation and invasion in vitro
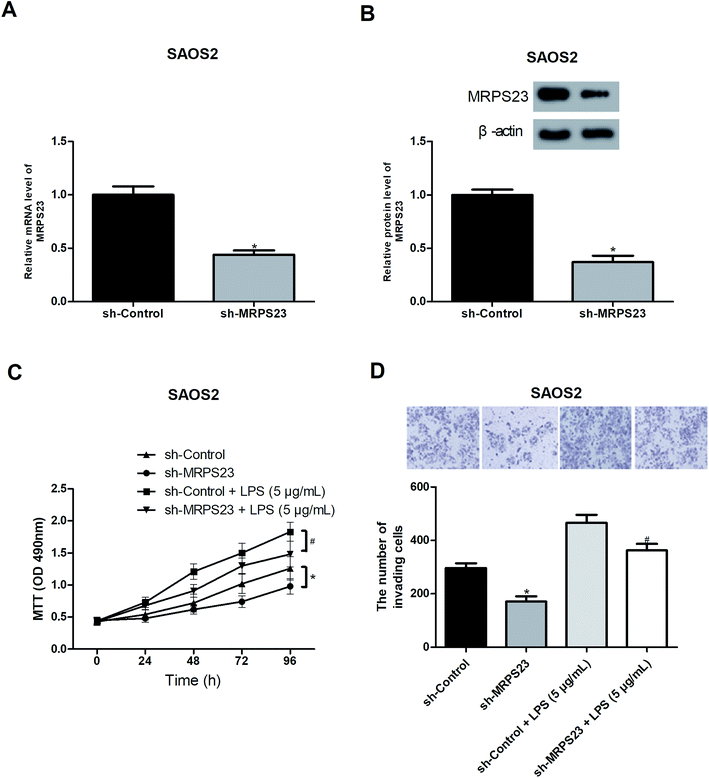
To further investigate the role of MRPS23 in OS, we decreased the expression of MRPS23 in SAOS2 cells via shRNA transfection. RT-PCR and western blot assays were performed to verify the transfection efficiency (Fig. 2A and B). Considering the association of MRPS23 with the mitochondrial functions, we investigated the change of mitochondrial membrane potential in OS cells after sh-MRPS23 transfection. As shown in Fig. S1,† down-regulation of MRPS23 induced a significant decrease in mitochondrial membrane potential in SAOS2 cells. LPS is known as an inducer of cell proliferation and invasion in various cancers.29,30 Therefore, we examined the effect of MRPS23 down-regulation on LPS-induced proliferation and invasion of OS cells. The MTT and transwell assays were performed to measure cell proliferation and invasion, respectively. As shown in Fig. 2C and D, LPS-treated SAOS2 cells showed enhanced proliferative and invasive abilities compared with corresponding control cells. However, these promoted effects were obviously prevented by MRPS23 down-regulation (Fig. 2C and D).Down-regulation of MRPS23 inhibits LPS-induced OS cell growth and metastasis in vivo
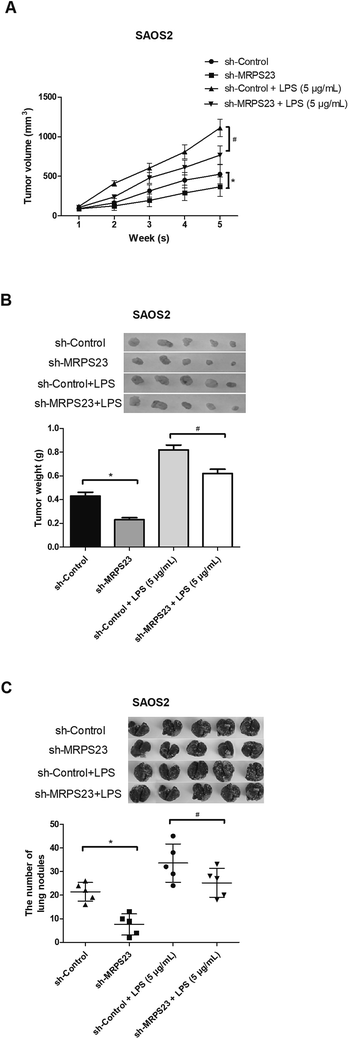
To determine whether MRPS23 down-regulation can suppress LPS-induced OS cell growth in vivo, SAOS2 cells transfected with sh-MRPS23 or sh-Control were treated with or without LPS and then injected into the left flank of mice. As shown in Fig. 3A, the growth rate of tumors was rapid in the SAOS2/LPS group but markedly slow in the SAOS2/sh-MRPS23 group in comparison with corresponding control groups. In addition, the average weight of tumors generated from LPS-treated SAOS2 cells was increased but greatly decreased in the SAOS2/sh-MRPS23 group in comparison with corresponding control groups (Fig. 3B). To further investigate the effect of MRPS23 down-regulation on OS cell metastasis in vivo, SAOS2 cells transfected with sh-MRPS23 or sh-Control were treated with or without LPS and then transplanted orthotopically into the bones of mice. As shown in Fig. 3C, more metastatic nodules were found in the SAOS2/LPS group while less metastatic nodules were detected in the SAOS2/sh-MRPS23 group, in comparison with corresponding control groups.Down-regulation of MRPS23 abrogates LPS-induced activity of the NF-κB signaling pathway in OS cells
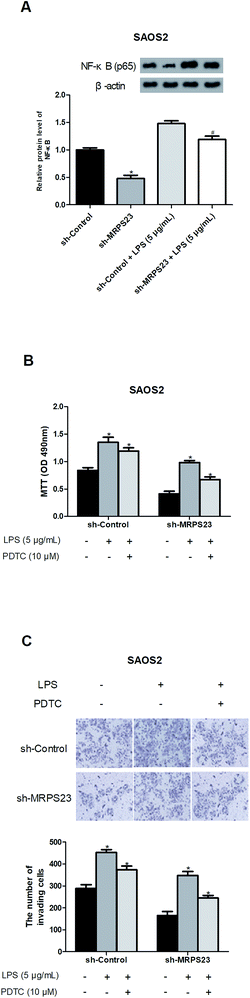
Previous studies have reported that LPS could promote cancer progression via regulating the NF-κB signaling pathway,31–33 so we investigated whether MRPS23 down-regulation inhibited LPS-induced NF-κB pathway. As expected, LPS obviously increased the expression of NF-κB in SAOS2 cells while MRPS23 down-regulation significantly blocked this effect (Fig. 4A). To further confirm implication of the NF-κB pathway in the suppressive effect of MRPS23 down-regulation on LPS-induced OS cell proliferation and invasion, PDTC (NF-κB inhibitor) was used to treat OS cells. The results indicated that LPS-induced OS cell proliferation and invasion were inhibited by MRPS23 down-regulation and these effects were enhanced after PDTC treatment (Fig. 4B and C).Discussion
Characterized by different degrees of mesenchymal differentiation, OS is a heterogeneous group of malignancies.34 With a high rate of recurrence and metastasis, OS causes the death of a majority of OS patients.35 Therefore, it is urgently required to explore more effective treatment alternatives.Previous studies have demonstrated the abnormal expression and potential value of MRPS23 in several types of cancers.26,27 The association of MRPS23 with cancer progression was first found in cervical cancer where overexpression of MRPS23 indicated high proliferative ability and oxygen consumption.28 MRPS23 has also been reported to exert a similar effect on hepatocellular cancer where MRPS23 functions as a powerful driver of tumor proliferation.36 In consistent with the previous findings, our study showed that MRPS23 was highly expressed in OS tissues and cell lines. Furthermore, down-regulation of MRPS23 inhibited OS cell proliferation and invasion. This in vitro result was further verified by our xenograft tumor assay which indicated that MRPS23 down-regulation suppressed OS cell growth and metastasis in vivo. All these observations suggested that MRPS23 might serve as a novel oncogene in cancer progression.
LPS is a strong inflammation stimulator and is associated with alteration of cytokine levels in tumor microenvironment.37 The exposure to LPS may induce tumor proliferation and facilitate invasion.29,30,32 For example, Wang et al. reported that hepatocellular cancer cell survival and proliferation were enhanced upon LPS stimulation.38 Similarly, Yang et al. demonstrated that LPS could induce cell proliferation in colorectal cancer.39 Consistently, Bedini et al. found that LPS exerted a promoting effect on glioblastoma cell proliferation.40 Moreover, LPS is also an important player in mediation of the epithelial–mesenchymal transition, a key process with enhanced migration and invasion during cancer progression.31,41 In this study, we found that LPS induced OS cell proliferation and invasion and this promoting effect was abrogated by MRPS23 down-regulation. We observed similar results in our in vivo experiments.
In regard to the mechanisms by which MRPS23 regulated LPS-induced OS progression, we focused on the NF-κB pathway. The NF-κB family consists of a group of transcription factors such as NF-κB1, NF-κB2, Rel A, Rel-B and Rel-C.42,43 These members participate in various cellular processes and regulation.44 A growing body of evidence has demonstrated that abnormal activation of the NF-κB pathway will cause aberrant expression of cancer-related genes, promote cell proliferation and accelerate tumor metastasis.45,46 In this study, we examined the role of the NF-κB pathway in the inhibitory effect of MRPS23 down-regulation on LPS-induced OS progression. The results showed that LPS increased the expression of NF-κB in OS cells while MRPS23 down-regulation significantly reversed this effect. To further confirm involvement of the NF-κB pathway, OS cells were treated with PDTC (NF-κB inhibitor). The results indicated that LPS-induced OS cell proliferation and invasion were suppressed by MRPS23 down-regulation and these effects were potentiated after PDTC treatment. Based on these findings, we reasonably suggested that the NF-κB pathway could be responsible for part of the mechanisms by which MRPS23 mediated LPS-induced OS progression.
In conclusion, this study demonstrated that MRPS23 was highly expressed in OS tissues and cell lines and was involved in the regulation of LPS-induced OS cell proliferation and invasion in vitro and in vivo. Furthermore, MRPS23 exerted these effects partly via the NF-κB pathway. Taken together, our study provided evidence supporting MRPS23 as a potential therapeutic target for OS treatment.
Materials and methods
Ethics statement
This study was performed in strict accordance with the NIH guidelines for the care and use of laboratory animals (NIH Publication No. 85-23 Rev. 1985) and was approved by the Ethics Committee of the First Affiliated Hospital of Henan University (Kaifeng, China).Patients and tissue samples
A total of 48 OS patients from the First Affiliated Hospital of Henan University (Kaifeng, China) were enrolled in the study. The OS tissues and the adjacent normal bone tissues were collected during surgery and then frozen in liquid nitrogen and stored at −80 °C for future experiments. All tissue specimens were used with informed consent from each patient and with approval of the Ethics Committee of Henan University.Cell lines and cell culture
Human OS cell lines (MG63 and SAOS2) and normal osteoblast cell line hFOB1.19 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen) and 1% penicillin/streptomycin and then incubated at 37 °C in a humidified atmosphere with 5% CO2.Immunohistochemistry (IHC)
Sliced tissues were washed with PBS containing 0.1% bovine serum albumin and then incubation with 3% hydrogen peroxide for 10 min. Subsequently, the slices were subjected to overnight incubation at 4 °C with primary anti-MRPS23 antibody (Cell Signaling Technology, Danvers, MA, USA), followed by incubation with HRP goat anti-mouse/rabbit IgG (Cell Signaling Technology) for 15 min at room temperature. Diaminobenzidine (DAB; Invitrogen) was used for color development. Representative images of tissues were captured using a light microscope (200×).Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissues or cells using TRIzol Reagent (Invitrogen) and then reversely transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The following primers were used: MRPS23, 5′-GGTTTGACGTATATGACGCCTT-3′ (forward) and 5′-CTCTAATCCGATCCTCGTGGTA-3′ (reverse); β-actin, 5′-CATGTACGTTGCTATCCAGGC-3′ (forward) and 5′-CTCCTTAATGTCACGCACGAT-3′ (reverse). The reaction conditions were as follows: 96 °C for 30 s, 40 cycles of 96 °C for 15 s, 62 °C for 1 min and 75 °C for 45 s. β-Actin was used as an internal control for normalization of the results. The relative mRNA expression was calculated by the 2−ΔΔCt method.Western blot analysis
Tissues or cells were lysed in lysis buffer. Cell lysates were collected after 5 min of centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. The concentration of lysate proteins was measured using the BCA Protein Assay Kit (Pierce, Rockford, IL, USA). An equal amount of protein was separated by 10% SDS-PAGE and then transferred onto PVDF membranes (Millipore, Billerica, MA, USA). After blocking in 5% non-fat milk, the membranes were incubated overnight at 4 °C with primary antibodies against MRPS23, NF-κB and β-actin. Subsequently, the membranes were washed three times with TBST and then incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. All antibodies were purchased from Invitrogen. Protein bands were detected using an enhanced chemiluminescence detection system (Cell Signaling Technology) and their intensity was analyzed using the Quantity One software (Bio-Rad, Hercules, CA, USA).
000g. The concentration of lysate proteins was measured using the BCA Protein Assay Kit (Pierce, Rockford, IL, USA). An equal amount of protein was separated by 10% SDS-PAGE and then transferred onto PVDF membranes (Millipore, Billerica, MA, USA). After blocking in 5% non-fat milk, the membranes were incubated overnight at 4 °C with primary antibodies against MRPS23, NF-κB and β-actin. Subsequently, the membranes were washed three times with TBST and then incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. All antibodies were purchased from Invitrogen. Protein bands were detected using an enhanced chemiluminescence detection system (Cell Signaling Technology) and their intensity was analyzed using the Quantity One software (Bio-Rad, Hercules, CA, USA).
Cell transfection
The shRNA sequences targeting MRPS23 (GCAGAGTCTTGGAGAAACA) were purchased from GeneCopoeia (Rockville, MD, USA). Cells were plated in 6-well plates at a density of 1 × 105 cells per well. After reaching 80% confluence, cells were transfected with sh-MRPS23 or sh-Control using Lipofectamine 2000 reagent (Invitrogen). 48 h later, western blot assays were performed to confirm the transfection efficiency.MTT assay
Cells were seeded in a 96-well plate at a density of 2 × 103 cells per well and cultured for different time. After treatment with LPS for 24 h, MTT (5 mg mL−1; Sigma, St. Louis, MO, USA) was added to each well at different time points. After removal of culture medium, DMSO (Sigma) was added to each well. The absorbance was measured at 490 nm using a microplate reader.Transwell assay
Cell invasion was detected using Matrigel-coated transwell chambers. Cells (1 × 105) were added to the upper chamber containing serum-free medium and the lower chamber was filled with culture medium containing 10% FBS. After incubation with LPS for 24 h, cells remaining on the upper surface of the filter were removed and cells invading to the lower surface of the filter were fixed and stained with 0.1% crystal violet. The number of invading cells from five random fields was counted under a microscope (200×).Measurement of mitochondrial membrane potential
Changes in the mitochondrial membrane potential were measured using 3,3′-tetraethylbenzimidazolylcarbo-cyanine iodide (JC-1). In brief, cells were seeded in a 96-well plate and allowed to reach about 70% density. Subsequently, JC-1 was added and cells were further incubated for 30 min at 37 °C and then washed three times with PBS. Fluorescence was measured using a flow cytometry (excitation: 488 nm and emission: 525 nm).In vivo xenograft tumor assay
Male BALB/c nude mice (4 to 6 weeks old) were obtained from Shanghai Laboratory Animal Center (Shanghai, China). All animal experiments were approved by the Animal Care and Use Committee of Henan University. For the tumor growth assay, mice were randomly divided into 4 groups (n = 5). 2 × 106 SAOS2 cells transfected with sh-MRPS23 or sh-Control with or without treatment of LPS (5 μg mL−1) were resuspended in PBS and then subcutaneously injected into the left flank of mice. Tumors were measured every week. Tumor volume was calculated by the following formula: volume = 0.5 × width2 × length. 35 days later, mice were sacrificed and tumors were harvested and weighed.For the tumor metastasis assay, mice were randomly divided into 4 groups (n = 5). 2 × 106 SAOS2 cells transfected with sh-MRPS23 or sh-Control with or without treatment of LPS (5 μg mL−1) were resuspended in PBS and then transplanted orthotopically into the bones of mice. 5 weeks later, mice were euthanized and their lungs were removed. The number of metastatic lung nodules was counted under a microscope.
Statistical analysis
All experiments were conducted three times and data were expressed as means ± standard deviation (SD). Statistical analysis was performed using SPSS 18.0 software. Student's t-test or one-way ANOVA was used for the comparison between different groups. P < 0.05 was considered statistically significant.Conflicts of interest
The authors declare that they have no conflicts of interest.Acknowledgements
None.References
- G. Pierron and E. Al, A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion, Nat. Genet., 2012, 44, 461–466 CrossRef CAS PubMed.
- L. Mirabello, R. J. Troisi and S. A. Savage, Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program, Cancer, 2010, 115, 1531–1543 CrossRef PubMed.
- D. Benayahu, I. Shur, R. Marom, I. Meller and J. Issakov, Cellular and molecular properties associated with osteosarcoma cells, J. Cell. Biochem., 2001, 84, 108–114 CrossRef CAS PubMed.
- M. J. Klein and G. P. Siegal, Osteosarcoma: anatomic and histologic variants, Am. J. Clin. Pathol., 2006, 125, 555–581 CrossRef PubMed.
- A. Panizosantos, I. Sola, M. Lozano, A. E. De and J. Pardo, Metastatic osteosarcoma presenting as a small-bowel polyp. A case report and review of the literature, Arch. Pathol. Lab. Med., 2000, 124, 1682–1684 CAS.
- L. Kager, A. Zoubek, U. Kastner, B. Kempfbielack, J. Potratz, R. Kotz, G. U. Exner, C. Franzius, S. Lang and R. Maas, Skip metastases in osteosarcoma: experience of the Cooperative Osteosarcoma Study Group, J. Clin. Oncol., 2006, 24, 1535–1541 CrossRef PubMed.
- G. Bulut, S. H. Hong, K. Chen, E. M. Beauchamp, S. Rahim, G. W. Kosturko, E. Glasgow, S. Dakshanamurthy, H. S. Lee and I. Daar, Small molecule inhibitors of ezrin inhibit the invasive phenotype of osteosarcoma cells, Oncogene, 2012, 31, 269–281 CrossRef CAS PubMed.
- J. Gill, M. K. Ahluwalia, D. Geller and R. Gorlick, New targets and approaches in osteosarcoma, Pharmacol. Ther., 2013, 137, 89–99 CrossRef CAS PubMed.
- C. L. Schwartz, R. Gorlick, L. Teot, M. Krailo, Z. Chen, A. Goorin, H. E. Grier, M. L. Bernstein and P. Meyers, Multiple drug resistance in osteogenic sarcoma: INT0133 from the Children's Oncology Group, J. Clin. Oncol., 2007, 25, 2057–2062 CrossRef PubMed.
- S. Ferrari and E. Palmerini, Adjuvant and neoadjuvant combination chemotherapy for osteogenic sarcoma, Curr. Opin. Oncol., 2007, 19, 341–346 CrossRef CAS PubMed.
- P. Zhang, Y. Yang, P. Zweidler-Mckay and D. P. Hughes, Critical role of notch signaling in osteosarcoma invasion and metastasis, Clin. Cancer Res., 2013, 19, 5256 CrossRef PubMed.
- M. D. Esposti, Bioenergetic Evolution in Proteobacteria and Mitochondria, Genome Biol. Evol., 2014, 6, 3238–3251 CrossRef CAS PubMed.
- K. Y. Hostetler, d. B. H. Van and L. L. van Deenen, The mechanism of cardiolipin biosynthesis in liver mitochondria, Biochim. Biophys. Acta, 1972, 260, 507–513 CrossRef CAS.
- N. S. Chandel, Evolution of Mitochondria as Signaling Organelles, Cell Metab., 2015, 22, 204–206 CrossRef CAS PubMed.
- D. D. Ritchie, Reactions of fungus mitochondria to environmental changes, Trans. N. Y. Acad. Sci., 2012, 15, 157 CrossRef.
- J. F. Dumas, D. Roussel, G. Simard, O. Douay, F. Foussard, Y. Malthiery and P. Ritz, Food restriction affects energy metabolism in rat liver mitochondria, Biochim. Biophys. Acta, 2004, 1670, 126–131 CrossRef CAS.
- R. Rizzuto, P. Pinton, D. D. Stefani, C. Giorgi, C. Mammucari, S. Marchi, A. Rimessi, A. Romagnoli, R. Siviero and E. Zecchini, Mitochondria, calcium signaling and cell death by apoptosis and autophagy, Biochim. Biophys. Acta, 2010, 1797, 4 CrossRef.
- D. R. Green, L. Galluzzi and G. Kroemer, Mitochondria and the autophagy-inflammation-cell death axis in organismal aging, Science, 2011, 333, 1109–1112 CrossRef CAS PubMed.
- A. M. Tolkovsky, L. Xue, G. C. Fletcher and V. Borutaite, Mitochondrial disappearance from cells: a clue to the role of autophagy in programmed cell death and disease?, Biochimie, 2002, 84, 233–240 CrossRef CAS PubMed.
- A. Salas, Y. G. Yao, V. Macaulay, A. Vega, A. Carracedo and H. J. Bandelt, A critical reassessment of the role of mitochondria in tumorigenesis, PLoS Med., 2005, 2, e296 CrossRef PubMed.
- S. Srinivasan, M. Guha, A. Kashina and N. G. Avadhani, Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection, Biochim. Biophys. Acta, 2017, 1858, 602–614 CrossRef CAS PubMed.
- N. Kenmochi, T. Suzuki, T. Uechi, M. Magoori, M. Kuniba, S. Higa, K. Watanabe and T. Tanaka, The human mitochondrial ribosomal protein genes: mapping of 54 genes to the chromosomes and implications for human disorders, Genomics, 2001, 77, 65–70 CrossRef CAS PubMed.
- U. Richter, T. Lahtinen, P. Marttinen, M. Myöhänen, D. Greco, G. Cannino, H. T. Jacobs, N. Lietzén, T. A. Nyman and B. J. Battersby, A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation, Curr. Biol., 2013, 23, 535–541 CrossRef CAS PubMed.
- J. Lee, M. Y. Seol, S. Jeong, C. R. Lee, C. R. Ku, S. W. Kang, J. J. Jeong, D. Y. Shin, K. H. Nam and E. J. Lee, A metabolic phenotype based on mitochondrial ribosomal protein expression as a predictor of lymph node metastasis in papillary thyroid carcinoma, Medicine, 2015, 94, e380 CrossRef CAS PubMed.
- M. Mints, M. Mushtaq, N. Iurchenko, L. Kovalevska, M. C. Stip, D. Budnikova, S. Andersson, L. Polischuk, L. Buchynska and E. Kashuba, Mitochondrial ribosomal protein S18-2 is highly expressed in endometrial cancers along with free E2F1, Oncotarget, 2016, 7, 22150–22158 CrossRef PubMed.
- Z. H. Zhang, D. D. Xie, S. Xu, M. Z. Xia, Z. Q. Zhang, H. Geng, L. Chen, D. M. Wang, W. Wei and D. X. Yu, Total glucosides of paeony inhibits lipopolysaccharide-induced proliferation, migration and invasion in androgen insensitive prostate cancer cells, PLoS One, 2017, 12, e0182584 CrossRef PubMed.
- M. A. Seol, J. H. Park, J. H. Jeong, J. Lyu, S. Y. Han and S. M. Oh, Role of TOPK in lipopolysaccharide-induced breast cancer cell migration and invasion, Oncotarget, 2017, 8, 40190–40203 CrossRef PubMed.
- B. Jung, E. H. Jang, D. Hong, I. H. Cho, M. J. Park and J. H. Kim, Aqueous extract of Psoralea corylifolia L. inhibits lipopolysaccharide-induced endothelial-mesenchymal transition via downregulation of the NF-κB-SNAIL signaling pathway, Oncol. Rep., 2015, 34, 2040–2046 CrossRef CAS PubMed.
- P. Xu, F. Cai, X. Liu and L. Guo, Sesamin inhibits lipopolysaccharide-induced proliferation and invasion through the p38-MAPK and NF-κB signaling pathways in prostate cancer cells, Oncol. Rep., 2015, 33, 3117–3123 CrossRef CAS PubMed.
- I. H. Cho, E. H. Jang, D. Hong, B. Jung, M. J. Park and J. H. Kim, Suppression of LPS-induced epithelial-mesenchymal transition by aqueous extracts of Prunella vulgaris through inhibition of the NF-κB/Snail signaling pathway and regulation of EMT-related protein expression, Oncol. Rep., 2015, 34, 2445 CrossRef CAS PubMed.
- Y. Wang, Q. Fu and W. Zhao, Tetramethylpyrazine inhibits osteosarcoma cell proliferation via downregulation of NF-κB in vitro and in vivo, Mol. Med. Rep., 2013, 8, 984–988 CrossRef CAS PubMed.
- B. C. He, L. Chen, G. W. Zuo, W. Zhang, Y. Bi, J. Huang, Y. Wang, W. Jiang, Q. Luo and Q. Shi, Synergistic antitumor effect of the activated PPARgamma and retinoid receptors on human osteosarcoma, Clin. Cancer Res., 2010, 16, 2235–2245 CrossRef CAS PubMed.
- M. L. Gatza, G. O. Silva, J. S. Parker, C. Fan and C. M. Perou, An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer, Nat. Genet., 2014, 46, 1051–1059 CrossRef CAS PubMed.
- K. Szuhai, A. M. Cleton-Jansen, P. C. Hogendoorn and J. V. Bovée, Molecular pathology and its diagnostic use in bone tumors, Cancer Genet., 2012, 205, 193–204 CrossRef CAS PubMed.
- H. Lyng, R. S. Brøvig, D. H. Svendsrud, R. Holm, O. Kaalhus, K. Knutstad, H. Oksefjell, K. Sundfør, G. B. Kristensen and T. Stokke, Gene expressions and copy numbers associated with metastatic phenotypes of uterine cervical cancer, BMC Genomics, 2006, 7, 1–15 CrossRef PubMed.
- M. Pu, J. Wang, Q. Huang, G. Zhao, C. Xia, R. Shang, Z. Zhang, Z. Bian, X. Yang and K. Tao, High MRPS23 expression contributes to hepatocellular carcinoma proliferation and indicates poor survival outcomes, Tumor Biol., 2017, 39, 1010428317709127 CrossRef PubMed.
- A. J. Schetter, N. H. Heegaard and C. C. Harris, Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways, Carcinogenesis, 2010, 31, 37–49 CrossRef CAS PubMed.
- L. Wang, R. Zhu, Z. Huang, H. Li and H. Zhu, Lipopolysaccharide-induced toll-like receptor 4 signaling in cancer cells promotes cell survival and proliferation in hepatocellular carcinoma, Dig. Dis. Sci., 2013, 58, 2223–2236 CrossRef CAS PubMed.
- P. Yang, Z. Li, H. Li, Y. Lu, H. Wu and Z. Li, Pyruvate kinase M2 accelerates pro-inflammatory cytokine secretion and cell proliferation induced by lipopolysaccharide in colorectal cancer, Cell. Signalling, 2015, 27, 1525–1532 CrossRef CAS PubMed.
- A. Bedini, M. Baiula, G. Vincelli, F. Formaggio, S. Lombardi, M. Caprini and S. Spampinato, Nociceptin/orphanin FQ antagonizes lipopolysaccharide-stimulated proliferation, migration and inflammatory signaling in human glioblastoma U87 cells, Biochem. Pharmacol., 2017, 140, 89–104 CrossRef CAS PubMed.
- D. Hong, S. Y. Jang, E. H. Jang, B. Jung, I. H. Cho, M. J. Park, S. Y. Jeong and J. H. Kim, Shikonin as an inhibitor of the LPS-induced epithelial-to-mesenchymal transition in human breast cancer cells, Int. J. Mol. Med., 2015, 36, 1601–1606 CrossRef CAS PubMed.
- M. D. Sanchezniño, A. B. Sanz, M. C. Izquierdo, J. Poveda and A. Ortiz, NF-κB Family, Encyclopedia Signal. Mol., 2012, pp. 1216–1222 Search PubMed.
- J. A. S. Baldwin, Series Introduction: The transcription factor NF-κB and human disease, J. Clin. Invest., 2001, 107, 3–6 CrossRef PubMed.
- S. Vallabhapurapu and M. Karin, Regulation and function of NF-kappaB transcription factors in the immune system, Annu. Rev. Immunol., 2009, 27, 693–733 CrossRef CAS PubMed.
- N. Sasaki, T. Morisaki, K. Hashizume, T. Yao, M. Tsuneyoshi, H. Noshiro, K. Nakamura, T. Yamanaka, A. Uchiyama and M. Tanaka, Nuclear factor-kappaB p65 (RelA) transcription factor is constitutively activated in human gastric carcinoma tissue,, Clin. Cancer Res., 2001, 7, 4136–4142 CAS.
- P. Viatour, M. P. Merville, V. Bours and A. Chariot, Phosphorylation of NF-κB and IκB proteins: implications in cancer and inflammation, Trends Biochem. Sci., 2005, 30, 43–52 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra08973f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |