 Open Access Article
Open Access ArticleGreen synthesis of silver nanoparticles: biomolecule-nanoparticle organizations targeting antimicrobial activity†
Anupam Roy‡
a,
Onur Bulut‡
bcd,
Sudip Some
e,
Amit Kumar Mandal
*e and
M. Deniz Yilmaz
 *df
*df
aLaboratory of Food Chemistry and Technology, Department of Chemical Engineering, Birla Institute of Technology, Mesra, Ranchi-835215, India
bDepartment of Molecular Biology and Genetics, Faculty of Agriculture and Natural Sciences, Konya Food and Agriculture University, 42080 Konya, Turkey
cDepartment of Biological Sciences, Middle East Technical University, 06800 Ankara, Turkey
dResearch and Development Center for Diagnostic Kits (KIT-ARGEM), Konya Food and Agriculture University, 42080 Konya, Turkey. E-mail: deniz.yilmaz@gidatarim.edu.tr
eChemical Biology Laboratory, Department of Sericulture, Raiganj University, Uttar Dinajpur-733134, India. E-mail: amitmandal08@gmail.com
fDepartment of Bioengineering, Faculty of Engineering and Architecture, Konya Food and Agriculture University, 42080 Konya, Turkey
First published on 21st January 2019
Abstract
Since discovery of the first antibiotic drug, penicillin, in 1928, a variety of antibiotic and antimicrobial agents have been developed and used for both human therapy and industrial applications. However, excess and uncontrolled use of antibiotic agents has caused a significant growth in the number of drug resistant pathogens. Novel therapeutic approaches replacing the inefficient antibiotics are in high demand to overcome increasing microbial multidrug resistance. In the recent years, ongoing research has focused on development of nano-scale objects as efficient antimicrobial therapies. Among the various nanoparticles, silver nanoparticles have gained much attention due to their unique antimicrobial properties. However, concerns about the synthesis of these materials such as use of precursor chemicals and toxic solvents, and generation of toxic byproducts have led to a new alternative approach, green synthesis. This eco-friendly technique incorporates use of biological agents, plants or microbial agents as reducing and capping agents. Silver nanoparticles synthesized by green chemistry offer a novel and potential alternative to chemically synthesized nanoparticles. In this review, we discuss the recent advances in green synthesis of silver nanoparticles, their application as antimicrobial agents and mechanism of antimicrobial mode of action.
Introduction
The increasing tendency of microbial infections, rapid emergence of drug-resistance to recent antibiotics and quick evolution through mutation necessitate development or modification of antimicrobial compounds and alternative treatments.1–4 Advanced research in nanotechnology recently comes with the development of nano-scale objects with prominent antimicrobial actions against multidrug resistant pathogens suggesting a platform to fight against bacterial mutation arch.5,6 Among the most advanced nanotechnological applications, metal nanoparticles provide the most effective results with their unique mode of actions.7Based upon the evidence of microcidal lethal effects of essential metals (indispensible for the biochemistry of life in all organism fulfilling cellular functions) in excess dose and nonessential metals even at very minute doses, the use of metals as antimicrobial agents has proved to have strong scientific evidences since antiquity.8–10 In some cases metal ion being tightly bound to antibiotic structure (e.g. bacitracin, bleomycin, streptonigrin, and albomycin) regulates the biocidal action,11 they have been attached to the antibiotic molecule (e.g. tetracyclines, aureolic acids, and quinolones) without causing a major change in antibiotic structure but enhancing the activity.12 Based on the microcidal nature, the synthesis of metal nanoparticles elicited high interest among the researchers and nanotechnologists. Efforts are made to prepare green synthesized nanoscale objects with different metals like copper,13,14 zinc,15 titanium,16 magnesium,17 gold18 and silver.19 Other green synthesized metals like gold with their low-dimensional structure20 finds several other application in varying fields of engineering and technology; whereas, synthesis of silver nanoparticle by green method finds its possible application in biomedical field specially in antimicrobial development (AgNPs).21
Nanoscale silver particles with high surface-area-to-volume ratio (size below 100 nm) are of prime interest due to high antimicrobial actions against both Gram-positive and Gram-negative bacteria,19,22 viruses and other eukaryotic micro-organisms23 as compared to other metals in their nano form. It also offers potential effects against multidrug susceptible as well as multidrug resistant strains like Pseudomonas aeruginosa, ampicillin-resistant Escherichia coli, erythromycin-resistant Streptococcus pyogenes, methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Staphylococcus aureus (VRSA).7 Due to their biocidal actions, several physical and chemical routes have been applied to synthesize AgNPs, and AgNPs have been tried to be applied in modifying surfaces, coating, fibre grafting, gel preparations etc.24–27 But the drawbacks including use of toxic precursor chemicals (such as sodium borohydride, potassium bitartrate, methoxypolyethylene glycol, and hydrazine), use of toxic solvents (such as sodium dodecyl benzyl sulphate and polyvinyl pyrrolidone), and generation of toxic by-products arise via traditional roots (physical and chemical methods) of silver nanoparticle synthesis. With the advancement of science, the synthesis of metal nanoparticles finds alternative eco-friendly routes. Here, a variety of biological agents (plant extracts, bacterial and fungal derived compounds) are used to convert metal salts to nanoparticles. These agents act as in vitro reducing and capping agents. These strategies are used for the synthesis of different metal nanoparticles like silver, copper, iron, etc.28,29
Metal nanoparticle structure and surface modification: clue to enhance the antimicrobial activity
Nanotechnology offers production, manipulation and the use of materials below 100 nm range. Due to large surface area to volume ratio, nanoparticles offer novel properties leading to widespread use in different field of applications. But this high surface area often brings various drawbacks associated with the surface phenomena.30 When reducing the overall surface energy, individual nanoparticles interact with each other via chemical bonds and physical attraction forces at interfaces, resulting in agglomeration. This agglomeration strictly depends on the individual nanostructure size, less the dimension of nanostructures strong the van der Waals attraction forces between nano-structured materials. So to obtain the stable nanoparticle dispersions, this huge surface energy arises from the individual nanostructure should be taken care of. Here comes the concept of stabilizing agents. If a stabilizing agent acts as an antimicrobial agent, coining antimicrobial effect can be achieved effectively.To get the effective engineered nanoparticles, better understanding of nanoparticle structure is necessary. In general sense, nanoparticle structures consist of three layers: (i) core, (ii) shell, and (iii) surface.31 The core of the nanoparticle can be synthesized using various methods, reaction conditions and precursors whereas shell is added if necessary. But during the preparation of one dimensional nanoparticle from single salt, the shell is generally composed of the same material with the core. And last, but not at least, most important is surface, which can be functionalized depending upon the need. The need is either to prevent the agglomeration or to give specific function to nanoparticles. The surface functionalization of nanoparticles is due to transient van der Waals interactions of used compounds with the nanoparticle surface.
Green synthesis: biomolecule-nanoparticle organization
Reduction of metal complexes in dilute solutions is the general method in the synthesis of metal colloidal dispersions. Generally accepted reducing agents such as hydrazine hydrate and sodium borohydride are not preferred due to their tendency in in situ reducing and capping, resulting in additional effects of undesired toxicity impregnated with nanoparticles. Thus the use of green materials like plant extracts, microbial extracts, and algal extracts as reducing agents possesses great advantages (Fig. 1). In general, biological extracts are mixed with the metal salt solutions.32 Effect of nature and concentration of the plant extract and salt, temperature, pH, time, radiation, and hypothermia are also studied.33 Besides the most often used metal is silver, gold, copper, and iron are also used.30 Bio compounds (including alkaloids, phenolic compounds, terpenoids, enzymes, co-enzymes, proteins, and sugars etc.) reduce metal salts from positive oxidation state to zero oxidation state. Size and size distribution of metallic nanoparticles strongly depend on the bio compounds present in the extract. The presence of a strong reductant in the extract promotes a fast reaction rate and favours the formation of smaller nanoparticles. If the biomolecules rapidly reduce the salt leading to continuous formation of new nuclei or secondary nuclei, a narrow size distribution can be observed. However, a slow reaction may also occur if the secondary nucleation is suppressed over primary one. Bio molecules serve a secondary option to form a monolayer on the surface of nanoparticles to prevent the agglomeration. But recent studies on antibiotic mediated synthesis of nanoparticles confirm the surface functionalization of nanoparticles with antibiotics enhancing biocidal activity.34,35 Thus, enhanced antimicrobial activity of extract mediated nanoparticles are due to the biomolecules attached on the surface of nanoparticles.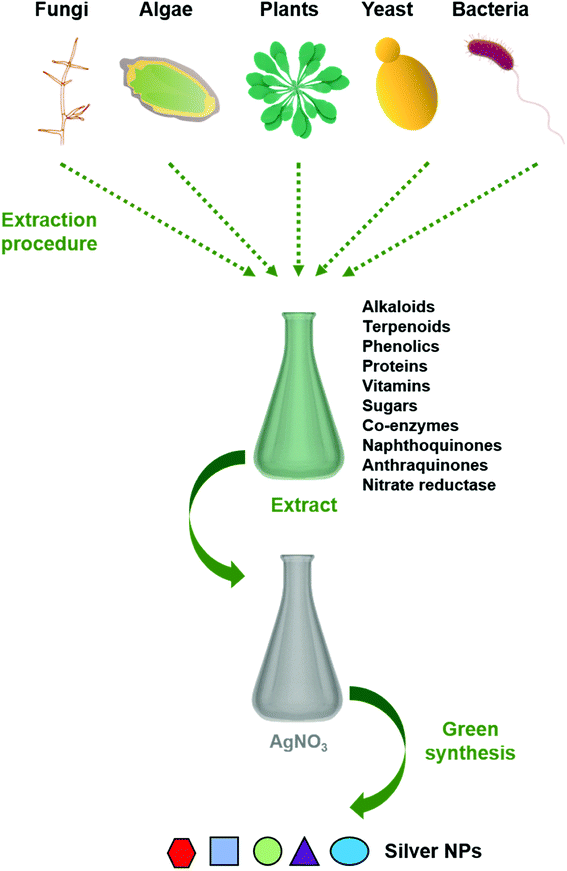 | ||
| Fig. 1 Schematic representation of the procedure for green synthesis of silver nanoparticles using various biological entities. | ||
Role of biological molecules in green nanoparticle synthesis and improving antimicrobial action
Bio molecules produced by living organisms (large macromolecules such as proteins, polysaccharides, lipids, and nucleic acids, as well as small molecules such as primary and secondary metabolites, and natural products) pose specific functions for the growth, development and sustaining in the environment. Biological molecules extracted from living systems show potential role and have become preferred choice in the synthesis of AgNPs, which are summarized in Table 1.| Biological entity | Size and morphology | Test microorganism | Method | Comment | Reference |
|---|---|---|---|---|---|
| Plants | |||||
| Elephantopus scaber | 37 nm; spherical | B. subtilis, L. lactis, P. fluorescens, P. aeruginosa, A. flavus, and A. penicillioides | Agar well diffusion method | 16–24 mm and 11–12 mm zones of inhibition against the tested bacterial and fungal strains, respectively | 36 |
| Green and black tea | 10–20 nm; spherical | Methicillin- and vancomycin-resistant S. aureus | Disk diffusion and broth dilution methods | 19–21 mm inhibition zones, and MIC = 8 μg ml−1 | 37 |
| Phyllanthus amarus | 30–42 nm; flower-like | E. coli, Pseudomonas spp., Bacillus spp., Staphylococcus spp., A. niger, A. flavus, and Penicillium spp. | Disk diffusion method | 6–11 mm and 5–8 mm zones of inhibition against the tested bacterial and fungal strains, respectively | 38 |
| Salvia leriifolia | 27 nm; spherical | P. aeruginosa, E. coli, C. freundii, E. aerogenes, A. baumannii, S. marcescens, K. pneumoniae, and S. pneumoniae | Disk diffusion method | 67.9% (P. aeruginosa), 76.5% (E. coli), 84.7% (E. aerogenes), 101.4% (A. baumannii), 25.3% (S. marcescens), 191.8% (K. pneumonia), and 141% (S. pneumoniae) inhibition levels | 39 |
| Psidium guajava | 20–35 nm; spherical | B. aryabhattai, B. megaterium, B. subtilis, A. creatinolyticus, E. coli, A. faecalis, S. cerevisiae, A. niger, and R. oryzae | Agar well diffusion method | 19–22 mm, 23–26 mm, 17–19 mm inhibition zones at 300 μg ml−1 concentration against Gram positive and negative bacteria, and fungi, respectively | 40 |
| Alpinia katsumadai | 12.6 nm; quasi-spherical | S. aureus, E. coli, and P. aeruginosa | Broth dilution method | MIC = 20 μg ml−1 for S. aureus and E. coli, and MIC = 40 μg ml−1 for P. aeruginosa. Inhibition of growth at 20 μg ml−1 for S. aureus and E. coli, and at 40 μg ml−1 for P. aeruginosa was 65%, 64% and 63% after 9 h incubation, respectively | 41 |
| Nelumbo nucifera | 12.9 nm; quasi-spherical | Streptococcus aureus, and P. aeruginosa | Broth dilution method | MIC = 10 μg ml−1 for both S. aureus and P. aeruginosa. The concentration of 10 μg ml−1 was found to be an effective bactericide, and no bacterial growth was observed within 10 h | 42 |
| Convolvulus arvensis | 28 nm; spherical | E. coli, S. aureus, and P. aeruginosa | Disc diffusion and broth macro-dilution methods | 17 mm zone of inhibition against E. coli at the highest dose. Growth of E. coli in the medium supplemented with bio-synthesized AgNPs was slowed down. Minimum biofilm eradication concentration was 20 μg ml−1 (S. aureus) and 50 μg ml−1 (P. aeruginosa) | 43 |
| Erythrina suberosa | 15–34 nm; spherical | B. subtilis, S. aureus, E. coli, P. aeruginosa, C. albicans, C. krusei, C. viswanathi, and T. mentagrophytes | Agar cup and broth micro-dilution methods | No zone of inhibition was observed against E. coli, B. subtilis, and C. viswanathi, whereas it was 16–24 mm against the rest of test microorganisms. Growth of inhibition was 16.27–99.26% and 36–82.27% against bacterial and fungal strains, respectively | 44 |
| Ocimum sanctum | 14.6 nm; spherical | E. coli | Disc diffusion and broth dilution methods | Antibacterial activity of AgNPs bio-synthesized using the plant extract and pure bioactive compound was compared. Similar zones of inhibition (10 mm) and MICs (150 μg ml−1) were observed | 45 |
| Cichorium intybus | Various sizes depending on the [metal]-to-[extract] ratio; spherical | E. coli, S. aureus, and P. aeruginosa | Disc diffusion and broth dilution methods | Antibacterial activity of AgNPs was tested against both reference strains (10–14 mm zones of inhibition and MICs = 4.7–8.4 pM) and clinical isolates (13–15 mm zones of inhibition and MICs = 4.4–5.2 pM) | 46 |
| Phlomis | 19–30 nm; spherical | E. coli, S. typhimurium, S. aureus, and B. cereus | Agar well diffusion method | 12–15 mm zones of inhibition against all test microorganisms | 47 |
| Artocarpus altilis | 25–43 nm; spherical | E. coli, S. aureus, and P. aeruginosa | Agar well diffusion method | 8–10 mm zones of inhibition were observed at the concentration of 100 μg ml−1 AgNPs | 48 |
| Pedalium murex | 10–150 nm; spherical | E. coli, K. pneumoniae, P. aeruginosa, M. flavus, B. subtilis, B. pumilus, and S. aureus | Disk diffusion method | Zones of inhibition at the highest concentration were 8.5–10.5 mm and 9–10 mm against the tested Gram negative and positive bacteria, respectively | 49 |
| Emblica officinalis | 15 nm; spherical | S. aureus, B. subtilis, E. coli, and K. pneumoniae | Disk diffusion method | Bio-synthesized AgNPs were more effective against Gram negative bacteria than Gram positive bacteria. 19–21 mm zones of inhibition were observed against Gram negative bacteria, whereas it was 15–17 mm against Gram positive bacteria | 50 |
| Eucalyptus globulus | 1.9–4.3 nm and 5–25 nm, with and without microwave treatment; spherical | ESBL producing, P. aeruginosa, E. coli, methicillin sensitive and resistant S. aureus | Agar well diffusion and broth dilution methods | 19–21 mm zones of inhibition, MICs = 27–36 μg ml−1, and MBCs = 30–42 μg ml−1 were observed against all test microorganisms. Additionally, ∼80% inhibition of biofilms formed by S. aureus and P. aeruginosa was observed at 30 μg ml−1 concentration | 51 |
| Musa paradisiaca | 23.7 nm; spherical | B. subtilis, S. aureus, P. aeruginosa, and E. coli | Agar well diffusion and broth dilution methods | MICs and MBCs of AgNPs were determined to be 1.7–6.8 μg ml−1 and 5.1–10.2 μg ml−1, respectively | 52 |
| Dalbergia spinosa | 18 nm; spherical | B. subtilis, S. aureus, P. aeruginosa, and E. coli | Disk diffusion and broth micro-dilution methods | 19–28 mm inhibition zones at 300 μg ml−1 concentration, and MIC = 11–18 μg ml−1 | 53 |
| Boerhaavia diffusa | 25 nm; spherical | Fish pathogens: A. hydrophila, P. fluorescens, and F. branchiophilum | Agar well diffusion method | MICs were determined to be 3.12 μg ml−1 (F. branchiophilum), 25 μg ml−1 (A. hydrophila) and 50 μg ml−1 (P. fluorescence) | 54 |
| Piper nigrum | 4–50 nm; hexagonal and spherical | Plant pathogens: C. freundii and E. cacticida | Agar well diffusion method | AgNPs bio-synthesized using leaf and stem extracts of the plant were compared. Both yielded similar zones of inhibition (8–9 mm) against the pathogens. AgNP-chloramphenicol combination further increased the activity | 55 |
| Mimusops elengi | 55–83 nm; spherical | MDR clinical isolates of K. pneumoniae, M. luteus and S. aureus | Disk diffusion method | Dose-dependent activity of AgNPs was observed. 11–18 mm zones of inhibition against all isolates at the highest concentration | 56 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Algae | |||||
| Green algae | |||||
| Ulva compressa; Cladophora glomerata | 66.3 nm and 81.8 nm; cubic | K. pneumoniae, P. aeruginosa, E. faecium, E. coli and S. aureus | Direct contact test | Biosynthesized AgNPs-polysulfone composite material suppressed the growth of all tested bacteria | 57 |
| Caulerpa serrulata | 10 nm; spherical–ellipsoidal | E. coli, S. aureus, Shigella sp., S. typhi, and P. aeruginosa | Agar well diffusion method | 14–21 mm zones of inhibition against all tested bacteria | 58 |
| Enteromorpha compressa | 4–24 nm; spherical | E. coli, P. aeruginosa, K. pneumoniae, S. aureus, S. paratyphi, A. terreus, A. flavus, A. ochraceus, A. niger, and F. moniliforme | Agar well diffusion method | 10.5–12 mm and 9.2–10.2 mm zones of inhibitions against bacterial and fungal strains, respectively | 59 |
| Spirogyra varians | 35 nm; quasi-spheres | S. aureus, B. cereus, S. enterica typhimurium, E. coli, L. monocytogenes, P. aeruginosa and Klebsiella | Agar well diffusion and broth macro-dilution methods | 11–20 mm zones of inhibition, MICs = 0.25–0.50 mg ml−1, and MBCs = 0.50–1 mg ml−1 | 60 |
| Caulerpa racemosa | 10 nm; spherical | S. aureus and P. mirabilis | Agar well diffusion method | Dose dependent antimicrobial activity was observed. 12–14 mm zones of inhibition at the highest concentration | 61 |
| Chlorella pyrenoidosa | 2–15 nm; spherical | K. pneumoniae, A. hydrophila, Acinetobacter sp., and S. aureus | Agar well diffusion method | The highest antibacterial activities were measured as 15–23% inhibition in the cases of Acinetobacter sp. and A. hydrophila, whereas K. pneumonia and S. aureus showed relatively lower inhibition | 62 |
| Enteromorpha flexuosa | 2–32 nm; spherical | B. subtilis, B. pumilus, E. faecalis, S. aureus, S. epidermidis, E. coli, P. aeruginosa, K. pneumoniae, A. niger, C. albicans, and S. cerevisiae | Disk diffusion and broth dilution methods | The antimicrobial activity of bio-synthesized AgNPs were compared to chemically synthesized AgNPs. Bio-synthesized AgNPs exhibited high antimicrobial activity (expect for P. aeruginosa and A. niger), which was slightly lower than chemically synthesized AgNPs | 63 |
| Brown algae | |||||
| Spatoglossum asperum | 22–60 nm; spherical | K. pneumoniae | Agar bioassay and disk diffusion methods | At 100 μg ml−1 concentration, 18 mm inhibition zone and complete inhibition of K. pneumoniae growth | 48 |
| Saccharina japonica | 14.77 nm; spherical | E. coli, P. aeruginosa, S. typhimurium, S. aureus, L. monocytogenes, B. cereus, C. albicans and A. brasiliensis | Agar well diffusion and broth dilution methods | High antimicrobial activity except for P. aeruginosa and A. brasiliensis. 14–22 mm zones of inhibition, MICs = 6–25 μl ml−1 and MBCs = 11–45 μl ml−1 | 64 |
| Ecklonia cava | 43 nm; spherical | E. coli and S. aureus | Agar well diffusion method and MTT assay | ∼12 mm zones of inhibition and EC50 = 15–16 μg ml−1 against the tested bacteria | 65 |
| Sargassum longifolium | N.A.; spherical and ellipsoidal | Clinical isolates of A. fumigatus, C. albicans, and Fusarium sp. | Disk diffusion method | Dose-dependent antifungal activity; 11–18 mm and 13–22 mm zones of inhibitions at the lowest and highest concentrations, respectively | 66 |
| Colpomenia sinuosa | 15–35 nm; spherical | E. coli and S. aureus | Tested on cotton fabrics | Cotton fabrics treated with bio-synthesized AgNPs exhibited high antimicrobial activity. However, it decreased by increasing number of washing cycles | 67 |
| Sargassum tenerrimum | 20 nm; spherical | B. cereus, E. coli, K. pneumoniae, P. mirabilis, P. aeruginosa, S. typhi, S. flexneri, S. aureus, and V. cholerae | Disk diffusion method | 14–18 mm zones of inhibition against all test microorganisms | 68 |
| Red algae | |||||
| Gelidium amansii | 27–54 nm; spherical | S. aureus, B. pumilus, E. coli, P. aeruginosa, V. parahaemolyticus, and A. hydrophila | Anti-microfouling assay | At the highest concentration, up to 99% reduction of microfouling | 69 |
| Spyridia fusiformis | 5–50 nm; spherical | E. coli, K. pneumoniae, S. aureus and P. aeruginosa | Disk diffusion method | At the highest concentration, AgNPs exhibited higher antimicrobial activity than the standard drug streptomycin; 21–26 mm zones of inhibition | 70 |
| Gracilaria birdiae | 20.2–94.9 nm; spherical | E. coli and S. aureus | Broth micro-dilution method | Effects of pH and extract concentrations were tested. At the optimum conditions, MICs were 34.3 μM and 81.2 μM for E. coli and S. aureus, respectively | 71 |
| Acanthophora spicifera | 33–81 nm; cubic | S. aureus, B. subtilis, Salmonella sp., E. coli, and C. albicans | Disk diffusion method | 18.75–23.5 mm zones of inhibition | 72 |
| Gracilaria crassa | 60–200 nm; spherical | E. coli, P. mirabilis, B. subtilis, and P. aeruginosa | Agar well diffusion method | 20–40 mm zones of inhibition; high antibacterial activity against E. coli and P. mirabilis when compared with streptomycin sulphate | 73 |
| Gracilaria corticata | 18–46 nm; spherical | C. albicans and C. glabrata | Disk diffusion method | 12 and 11 mm zones of inhibition against C. albicans and C. glabrata, respectively | 54 |
| Pterocladia capillacea and Jania rubens | 7–18 nm and 5–20 nm; spherical | E. coli and S. aureus | Tested on cotton fabrics | Cotton fabrics treated with bio-synthesized AgNPs exhibited high antimicrobial activity. However, it decreased by increasing number of washing cycles | 74 |
| Porphyra vietnamensis | 13 nm; spherical | E. coli and S. aureus | Broth dilution method | 100% and 60% inhibition of growth at 5 μg ml−1 and 15 μg ml−1 for E. coli and S. aureus, respectively | 75 |
| Blue-green algae | |||||
| Nostoc sp. | 51–100 nm; spherical | R. solanacearum, X. campestris, A. niger, and T. harzianum | Agar well diffusion method | 23–25 mm zones of inhibition against the bacterial strains, poor antifungal activity | 76 |
| Synechococcus sp. | 140 nm; spherical | B. subtilis, S. aureus, and E. coli | Growth kinetic monitoring | In the presence of bio-synthesized AgNPs, growth rates of the test strains were significantly reduced | 77 |
| Anabaena sp. | 24.13 nm; irregular | B. megaterium, E. coli, B. subtilis, S. aureus, P. aeruginosa, and M. luteus | Agar well diffusion method | 16–20 mm and 16–22 mm zones of inhibitions against tested Gram positive and negative bacteria, respectively | 78 |
| Limnothrix sp. | 31.86 nm; elongated | B. megaterium, E. coli, B. subtilis, S. aureus, P. aeruginosa, and M. luteus | Agar well diffusion method | No antimicrobial activity | 78 |
| Synechocystis sp. 48-3 | 14.64 nm; irregular | B. megaterium, E. coli, B. subtilis, S. aureus, P. aeruginosa, and M. luteus | Agar well diffusion method | 14–18 mm and 15–20 mm zones of inhibitions against tested Gram positive and negative bacteria, respectively | 78 |
| Anabaena doliolum | 10–50 nm; spherical | Clinical isolates of MDR K. pneumoniae, E. coli, and S. aureus | Disk diffusion method | Dose-dependent antimicrobial activity; 17–18 mm and 33–36 mm zones of inhibitions at the lowest and highest concentrations, respectively | 79 |
| Nostoc commune | 15–54 nm; spherical | E. coli and grains-borne fungi | Broth dilution method | MIC = 0.012 mg ml−1 and MBC = 0.016 mg ml−1 for E. coli. The antifungal activity was evaluated by sterilizing various grain seeds with bio-synthesized AgNPs prior to cultivation. Fungi formation was significantly decreased on AgNPs-treated seeds when compared to the standard chemical treatment | 80 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Fungi and yeasts | |||||
| Penicillium polonicum | 10–15 nm; spherical | MDR Acinetobacter baumannii | Agar well diffusion and broth micro-dilution methods | 21.2 mm zone of inhibition, MIC = 15.62 μg ml−1, and MBC = 31.24 μg ml−1 | 81 |
| Raphanus sativus | 4–30 nm; spherical | Methicillin resistant B. subtilis, S. aureus, E. coli, and S. marcescens | Disk diffusion method | More effective against tested Gram negative strains; at the highest concentration, 12–23 mm and 34–36 mm zones of inhibition against Gram positive and negative strains, respectively | 82 |
| Penicillium aculeatum | 4–55 nm; spherical | E. coli, P. aeruginosa, S. aureus, B. subtilis, and C. albicans | Disk diffusion method | 12–16 mm and 18 mm zones of inhibition against bacterial and fungal strains, respectively | 83 |
| Pleurotus ostreatus | 10–40 nm; spherical | B. subtilis, B. cereus, S. aureus, E. coli, and P. aeruginosa | Disk diffusion and broth micro-dilution methods | Bio-synthesized AgNPs lower than 100 μg did not exhibit any antimicrobial activity. 11–14 mm zones of inhibition at the highest concentration, MICs = 13.21–18.96 μg ml−1 | 84 |
| Candida lusitaniae | 2–83 nm; variable | S. aureus, and K. pneumoniae | Broth micro-dilution assay | At 50 μg ml−1 bio-synthesized Ag/AgCl NP concentration, 83–85% inhibition of growth was observed for the test strains | 85 |
| Cryptococcus laurentii | 35–400 nm; spherical | Phytopathogenic fungal strains: B. cinerea, P. expansum, A. niger, Alternaria sp., and Rhizopus sp. | Agar well diffusion and broth micro-dilution methods | 7.9–11.4 mm zones of inhibition, and MICs = 4 mg l−1 against all tested fungal strains | 86 |
| Rhodotorula glutinis | 15–220 nm; spherical | Phytopathogenic fungal strains: B. cinerea, P. expansum, A. niger, Alternaria sp., and Rhizopus sp. | Agar well diffusion and broth micro-dilution methods | 9.7–15.1 mm zones of inhibition, and MICs = 2 mg l−1 against all tested fungal strains | 86 |
| Arthroderma fulvum | 15.5 nm; spherical | Candida spp., Aspergillus spp., and Fusarium spp. | Broth micro-dilution method | High antifungal activity, especially against Candida spp. MICs were 0.125–0.500 μg ml−1, 1–2 μg ml−1, and 2–4 μg ml−1 for Candida spp., Aspergillus spp., and Fusarium spp., respectively | 87 |
| Ganoderma applanatum | 133 nm; spherical | E. coli, B. subtilis, S. epidermidis, V. cholerae, S. aureus, and S. flexneri | Agar well diffusion method | 15–19 mm and 26–28 mm zones of inhibition against the tested Gram negative and positive bacteria, respectively | 88 |
| Saccharomyces cerevisiae | 5–50 nm; spherical | Fluconazole susceptible and resistant C. albicans strains | Broth micro-dilution method | MICs for both susceptible and resistant strains were in the range of 2–4 μg ml−1 | 89 |
| Guignardia mangiferae | 5–30 nm; spherical | E. coli, P. mirabilis, K. pneumoniae, P. aeruginosa, S. aureus, S. epidermidis, E. faecalis, and B. subtilis | Agar well diffusion and broth micro-dilution methods | Various pH conditions (4–10) were used to optimize the bio-synthesis of AgNPs with high antimicrobial activity. At optimum pH of 7.0, zones of inhibitions and MICs were 6–13 mm and 3.12–12.5 μg ml−1 against all test strains, respectively | 90 |
| Curvularia lunata | 64.3 nm; spherical | E. coli, P. aeruginosa, S. paratyphi, B. subtilis, S. aureus, and B. cereus | Disk diffusion assay | The antibacterial activity of standard drugs was enhanced in the presence of bio-synthesized AgNPs. The highest fold increase was observed for erythromycin and carbenicillin against E. coli, ampicillin against S. paratyphi, erythromycin against B. subtilis | 90 |
| Penicillium sp. | 25 nm; spherical | E. coli, P. aeruginosa, K. pneumoniae, S. typhimurium, and E. aerogenes | Agar well diffusion method | 13–21 mm zones of inhibition against all test strains | 91 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Bacteria | |||||
| Phenerochaete chrysosporium | 34–90 nm; spherical and oval | P. aeruginosa, K. pneumoniae, S. aureus, and S. epidermidis | Agar well diffusion method | More effective against Gram negative bacteria; 11–13 mm and 17–18 mm zones of inhibition in the cases of Gram positive and negative bacteria, respectively | 92 |
| Bacillus endophyticus | 4.8–6.6 nm; spherical | C. albicans, E. coli, S. typhi, and S. aureus | Agar well diffusion method | 11–19 mm and 15 mm zones of inhibitions against the tested bacterial strains and fungal strain, respectively | 93 |
| Acinetobacter baumannii | 37–168 nm; spherical | MDR E. coli, P. aeruginosa, and K. pneumoniae | Broth micro-dilution method | MICs = 1.56–3.125 μg ml−1 against all tested strains; additionally, bio-synthesized AgNPs reduced the bacterial attachment at the rate of 66–86% | 94 |
| Weissella oryzae | 150 nm; spherical | V. parahaemolyticus, B. cereus, B. anthracis, S. aureus, E. coli, and C. albicans | Disk diffusion method | 12–26 mm and 25 mm zones of inhibitions against the tested bacterial strains and fungal strain, respectively. Additionally, 5–6 μg of bio-synthesized AgNPs significantly inhibited the biofilm formation | 95 |
| Bacillus thuringiensis | 10–30 nm; spherical | E. coli | Live/dead bacterial viability fluorescence assay | Viability of the bacterium was reduced to 45% in the presence of 9.2 μg ml−1 bio-synthesized AgNPs | 96 |
| Pseudomonas deceptionensis | 127 nm; spherical | S. aureus, S. enterica, V. parahaemolyticus, C. albicans, and B. anthracis | Agar well diffusion method | The antimicrobial activity was maximum for V. parahaemolyticus, followed by C. albicans, S. aureus, S. enterica, and B. anthracis | 97 |
| Ochrobactrum anthropi | 38–85 nm; spherical | S. typhi, S. paratyphi, V. cholera, and S. aureus | Agar well diffusion method | 14–15 mm zones of inhibition against all test strains | 98 |
| Serratia nematodiphila | 10–31 nm; spherical | B. subtilis, K. planticola, and P. aeruginosa | Agar well diffusion method | Dose dependent antimicrobial activity; 10–14 mm and 13–18 mm zones of inhibition against all test strains at the lowest and highest concentrations, respectively | 99 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Actinomycetes | |||||
| Streptomyces parvulus | 100 nm; spherical | E. coli, S. aureus, M. smegmatis, C. albicans, C. atlantica, C. valderwaltii, and methicillin-resistant S. aureus | Agar well diffusion and broth micro-dilution methods | 20–34 mm and 14–24 mm zones of inhibition against test bacterial and fungal strains respectively. MICs were determined to be 1.95 μM (MRSA), 1.95 μM (S. aureus), and 31.25 μM (E. coli) | 100 |
| Streptomyces seoulensis | 121 nm; spherical | E. coli, S. aureus, M. smegmatis, C. albicans, C. atlantica, C. valderwaltii, and methicillin-resistant S. aureus | Agar well diffusion and broth micro-dilution methods | No antimicrobial activity against M. smegmatis and C. albicans; 13–23 mm zones of inhibition against the other strains. MICs were determined to be 250 μM (MRSA), 250 μM (S. aureus), and 62.5 μM (E. coli) | 100 |
| Streptomyces owasiensis | 160 nm; spherical | E. coli, S. aureus, M. smegmatis, C. albicans, C. atlantica, C. valderwaltii, and methicillin-resistant S. aureus | Agar well diffusion and broth micro-dilution methods | 15–29 mm and 15–28 mm zones of inhibition against test bacterial and fungal strains respectively. MICs were determined to be 125 μM (MRSA), 62.5 μM (S. aureus), and 125 μM (E. coli) | 100 |
| Streptacidiphilus durhamensis | 8–48 nm; spherical | S. aureus, B. subtilis, E. coli, P. aeruginosa, K. pneumoniae, P. mirabilis | Disk diffusion and broth micro-dilution methods | 6–10 mm zones of inhibitions against the test strains (except for S. infantis); MICs = 6.25–50 μg ml−1, MBCs = 50–100 μg ml−1. The synergistic effect of bio-synthesized AgNPs with various standard drugs was also observed | 101 |
| Streptomyces rochei | 22–85 nm; spherical | B. subtilis, S. aureus, P. aeruginosa, B. cereus, S. typhimurium, E. coli, V. fluvialis, and V. damsela | Agar well diffusion method | Bio-synthesized AgNPs by one of the isolates exhibited the maximum zones of inhibition (16–19 mm) against all tested microorganisms. Synergistic effect of AgNPs with standard drugs was also observed | 102 |
| Streptomyces sp. | 20–45 nm; various | C. albicans, Candida tropicalis and Candida krusei | Agar well diffusion method | 16–20 mm zones of inhibition against all test strains | 103 |
| Nocardiopsis sp. | 30–90 nm; spherical | E. coli, B. subtilis, E. hirae, P. aeruginosa, S. flexneri, S. aureus, A. niger, A. brasiliensis, A. fumigates, and C. albicans | Agar well diffusion and broth micro-dilution methods | 22–33 mm and 23–28 mm zones of inhibition, MICs = 7–18 μg ml−1 and 13–18 μg ml−1 against tested bacterial and fungal strains, respectively | 104 |
| Streptomyces albidoflavus | 10–40 nm; spherical | B. subtilis, M. luteus, E. coli, and K. pneumoniae | Agar well diffusion method | The highest zone of inhibition (36 mm) was observed in the case of K. pneumoniae. 22–28 mm zones of inhibition against the rest of test strains | 105 |
Most frequently used bio molecules originate from plant extracts.32,106,107 Apart from the plant extract, bio molecules from microbial (bacteria, fungi, yeasts and actinomycetes), algal and cyanobacterial origin have also been successfully utilized for the synthesis of nanoparticles.33,34,108,109 These biological molecules extracted from different sources are externally added to metal salts to form nanoparticles. To synthesize AgNPs, biological extract in situ reduces silver salts (Ag+) to metallic silver, Ag0. In the process of nanoparticle synthesis in a green way, biological molecules not only reduce the metal salts but also cover the formed nanoparticles or acts as in situ reducing and capping agent. This capping is advantageous over as it acts as multifunctional way; (i) prevents the agglomeration of the nanoparticles, (ii) reduces the toxicity,35,110 and (iii) improves antimicrobial action.3–5,106 If these coating agents themselves show antimicrobial action, a synergistic effect of metal nanoparticles and the capped bio molecules may be found. Here AgNPs offer action due to its size and bio molecule attached over nanoparticle surface for its own antimicrobial action. This is the possible explanation of low antimicrobial action of biologically synthesized nanoparticles in many cases. So, for synthesizing silver nanoparticles in a greenway, research is focusing to synthesize AgNPs using bio molecules which have antimicrobial action of their own.
Synthesis of silver nanoparticles by microbes
Microbes are being used as a promising biological source for synthesizing metal nanoparticles. But all the organisms do not show ability to convert metals to its nano forms. Microbial synthesis of silver nanoparticles can be achieved either by intracellular scheme or extracellular scheme.111 In intracellular scheme, an accumulation of silver inside the cell nucleates the silver nanoparticle synthesis and proceeds with the microbial growth. Nanoparticles with live cells are harvested after optimum time of bacterial growth. The harvested cells, thus, require special treatment to release the synthesized nanoparticles. In extracellular synthesis process, extracellular secretes of the bacterial populations are separated and used for the synthesis. Microorganisms generally show resistance to silver ions are generally tend to produce AgNPs effectively. Resistance mechanism of individual microbes also differs with organisms. Bacillus licheniformis use the synthesis of silver nanoparticles at 1 mM concentration without undergoing cell death. Increasing the concentration (crosses the threshold level) to 10 mM, the organism undergoes cell death within minutes.107,108,112,113 A silver resistant bacterium screened from silver mine accumulated AgNPs (size 35–46 nm) in their periplasmic space.109,114 Reduction of Ag+ ions is proposed to come from the combinations of bio molecules (enzymes/proteins, amino acids, polysaccharides, and vitamins etc.) found in these extracts. But the exact mechanism is not till understood. As shown in Fig. 2, mostly accepted mechanism is based on the presence of enzyme “nitrate reductase”.107,109,110,112,115 The enzyme converts nitrate (NO3−) to nitrite (NO2−). During this reduction process where NO3− is converted into NO2−, the electron is transferred to the silver ion (Ag+) which then reduced to metallic silver (Ag0)3,114 showed direct evidence of it by using purified nitrate reductase from the organism Fusarium oxysporum for synthesizing silver nanoparticles. Besides, functional groups on bacterial cell wall also cause the reduction of Ag+ to Ag0 without the presence of nitrate reductase.112 Apart from bacteria, fungi can also be used for the synthesis of nanoparticles and have advantageous over productivity due to the huge presence of enzymes, proteins, and reducing components on its cell surface.113,116 Fungi can trap Ag+ onto their cell surfaces causing subsequent reduction using enzymes (like naphthoquinones and anthraquinones) of fungal systems.114,115,117,118 The use of yeast in synthesizing silver nanoparticles did not get success. Although a very few reports are also available. Formation of silver nanoparticles is thought to be due to the combinational effect of bio molecules found in extract. This is also true for the case of algae. Presence of enzymes serves actinomycetes to synthesize silver nanoparticles. Reduction of metal ions is thought to be occurred via intracellular interactions.117,119–122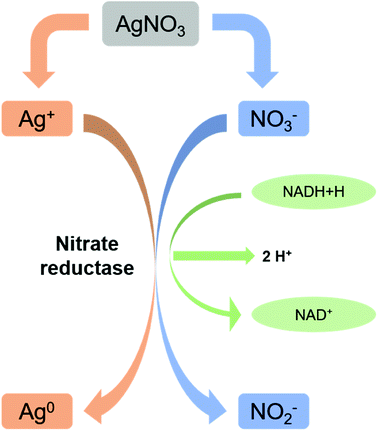 | ||
| Fig. 2 Nitrate reductase mediated synthesis of AgNPs (this figure has been adapted from ref. 119 with permission from Springer). | ||
Synthesis of silver nanoparticles by plants
Slow rate of nanoparticle synthesis using microbes makes plant extracts as preferred choice due to its simplicity, efficiency and viability. The ability of plants to accumulate and detoxify heavy-metals is well proved.118,123 Generally it is used as bio reductant in the process of synthesizing silver nanoparticles. A plant extract contains a number of metabolites and reductive biomolecules responsible for the reduction of metal ions. This includes terpenoids, flavones, ketones, aldehydes, amides, carboxylic acids, carbohydrates, proteins, and vitamins.124 Among many possible natural products, polysaccharides represent an excellent scaffold for this purpose.125 The use of polysaccharides like starch and chitosan for the synthesis of AgNPs has been reported in recent years. Hydroxypropyl starch is used for in situ reduction and capping for the synthesis of stable silver nanoparticles of 6 to 8 nm diameter. No aggregation was observed for more than 6 months due to its effective capping.126,127 Diaminopyridinyl (DAP)-derivatized heparin, a linear acidic mammalian polysaccharide has been used in the preparation of AgNPs.128 The use of hyaluronic acid, a high molecular weight polysaccharide for the synthesis of AgNPs and gold nanoparticles (AuNPs) was also reported.129 Synthesis of AuNPs using (1→3)-, (1→6)-α, β-D-glucan of an edible mushroom130 and a gum polysaccharide of Cochlospermum religiosum (katira gum)131 have been reported.Among the different strategies of green synthesis, plants extracts mediated synthesis has further practical applications. This method is comparatively faster than other biogenic route and the availability of raw material is cheap and sustainable. Apart from this, use of antimicrobial plant extract as in situ reducing and capping agent, helps the development of engineered nanoparticles with enhanced antimicrobial activity. The antimicrobial action of the nanoparticles often depends upon its size. This size is well protected by the action of capping agent. Now interestingly, if the capping agents itself have antimicrobial activity, it might offer enhanced antimicrobial action. Plants with antimicrobial action can be thus successfully used to develop silver nanoparticles with enhanced antimicrobial action.
Factors affecting the antimicrobial activity of silver nanoparticles
In the recent years, silver nanoparticles have gained much attention due to their unique antimicrobial properties. They are considered as a novel and potential alternative to standard antibiotic drugs since they have a great potential against the increasing multidrug resistance in pathogenic bacteria and fungi. Development and synthesis of these nanomaterials for use as an alternate antibiotic therapy straightly depend on some physical and chemical properties such as their size, shape, concentration and zeta potential.132,133 Therefore, the properties of nanoparticles should be considered and given more attention when designing antimicrobial AgNPs.One of the most important physico-chemical properties that affects the antimicrobial activity is the size. Typically, smaller nanoparticles have relatively increased stability and enhanced antimicrobial activity. This is due to the larger surface area of smaller nanoparticles, which provides a higher interaction area and ascendant intracellular penetration.22,134 In a broad sense, the size of nanoparticles should be lower than 50 nm in order to have an effective antimicrobial activity. In particular, nanoparticles in the range of 10–15 nm sizes have superior antimicrobial activity.135 Panáček et al. synthesized a wide range of AgNPs with narrow size distributions via reduction of Ag(NH3)2+ complex cation by four saccharides, namely glucose, galactose, maltose, and lactose.136 The colloidal AgNPs synthesized by using maltose and lactose had higher antibacterial and bactericidal activity against tested Gram-positive and Gram-negative bacteria than those synthesized by using glucose and galactose. This might be directly related to nanoparticle sizes, since the AgNP sizes were smaller for disaccharides (maltose and lactose) than monosaccharides (glucose and galactose). AgNPs synthesized by using maltose had the smallest size distribution and the highest antimicrobial activity. In contrast, the largest AgNPs, 50 nm, synthesized by using galactose exhibited the lowest antimicrobial activity. In another study, Li et al. showed that the antibacterial activity of AgNPs decreased with increase of the nanoparticle size.137 The highest antibacterial effect was observed in the case of smallest nanoparticles, 5 nm, in which attachment of the nanoparticles onto the cell membrane led to membrane damage, increased membrane permeability and finally cell death. To this end, Ivask et al. tested toxic effect AgNPs with different sizes on various types of organisms and cells.138 It was concluded that AgNPs in the range of 20–80 nm were toxic due to release of Ag+ ions. On the other hand, 10 nm and smaller nanoparticles were more toxic due to more efficient cell–particle interaction leading to higher intracellular bioavailability of AgNPs.
Shape is also a crucial physico-chemical property regarding the antimicrobial activity. AgNPs with different shapes have been shown to exhibit different antimicrobial activity due to displaying various degrees of interaction with the cell membrane and hence causing the membrane damage. In this context, the antimicrobial activity of AgNPs with different shapes has been compared. El-Zahry et al. compared the antibacterial activity of spherical, triangular and hexagonal shaped AgNPs against Gram negative bacterium E. coli by using conventional disk assay and surface enhanced Raman spectroscopy (SERS).139 Both methods demonstrated that hexagonal AgNPs exhibited the highest antibacterial activity whereas triangular AgNPs displayed no activity. Recent research revealed that the antimicrobial activity is highest in the case of truncated triangular AgNPs or similar geometries such as hexagonal and octahedral shapes.140–144 Conversely, some researchers reported that the shape of AgNPs does not have any effect on the antimicrobial activity. Actis et al. tested spherical, triangular and cubic AgNPs against methicillin-susceptible and -resistant S. aureus. Surprisingly, the nanoparticle shape had no significant effect on the antibacterial activity.145 The bacterial viability was completely reduced at the highest nanoparticle concentration, whereas the difference between different shaped AgNPs at lower concentrations was not remarkable. Despite many studies proposed the shape dependent antimicrobial susceptibility of AgNPs, none of them provided a mechanistic basis, therefore exact mechanism of the shape dependent antimicrobial activity is still unknown.
Another important factor is the nanoparticle concentration.146,147 This property is directly correlated to microbial species.56,79 Kim et al. reported that the antibacterial activity of AgNPs was more effective against E. coli than against S. aureus.148 The growth of E. coli was inhibited at lower nanoparticle concentrations, while the inhibitory effect of same concentration on S. aureus was inadequate. Pazos-Ortiz et al. reported the dose-dependent antimicrobial activity of a composite of AgNPs embedded in poly-epsilon-caprolactone nanofibers and tested against both Gram-positive and Gram-negative bacteria.149 It was found that beginning from the 12.5 mM nanoparticle concentration, AgNPs gradually displayed a significant antibacterial effect on the different bacterial strains. Similarly, Gurunathan et al. demonstrated the dose-dependent bactericidal activity of AgNPs at concentrations ranging from 0.1 to 1.0 μg ml−1 against two Gram-negative and two Gram-positive bacterial strains.22 Consistently, the cell viability was reduced as the AgNP concentrations increased, and no growth was observed at MIC values for each strain. In the case of Gram-negative bacteria, 0.5–0.6 μg ml−1 caused nearly 95% reduction in the bacterial population. However, same concentrations of AgNPs led to only 50% reduction in the Gram-positive bacterial population (Fig. 3). Therefore, the bactericidal concentration is specific for each bacterial strain.
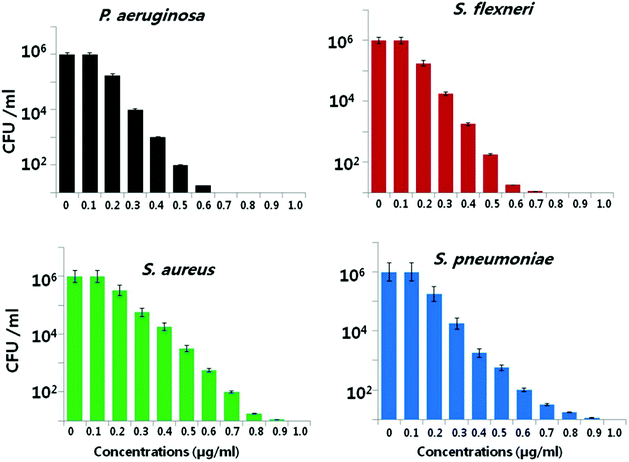 | ||
| Fig. 3 Bactericidal effect of green synthesized AgNPs on different bacterial strains. Dose-dependent activity of AgNPs synthesized using Allophylus cobbe leaves. The bacterial strains were incubated at various AgNP concentrations ranging from 0.1 to 1.0 μg ml−1 and bacterial rate survival was estimated by colony forming unit (CFU) assay at 4 h (this figure has been adapted from ref. 22 with permission from Springer). | ||
Zeta potential is another physico-chemical property that has an effect on antimicrobial activity since the interaction between nanoparticles and the cell membrane is based on electrostatic adhesion.150–152 In their study, El Badawy et al. synthesized AgNPs having various zeta potentials and investigated the antibacterial activity against Bacillus species.153 They found a direct correlation between the nanoparticle surface charge and antibacterial activity. Gram-positive Bacillus strains were more susceptible to positive AgNPs, and the more negative AgNPs were the least toxic. Bacterial resistance to the negative AgNPs was estimated to be caused by the repulsion between negatively charged functional groups of biomolecules located on the cell surface and negatively charged nanoparticle surface. Similarly, Ashraf et al. synthesized positively charged and polyhexamethylene biguanide (PHMB) functionalized AgNPs.154 The cationic AgNPs exhibited superior bacteriostatic and bactericidal against E. coli. The bactericidal activity of PHMB was around 10 μg ml−1; but when used in combination with AgNPs, bacteriostatic and bactericidal activities were lowered down to 0.075 and 0.150 μg ml−1, respectively. In this regard, environmental conditions have a significant influence on the nanoparticle activity and stability.155 At low pH, the nanoparticle surfaces are positively charged, which favours the interaction between the nanoparticle surface and the negatively charged groups of the bacterial cell surface.156 Peretyazhko et al. showed the oxidative dissolution of AgNPs through the interaction of Ag+ with dissolved oxygen and protons, and solubility of AgNPs was affected by the environment which was higher in acetic acid than in the neutral water.157 Such an alteration in aquatic chemistry might promote release of Ag+ from the nanoparticles and hence lead to increased antimicrobial activity.40 Additionally, Maillard et al. conducted biophysical analysis including surface pressure, zeta-potential and DLS measurements to study interaction of AgNPs with lipid membranes.158 Surprisingly, negatively charged green synthesized AgNPs could have interacted with the membrane via electrostatic attraction with the polar heads of the lipids, and displayed higher antibacterial activity as compared to AgNPs synthesized using citrate. This was presumed to be due to aromatic and hydrophobic moieties of capping agents used during green synthesis procedure, indicating the advantages of green chemistry on the antimicrobial activity of silver nanoparticles.
The reducing and capping materials of the green synthesized AgNPs are derived from the biological extracts containing various naturally occurring compounds and biomolecules such as alkaloids, terpenoids, phenolics, vitamins, co-enzymes, carbohydrates, enzymes and proteins. Therefore, these molecules increase the probability of attachment and action of AgNPs on the microbial cells.
Mechanism of antimicrobial action
Although antimicrobial activity of AgNPs alone or in combination with standard antibiotic drugs has been proved against a wide range of microorganisms including Gram-negative and Gram-positive bacteria, and fungi, still a little is known about the precise mechanism of their mode of antimicrobial action. Nevertheless, there have been extensive research in order to elucidate their mode of action, and three well-defined mechanisms have been proposed so far: (i) cell wall and membrane damage, (ii) intracellular penetration and damage, and (iii) oxidative stress159–161 (Fig. 4).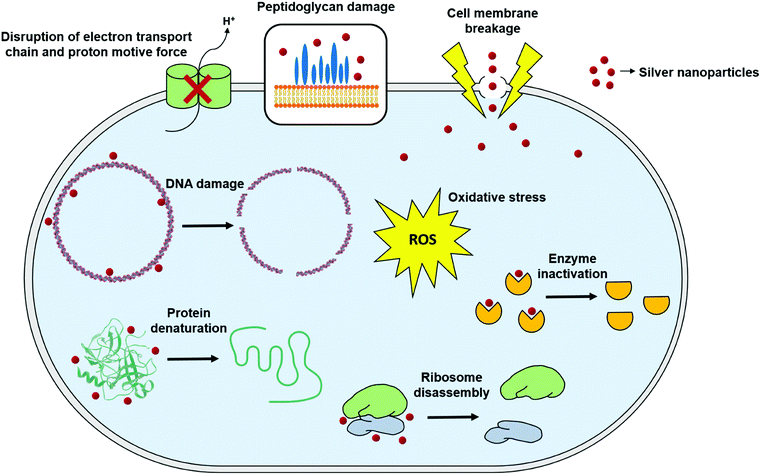 | ||
| Fig. 4 General mechanisms for antimicrobial mode of action of silver nanoparticles (this figure has been adapted from ref. 159 with permission from Frontiers). | ||
Cell wall and membrane damage
Components of the cell wall and membrane exert different adhesion pathways for the nanoparticles. One of the functions of the cell wall and membrane is to protect the microorganism against the environmental threats and to maintain homeostasis while still permitting the transport of nutrients inside the cell. Bacterial classification is based on the differences of cell wall structures; Gram-negative cell wall, also known as cell envelope, contains at least two layers of lipopolysaccharides whereas Gram-positive cell wall is typically thicker and primarily composed of a single type of molecule, peptidoglycans.162,163 Most of AgNPs exhibit higher antibacterial activity against Gram-negative bacteria than Gram-positive bacteria, since the peptidoglycan layer, made of glycan strands cross-linked by short peptides and anionic glycopolymers called teichoic acid, in the Gram positive bacterial cell wall provides a natural barrier which prevents penetration of the nanoparticles. On the other hand, Gram-negative bacteria have thinner cell wall and less peptidoglycan. Despite the key component of the Gram-negative bacterial cell wall, lipopolysaccharides (LPS), has a significant contribution to durability and integrity of the membrane, its negative charge facilitates adhesion of nanoparticles.7,62,73,143,164–166 The interaction between AgNPs and microorganisms starts with adhesion of AgNPs to the microbial cell wall and membrane, which is based on electrostatic attraction between the negatively charged microbial cell membrane and positively or less negatively charged AgNPs.167 The interaction of AgNPs with bacterial cells is also shown by a significant decrease in zeta potential of cell surface in both Gram-positive and Gram-negative bacteria.142,168 Upon such an attraction and interaction, morphological changes in the membrane structure are triggered by the nanoparticle, and thereby leading to disruption of membrane permeability and respiratory functions via membrane depolarization, and finally disruption of the cell integrity and cell death.169–172 As a result of the increased membrane permeability and disruption of the cell wall, the cellular content including proteins, enzymes, DNA, ions, metabolites and energy reservoir also leaks into the environment.48,137,142,169,173 Therefore, disintegration of the cell wall by the nanoparticle adhesion is assumed to be primary mechanism of the antimicrobial action.174,175In addition, AgNPs have been shown to cause irregular pit formations on the cell wall, which allowing the nanoparticles enter the periplasmic space and finally inside the cell176 (Fig. 5). After a brief contact of AgNPs with bacterial cells, peripheral damages and dense pits on the cell surface can be visualized by advanced imaging techniques such as transmission electron microscopy (TEM), scanning electron microscopy (SEM) and atomic force microscopy (AFM).101,164,175 Gopinath et al. investigated the structural changes in bacterial cell morphology caused by AgNPs by field emission scanning electron microscopy (FE-SEM).164 The untreated bacterial cells showed smooth and undamaged structures (Fig. 6A–D), whereas the cells subjected to 60 min of AgNP treatment were drastically damaged, lost their membrane integrity and the original morphological structures (Fig. 6E–H). They also examined the leakage of intracellular content from the AgNP treated bacterial cells using FITC, a fluorescent dye used as a marker for labelling proteins. Supernatants obtained from AgNP treated bacterial cell cultures and stained with FITC displayed fluorescent signal, indicating the protein leakage from the cell. Similarly, Ansari and Alzohairy evaluated morphological and ultra-structural changes of MRSA cell by SEM and TEM.174 They found that untreated MRSA cells were round-shaped and had smooth and intact cell walls. However, the cells were wrinkled and damaged upon AgNP treatment, indicating the disruption and disorganization of cell membrane and wall. TEM imaging further revealed that large translucent zones formed in the cytoplasm caused by local or complete disassociation of the cell membrane and the cell wall. As a result of the leakage of intracellular content, the cell membrane shrink occurred and led to death. Ahmad et al. also showed the physico-chemical and morphological changes in AgNP-treated cells by zeta potential and SEM analyses.142 Further, they investigated whether the intracellular content of cells would leak into the environment due to the AgNP-induced cell wall and membrane damage. For this, after 4 h of treatment, the cells were harvested by centrifugation, and leakage of nucleic acids and proteins into the medium was determined by measuring absorbance of the supernatant at 260 and 280 nm. With the increasing concentrations of AgNPs, absorbance at 260 and 280 nm also increased whereas absorbance of the control cells was zero.
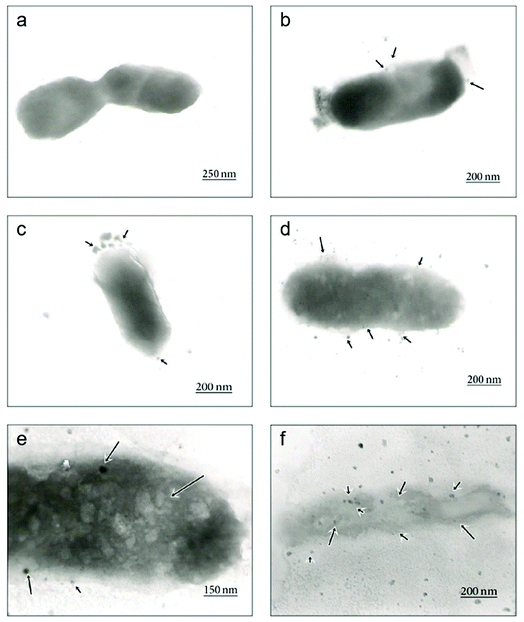 | ||
| Fig. 5 Time-dependent TEM images of E. coli cells treated with biosynthesized AgNPs using Aspergillus niger extract. (a) The untreated cell; (b) adhesion of AgNPs on the E. coli cell wall at 1 h; (c) disruption of the cell membrane by formation of pits and gaps at 5 h; (d) complete disruption of the cell wall and membrane; (e) penetration of AgNPs into the E. coli cell at 8 h; (f) disintegration of the cell and cell lysis at 12 h (this figure has been adapted from ref. 176 with permission from Hindawi). | ||
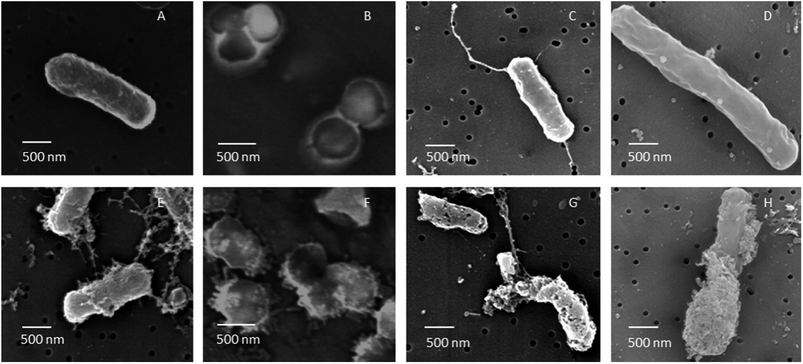 | ||
| Fig. 6 FE-SEM images of B. cereus, S. aureus, E. coli and P. aeruginosa cells untreated (A–D) and treated (E–H) with AgNPs. After 60 min of treatment with AgNPs biosynthesized using soil derived Pseudomonas putida, all cells were subjected to severe membrane damage (this figure has been adapted from ref. 164 with permission from Elsevier). | ||
Furthermore, it was proposed that AgNPs could interact with protein parts of the outer membrane, constitute complexes with oxygen, phosphorous, nitrogen or sulphur atom containing electron donors, and cause irreversibly changes in the cell wall structure.177 The interaction of protein thiol groups with silver ions and AgNPs is well-defined, and thiol groups in the enzymes of bacterial respiratory chain have been shown to be possible sites for Ag+ binding.118,178–180 In the proteomic study of Lok et al., it was shown that the interaction of AgNPs with sulphur containing membrane-bound proteins and enzymes led to inactivation of these molecules.181 Additionally, bacteria started to express a series of envelope proteins, heat shock proteins and periplasmic components responsible for protecting the cell against the entry of foreign substances as a stress response. Gomaa investigated the effect of AgNPs on the enzymatic activity of respiratory chain dehydrogenases of E. coli and S. aureus cells using a colorimetric method.169 Iodonitrotetrazolium chloride (INT) is a colourless compound and reduced by the bacterial respiratory chain dehydrogenases to a dark red water-insoluble iodonitrotetrazolium formazan (INF) under normal conditions. Decrease of the enzymatic activity of respiratory chain dehydrogenases in AgNP-treated cells was determined by increase of the absorbance of INF at 490 nm, whereas untreated cells increased the activity with time. These results demonstrate that AgNPs drastically damage the cell membrane and peptidoglycan, enter the cell, and inhibit the respiratory chain dehydrogenases leading to cell death. McQuillan and Shaw suggested another possible antimicrobial mechanism in which Ag+ released from AgNPs may enter the cell by cation selective membrane transport proteins called porins, and cause damage inside the cell.182
Intracellular penetration and damage
Depending on the degree of membrane damage, AgNPs can penetrate inside the cell and affect crucial and vital functions of the cell by interacting with DNA and proteins.183 One of the proposed mechanism for antimicrobial activity of AgNPs is based on silver ion release from the nanoparticles, which has an adverse effect on both DNA and proteins.184,185 Feng et al. showed that silver ions led to transformation of the bacterial DNA from the natural relaxed state to a condensed state in which the DNA molecule loses its replication ability.165 Furthermore, Energy Dispersive X-ray Analysis demonstrated the sulphur existence which indicates that silver ions interacted with thiol groups of proteins and led to inactivation of enzymatic activity. Besides the conformational change of DNA, AgNPs and silver ions can also cause degradation and or denaturation of DNA. Vishnupriya et al. studied the interaction of AgNPs with E. coli, and penetration inside the cell and intracellular damage using Raman spectroscopy along with the scattering-based hyperspectral imaging (HSI) at single-cell level.186 Time-dependent HSI demonstrated accumulation of AgNPs on the bacterial cell due to high affinity of silver for sulphur and amine groups of proteins located on the outer membrane, and then uptake of AgNPs by the cell. After penetration of AgNPs, DNA degradation was demonstrated by the increase in Raman peak intensities of adenine, guanine and cytosine bases. Researchers also suggested that relatively less increase in the thymine base is due to lack of exocyclic nitrogen in this base, whereas the interaction of AgNPs with adenine, guanine and cytosine is based on the exocyclic nitrogen present in these bases. It has been also shown that silver ions are physically attracted to DNA and interact with nucleoside part of the nucleotide. As a result, the base pairing between the complementary strands is disrupted by breaking the hydrogen bonds.184,187,188 Similarly, Chowdhury et al. studied effects of AgNPs on DNA by treating a plasmid DNA with various concentrations of AgNPs.189 They showed the dose-dependent AgNP-induced DNA strand breaks. With the increase in AgNP concentration, degradation of the supercoiled plasmid DNA to relaxed circle, linear form and finally to smaller linear fragments was observed by the agarose gel electrophoresis. Small fragments formed as a result of AgNP-induced DNA break were seen as extra lower bands in the gel (Fig. 7).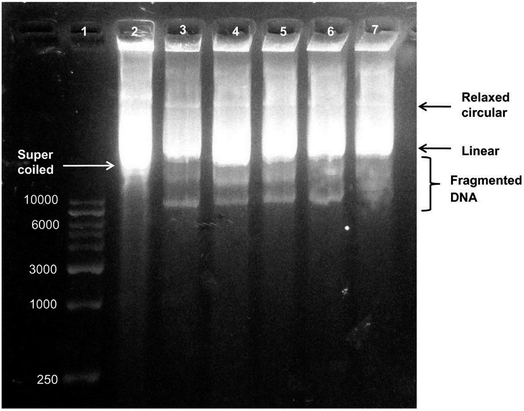 | ||
| Fig. 7 Agarose gel electrophoresis of biosynthesized AgNP-treated plasmid DNA. Lane 1, DNA marker. Lane 2, untreated plasmid DNA in supercoiled form. Lane 3, plasmid DNA treated with 0.51 μg of AgNPs showing a decrease in supercoiled DNA form. Lane 4, plasmid DNA treated with 1.02 μg of AgNPs. Lane 5, plasmid DNA treated with 2.55 μg of AgNPs. Lane 6 plasmid DNA treated with 0.51 μg of AgNPs showing degradation of DNA bands. Lane 7, plasmid DNA treated with 5.1 μg of AgNPs showing the highest degree of DNA degradation (this figure has been adapted from ref. 189 with permission from Springer). | ||
Radzig et al. compared the AgNP and silver ion susceptibility in E. coli wild-type and mutant strains.190 They found that E. coli strains carrying mutations in the genes responsible for repair of oxidative damage in the DNA (mutM, mutS, mutT and mutY) and in the genes coding for repair enzymes (nth and xthA) were more susceptible to AgNPs. Similarly, Dutta et al. conducted gene expression analyses upon AgNP treatment, and reported that genes coding for copper and magnesium transporters, proteins involved in sulfur transport, metal ion influx, antibiotic resistance and antioxidants were up-regulated.191 All these findings suggest that DNA damage caused by AgNP treatment might be also mediated by oxidative damage induced by AgNPs and intracellular antioxidant balance have an important role in the antibacterial effect of AgNPs and silver ions.
The intracellular effect of AgNPs is not limited to DNA damage. Recent proteomic studies have demonstrated effects of AgNPs on proteins and protein synthesis as well.181 As previously mentioned, AgNPs and Ag+ ions released from AgNPs have a tendency to react with thiol groups of proteins. Thiol or thiolate groups are found in the amino acid cysteine as the functional group. Cysteine is one of the least abundant amino acids, but commonly acts as a highly conserved residue within functional sites in proteins. Therefore, it is very important in biological reactions due to its high affinity metal binding property, nucleophilic role in catalytic reactions, and ability to form disulfide bonds which is vital for folding and three-dimensional structure of proteins.192,193 Yan et al. found that the expression of many membrane proteins was significantly up-regulated or down-regulated in P. aeruginosa as a result of AgNP treatment.194 These proteins are essentially responsible for ion binding, transport, flagellum assembly, pore formation, antibiotic resistance, and membrane stabilization. The most notably up-regulated proteins were found to function in ATP synthesis, phospholipid synthesis, and transmembrane transport. In addition, a series of metal transporters including copper, iron, zinc and magnesium transporters were repressed, which might assist transport of AgNPs into the cell. Wigginton et al. also conducted a proteomic study to identify interaction between E. coli proteins and AgNPs.195 Surprisingly, they found that 65% of E. coli proteins interacted with AgNPs were enzymes such as tryptophanase (TNase) and alcohol dehydrogenase while the rest was non-enzymatic proteins such as membrane porins, chaperonins, periplasmic peptide-binding proteins. Among the enzymatic proteins, TNase exhibited relatively higher affinity for AgNPs. Thereby, interaction of AgNPs with purified TNase was also investigated. TNase was found to be fragmented and lost its catalytic activity upon interaction, which might be due to the fact that the active site of TNase is in the vicinity of binding site of AgNPs. Similarly, Yuan et al. found that AgNPs repressed production of some proteins in nitrifying bacterium Nitrosomonas europaea.196 Among these proteins, GroEL, DnaK and ClpB which are chaperon proteins responsible for protein biosynthesis under stress conditions, and elongation factors Tu and G which are involved in binding of tRNAs and mRNAs to ribosome for translation, were the most notable proteins. Based on the fact that AgNPs can also repress the production of ammonia monooxygenase and hydroxylamine oxidoreductase which catalyse two critical reactions of nitrification, it can be concluded that AgNPs displayed the antibacterial effect on N. europaea by inhibiting protein biosynthesis and nitrification.197 The proteomic studies of bacteria–AgNP interaction also revealed that ribosomal subunits were also affected by the nanoparticles, leading to termination of the protein biosynthesis.198
Oxidative stress
Reactive oxygen species (ROS) are oxygen containing molecules having a strong redox potential. Under normal conditions, ROS production and the antioxidant capacity in the cell are balanced. However, in the case of imbalance between the antioxidant mechanism and excessive production of ROS, the redox balance of the cell favours oxidation, leading to oxidative stress.199 Nevertheless, oxidative stress is a normal cellular process involved in many aspects of cellular signalling, though excessive oxidative stress can be harmful. The literature revealed that nanoparticle challenge triggers the cellular oxidative stress.48,200,201 In order to overcome this stress, cells exhibit protective responses which include enzymatic or non-enzymatic defence mechanisms.201–203 When the oxidative stress overcomes the defence mechanisms, the cell wall and biomolecules such as proteins, lipids, and DNA are subjected to damage caused by ROS and free radicals such as hypochlorous acid (HOCl), hydrogen peroxide (H2O2), hydroxyl radical (·OH), superoxide anion (O2−) and singlet oxygen (1O2). DNA damages include deletions, mutations, single-breaks, double-strand breaks, adduct formation, and cross-linking with proteins. Studies confirmed oxidation-induced DNA fragmentation following exposure to metal oxide nanoparticles.130,161,204 In response to DNA insult, cells attempt to repair the damaged DNA. Repair failure may lead to cell death. It is most likely that in the bacterial cell where the antibiotics which were effluxed out from the cell wall of the bacteria could first approach to the bacterial cell membrane with the aid of nanoparticles because of nanoparticle-induced lipid peroxidation, thereby both the nanoparticles and antibiotics could damage the intracellular DNA and proteins.193,205 In this respect, Das et al. evaluated the ROS-mediated antibacterial activity of AgNPs against multidrug resistant E. coli and S. aureus, and found that ROS generation significantly contributed to the antibacterial action.206 Elevated ROS levels caused the membrane damage by increasing the permeability, which finally resulted in disruption of electron transport chain and leakage of the cellular content. Gurunathan et al. also tested AgNPs in combination with standard antibiotic drugs against bacteria.22 The synergistic effect was not only observed in the antibacterial effect, but also led to enhanced ROS generation capacity. Even though, sub-lethal concentrations of both antibiotic drugs and AgNPs produced low amount of ROS, use of AgNPs combined with antibiotic drugs such as ampicillin and vancomycin resulted in significantly higher production of ROS (Fig. 8). The increased antibacterial activity in the presence of enhanced ROS production demonstrated that AgNP-induced ROS production is vital for antimicrobial activity.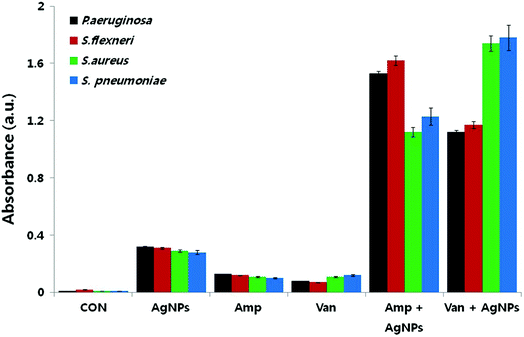 | ||
| Fig. 8 Synergistic effect of biosynthesized AgNPs and standard antibiotic drugs on ROS generation. Results of ROS measurement after 12 h treatment of the bacterial cells with AgNPs alone, antibiotics alone, and combinations of AgNPs with antibiotics (this figure has been adapted from ref. 22 with permission from Springer). | ||
Similarly, Das et al. tested AgNPs against multidrug resistant P. aeruginosa cells, and also investigated whether the antimicrobial activity of AgNPs was mediated by ROS formation. For this purpose, they used a fluorescence assay which is based on transformation of non-fluorescent 2′,7′-dichlorofluorescein diacetate (H2-DCFDA) to highly fluorescent 2′,7′-dichlorofluorescein (DCF) upon oxidation. AgNP-treated P. aeruginosa cells became DCF positive, indicating the intracellular ROS formation. Further, they used a potent ROS inhibitor (NAC) to investigate role of ROS formation in the cell death. It was observed that P. aeruginosa cells pre-treated with NAC were resistant to AgNP-induced toxicity and restored the cell viability more than 80%. Ramalingam et al. also used H2-DCFDA assay to investigate AgNP-induced ROS generation in E. coli and P. aeruginosa.168 Although ROS were also generated in the control cells, AgNP-treatment resulted in more intracellular ROS generation (Fig. 9A). The dose-dependent ROS generation was observed in both bacterial strains (Fig. 9B). ROS generation levels were approximately 70–80% at 2.7–4.5 μg ml−1 AgNP concentration interval as compared to the positive control, 1 mM H2O2 treated cells. This result is indicating that even AgNP-induced ROS production plays an important role, it is not effective alone, and some others factors such as cell membrane and wall damage are also involved in cell death. Moreover, they verified the ROS generation by measuring glutathione (GSH) concentration. GSH is a thiol-containing tripeptide and mainly present in a reduced form to protect cells from the deleterious effects of oxidative stress acting as an antioxidant. However, GSH is oxidized to disulfide (GSSG) in the case of oxidative stress, and this oxidation reaction might cause the cell death. Therefore, intracellular GSH concentration was measured to show the AgNP-triggered oxidative stress. As shown in Fig. 9C, GSH concentration significantly reduced in both E. coli and P. aeruginosa after treatment with 4.5 and 2.7 μg ml−1 of AgNPs, respectively. Thereby, GSH depleted cells are subjected to oxidative stress due to imbalance between ROS generation and antioxidant defence systems, leading to the cell death. The cell death was verified by epifluorescence microscopy, as well. For this, cells were stained with SYTO 9 and propidium iodide (PI). Red fluorescent membrane impermeant PI stains only dead cells by penetrating through the disrupted cell membrane, while green fluorescent membrane permeant SYTO 9 s quenched by PI and stains both dead and live cells. Therefore, in a bacterial population, live cells are stained with green fluorescent colour as the dead cells exhibit red fluorescence. Untreated E. coli and P. aeruginosa samples display only green fluorescent indicating the viable cells. On the other hand, AgNP-treated cells stained red color demonstrating that the cell membrane and damage led to cell death (Fig. 9D). These results suggest that the oxidative stress induced by elevated ROS production plays an important in the antimicrobial potential of AgNPs.
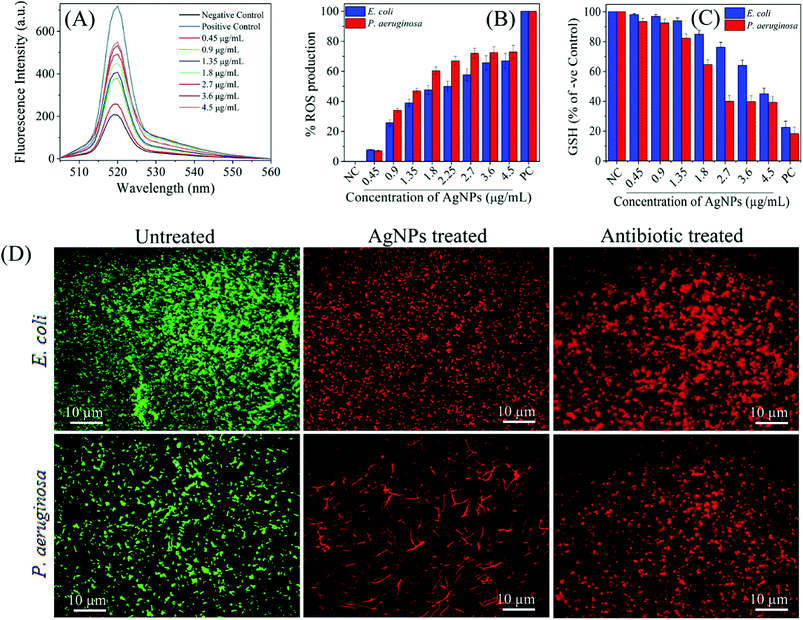 | ||
| Fig. 9 (A) H2-DCFDA assay for investigation biosynthesized AgNP-induced intracellular ROS generation; (B) the dose-dependent ROS generation in AgNP-treated in E. coli and P. aeruginosa; (C) relative GSH concentration in the treated cells as compared to positive control (PC), 1 mM H2O2; (D) fluorescence microscopy images of untreated, AgNP-treated, and gentamicin-treated E. coli and P. aeruginosa cells. SYTO 9 (green fluorescent) stained cells are intact and live, while PI (red fluorescent) stained cells are dead due to disruption of the cell membrane. As control, gentamicin, a standard antibiotic drug known to cause cell membrane damage, treated cell also displayed red fluorescent colour (this figure has been adapted from ref. 168 with permission from American Chemical Society). | ||
It is also possible that ROS generation mediated by Ag+ ions released from AgNPs can cause dysfunction of the bacterial electron transport chain and proton motive force as a result of inhibition of enzymes involved in the reactions.45,207 In addition to breakdown of membrane functions, Kim et al. found that AgNP-mediated ROS generation caused protein leakage by increasing the membrane permeability.208 Protein leakage from the cells treated with AgNPs finally resulted in cell death. Compared with S. aureus membrane, significantly higher amount of protein leakage occurred through the E. coli membrane. This result is consistent with the previous reports, and indicating that antibacterial activity is related to cell wall content of the microorganism. Additionally, they showed that activity of lactate dehydrogenase, an important respiratory enzyme, significantly decreased due to AgNP induced oxidative stress, indicating the effect of ROS on respiratory chain activity.
Deleterious effects of oxidative stress is not limited to cell membrane and electron transport chain damage, but it also has adverse effects on proteins, DNA, and other intracellular systems.209 Quinteros et al. showed that time-dependent ROS generation was induced after AgNP treatment.210 Their further analyses revealed that AgNP-induced oxidative stress caused an increase of 60% in the oxidation of DNA, proteins and lipid in E. coli. Thereby, ROS generation might be considered to have an important contribution to the antimicrobial action. Similarly, Yuan et al. confirmed increased ROS generation in bacterial cells upon AgNP treatment.173 They also measured the level of malondialdehyde (MDA) that is a product of a lipid peroxidation and serves as an indicator of lipid peroxidation oxidative stress, and found that MDA levels in AgNP-treated bacterial cells significantly increased.
Besides the direct effects on cell wall and components, oxidative stress also alters the gene expression. In Pseudomonas cells treated with AgNPs, translation of ribosomal proteins S2 and L9, keto-hydroxyglutarate aldolase (KHGA), alkyl hydroperoxide reductase C (AhpC) and thiol-specific antioxidant (TSA) was found to be up-regulated. In addition to their structural functions, the ribosomal proteins S2 and L9 are also involved in the regulation of protein biosynthesis. AhpC and TSA are members of an antioxidant family of enzymes called peroxiredoxins that protect the cell from oxidative stress by regulating the intracellular peroxide levels. KHGA is involved in sugar metabolism by metabolizing sugar acids, hexonates, and hexuronates to pyruvate and glyceraldehyde-3-phosphate, thereby prevents the accumulation of toxic intermediates. All together, the up-regulation of these proteins demonstrates a cellular response to AgNP-induced oxidative stress.211
It has been also found that AgNPs can act on bacterial cell signaling and signal transduction systems which continuously monitor the changing environmental and intracellular conditions and mediate the adaptive responses to changes in these conditions.212 Although various modification systems such as glycosylation, methylation and acetylation are involved in cell signalling, the bacterial signal transduction is mainly mediated by phosphorylation.213 The phosphorylation and dephosphorylation cycle of proteins has an important role on bacterial growth, metabolism, and cellular activities including cell cycle and DNA replication. In the case of inhibition of this cycle, all enzymatic activities would end up and lead to inhibition of the bacterial growth. Based on this fact, Shrivastava et al. investigated the phosphotyrosine profile of S. aureus and E. coli proteins to reveal the effect of AgNPs on bacterial signal transduction pathways.214 They found that AgNPs modulated the signal transduction by causing dephosphorylation of tyrosine residues on bacterial protein substrates which finally resulted in inhibition of the bacterial growth. There was no change in the profile of tyrosine phosphorylated proteins in S. aureus, whereas dephosphorylation of two peptides of E. coli and Salmonella typhi significantly changed. However, these are the initial findings, and proteomic characterization of the dephosphorylated peptide substrates and further research should be performed to elucidate the precise effect of AgNPs on signal transduction.
Option to decode bacterial lipopolysaccharide Achilles
Metal nanoparticles are thought to pose antimicrobial activity due to its small structures. Most important antimicrobial silver nanoparticles interacts with peptidoglycan cell wall215,216 and the plasma membrane;217 bacterial (cytoplasmic) DNA;218 and bacterial proteins,214,216 especially enzymes involved in vital cellular processes such as the electron transport chain. But in green synthesis method have advantageous for surface functionalization of AgNPs as the green constituents acts as both reducing and capping agent. These constituents can be effectively passed as an adjacent molecule coated over the nanoparticle. The uniqueness of nanoparticles is that it acts first at the defensive walls rather than attacking the bacteria itself. Once the damages complete, it can now interact with bacterial (cytoplasmic) DNA, bacterial proteins and enzymes.A recent finding reports the crystal structure of the unique integral membrane LPS translocon LptD–LptE complex in Gram negative bacteria. According to this report, LptD forms a novel 26-stranded largest β-barrel and LptE adopts a roll-like structure located inside the barrel of LptD to form an unprecedented two-protein ‘barrel and plug’ architecture.219 This bacterial molecular bricks is the key behind the antimicrobial resistance.
Hydrophilic O-antigen and the core oligosaccharide of the LPS may pass through the barrel and the lipid A of the LPS may be inserted into the outer leaflet of the outer membrane through a lateral opening between strands β1 and β26 of LptD.
Keeping this in view and the prevalence report of potential effects of AgNPs against multidrug susceptible as well as multidrug sensitive strains like Pseudomonas aeruginosa, ampicillin-resistant Escherichia coli, erythromycin-resistant Streptococcus pyogenes, methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Staphylococcus aureus (VRSA) suggests evidences of bacterial defensive architecture breakdown. Thus, green synthesis of silver nanoparticles using a strong antimicrobial agent as both reducing and capping agent may have synergistic effect. Nanoparticles can breakdown the defensive barrier and helps to insert attached drug molecule over the surfaces.
Besides, target nanoparticle surfaces can be charged that also provide ease attachment of nanoparticles over the defensive brick architecture. But to get the optimum results research should be focused over the antimicrobial pure bio compound based synthesis. Even biological core–shell may also prove advantageous where single metal in core and dual drug on the surface can be easily prepared. In that case the drugs and the metal should follow an ascending order of reduction potential.
Green synthesized silver nanoparticles controlling antifungal activity
Apart from bacterial pathogenesis, high rates of morbidity and mortality caused by fungal infections is also a severe concern to mankind. The problems is ever increasing as a limited new antifungal targets are explored and fungi are also developing resistance mechanism through mutation arch. In such scenario, silver nanoparticles which already been proved to exert antibacterial action are often tried against fungal infections. Due to limited antifungal arsenal, nanoparticles thought to serve as a carriers of antifungal. More interestingly, the if the reducing agent itself serves antifungal action and deposits over the nanoparticle surface as capping agent, it would offer synergistic antifungal action. Antifungal drug molecules can also be successfully adsorbed on the surface of biogenic silver thus can offer a efficient strategy for antifungal drug delivery.220This strategy was successfully implemented by Kumar and co-workers.221 They initially synthesized spherical and monodispersed silver nanoparticles of 9.8 nm size using the cell free supernatant of Delftia sp. strain KCM-006. This AgNPs also exhibited antifungal activity. Further they conjugated antifungal drug miconazole to biogenic silver nanoparticles surface. This strategy exhibited significant fungicidal activity, inhibited of ergosterol biosynthesis and biofilm inhibition by increasing ROS levels.
Similar strategy was followed by Tutaj and co-workers.222 They proceeded their synthesis in an alkaline environment using Amphotericin B (AmB) as reducing and coating agent. This alkaline environment prevented aggregation of AmB itself and promotes to develop a monodisperse, AmB-coated silver nanoparticles (AmB-AgNPs) with the diameter ∼ 7 nm. Similarly, Ahmad et al. who successfully developed a amphotericin B-conjugated biogenic silver nanoparticles with size of 424–433 nm. They have successfully used amphotericin B as in situ reducing and capping agent for silver nanoparticles. Amphotericin B-conjugated biogenic silver nanoparticles offered synergistic antifungal activity against selected fungal strains i.e. C. albicans and C. tropicalis.223
Apart from such drug delivery example, green synthesized AgNPs are also reported to exhibits antifungal activity due to its bio-coating in combination with its size. Ishida et al., reported antifungal activity green synthesized AgNPs from the fungus Fusarium oxysporum. Green synthesized AgNPs showed a high level of antifungal activity against Candida and Cryptococcus, with minimum inhibitory concentration values ≤ 1.68 μg ml−1 for both genera.224 It has been investigated using disc diffusion method that colloidal silver has strong antifungal activity against three species of Candida like C. albicans, C. glabrata and C. tropicalis.225 Silver nanoparticles synthesized from Pilimelia columellifera subsp. pallida SL19 strain demonstrated antifungal activity against fungi causing superficial mycoses viz., C. albicans and M. furfur. The lowest MIC of AgNPs was noticed against M. furfur (16 μg ml−1).226 Mentha pulegium aqueous extract mediated synthesized AgNPs also showed considerable antifungal activity against fluconazole resistant Candida albicans.227 It has been established by food poisoning technique that the growth of following fungi like Bipolaris spicifera, Fusarium oxysporum, Aspergillus niger and Curvularia lunata is inhibited by NPs.228 Therefore AgNPs may be used as good antifungal drug to control several pathogenic fungi.
Green synthesized silver nanoparticles are found to effective against plant pathogenic fungi. The colonies of plant pathogenic fungi Bipolaris sorokiniana and Magnaporthe grisea are inhibited by applying nanosilver and ionic silver in vitro condition.229 Green synthesized AgNPs are much effective as broad spectrum fungicide against Alternaria alternata and Botrytis cinerea. The hyphae and conidia of these fungi are damaged by applying the NPs.230 The NPs also have strong synergistic activity with other fungicides against several plant pathogenic fungi like Fusarium graminearum (cause head blight of wheat), Fusarium oxysporum (cause tomato wilt), Fusarium solani (cause potato rot), and Penicillium expansum (cause apple rot).231 Elgorban et al. showed that nano silver has strong inhibitory effect on the growth of Rhizoctonia solani.232 The growth of some wood degrading fungi like Gloeophyllum abietinum, G. trabeum, Chaetomium globosum, and Phanerochaete sordida is inhibited by biogenic synthesized AgNPs.116 Thus apart from the bacterial control, green synthesized silver nanoparticles can be used in fungal control.
Green synthesized silver nanoparticles against Leishmania
Leishmaniasis, disease caused by parasites of the Leishmania type, is an emerging and uncontrollable disease, characterized as a category-1 disease by WHO.233 After malaria, it is the most prevalent infectious disease in terms of fatality and geographical distribution.234 The most common forms of leishmaniasis are cutaneous, characterized by skin sores, and visceral leishmaniasis which affects several internal organs like spleen, liver etc. Limited availability of antileishmanial drugs, its high cost and continuum resistance over the available drugs makes the situation fatal. To overcome such, green synthesis based drug delivery strategy is seen to open a newer avenue of treatment. The parasite is very much sensitive to AgNPs for the generation of ROS. It is reported that NPs have synergistic antiprotozoal activity against Leishmania tropica under UV light.235 As mentioned in the earlier section, the reducing and coating or stabilizing agent can also impart functional activity to AgNPs. Additionally, this coated surface of the biogenic nanoparticles can also act as a active site for the targeted drug for drug delivery option.Ahmad et al. used this strategy and they initially synthesized spherical AgNPs (10–20 nm) using aqueous extract of Isatis tinctoria as reducing and capping agents. Later they have successfully adsorbed amphotericin B over the previously formed biogenic AgNPs surface.234 They observed a enhanced parasites activity of amphotericin B adsorbed silver nanoparticle (IC50 value 2.43 μg ml−1) than biogenic nanoparticle (IC50 value of 4.2 μg ml−1) against Leishmania tropica234 at promastigotes stage. Kalangi et al. has explored that miltefosine doped green synthesized AgNPs offer enhanced antileishmanial effect. They concluded their finding by reporting that green synthesized spherical AgNPs of 35 nm size at a concentration of (50 μM) didn't show antileishmanial effect on promastigote stage of Leishmania parasite. But after applying in combination with miltefosine (12.5 μM and 25 μM), 2 fold magnification in antileishmanial effect was observed.236 Spherical AgNPs with a size of 12.82 ± 2.5 nm was synthesized using aqueous leaf extract of Euphorbia prostrata. This biogenic nanoparticles offered parasitic activity against Leishmania donovani with a IC50 value of 3.89 and 14.94 μg ml−1 in amastigotes and promastigotes stage.237 Similar study by Ahmad et al. reported promising antileishmanial activity (IC50 value 4.37) of green synthesized spherical AgNPs (3–8 nm) using phytochemicals from Sargentodoxa cuneata.238 Lopes et al. reported that green synthesized AgNPs using tannic acid (TA) officered antileishmanial activity.239 El-khadragy et al. reported clinical efficacy of biosynthesize AgNPs against Leishmania in in vivo rat model study. Biosynthesized spherical silver nanoparticle (116 nm) using Moringa oleifera extract resulted in a significant reduction in the average size of leishmaniasis cutaneous lesions compared with untreated mice.240 Green synthesized spherical Ag-NPs of 26 nm using Olax nana Wall. ex Benth. (family: Olacaceae) aqueous extract offered activity against Leishmania tropica (KMH23) promastigotes (IC50 = 12.56 and 21.52 μg ml−1) and amastigotes (17.44 and 42.20 μg ml−1).241 Another study on green synthesized silver nanoparticle from Fusarium oxysporum exhibited strong activity against Leishmania amazonensis promastigote and amastigote forms.242 Ullah et al. showed that biogenic AgNPs are able to produce high level of nitric oxide (NO) in L. infantum infected macrophage cell. The production of NO radical is able to reduce the pathogenicity by killing the parasite.243 So green synthesized silver nanoparticle could be milestone in disease management of Leishmaniasis.
Cytotoxicity of AgNPs
Conversion of metal to its nanoform brings risk of toxicity. This is also true for the case of silver and depends upon the sizes,244 shapes245 and density of the AgNPs.110 The toxicity concern of silver nanoparticles is suitably reduced with green synthesis method. The primary reason behind the nominal toxicity lies upon its coating which are generally biocompatible in nature. Although the primary role of the coating is to stabilize the nanoparticle and prevent agglomeration; but the biocompatible behaviour of coating makes green synthesized AgNPs suitable in different bio medicinal applications.Early report suggests that stabilization of AgNPs by different polymers reduce the toxicity of AgNPs tested against mouse skin fibroblasts (L929), human hepatocarcinoma cells (HepG2), and mouse monocyte macrophages (J774A1). Polymer coated AgNPs at a concentration 1.5 ppm exhibited hemocompatible nature. It can be noted that materials with a hemolysis ratio less than 5% were regarded as hemocompatible.246 A number of reports reveals that coating over the silver nanoparticle reduces its toxicity.247–250
In green synthesis, reduction and stabilization of the silver nanoparticles are achieved by the biocompatible material and hence the toxicity concern is reduced in most of the cases. Green synthesis using biopolymer i.e. glucan gave a glucan capped spherical AgNPs of 2.445 ± 1.08 nm. This nanoparticles exhibited only 0.68% hemolysis to human RBCs at its LD50 dosage.130 Thus, AgNPs–glucan conjugates were found to be compatible with human RBCs at its LD50 dosage. In an another study, polysaccharide capped AgNPs of 2.78 ± 1.47 nm were synthesized from a hetero polysaccharide isolated from Lentinus squarrosulus (Mont.). This nanoparticles also gave compatible nature with human RBCs at its LD50 dosage.131 Similarly phyto-constituents also gave similar results of toxicity tested on human cell. AgNPs prepared using Croton bonplandianum leaves extract at its MIC dosage showed only 0.02% hemolysis.251 AgNPs (spherical, 9.8 mm size) from cell free supernatant of Delftia sp. strain KCM-006 which was further used as a carrier of antifungal drug exhibited non-toxic nature up to a concentration of 20 μM. Biocompatibility level was assessed determining cell viability and immunocytochemistry analysis against different normal cell lines including Chinese hamster ovary cells (CHO), human lung cell line (MRC5) and human vascular endothelial cells (HUVEC).221 Silver nanoparticles synthesized from Anethum graveolens leaf extract also confer biocompatible nature.236 Aqueous extract of Rosa damascena mediated silver nanoparticle of 15–27 nm size also exhibited compatibility against erythrocytes.252 In another study, phytosynthesized AgNPs from aqueous extract of Salacia chinensis (SC) bark offer full biocompatibility when treated against normal human fibroblasts and blood erythrocytes.253 Thus the biocompatibility achieved through green synthesis suggests its possible use in varying field of biomedical application.
Conclusion
Based upon the previous discussion it can be said that the synthesis of nanoparticles may serve as a future direction in biomedical nanotechnology in developing antimicrobial compounds. The previously used metal antimicrobial may come with lots of possibilities with the advancement of green nanotechnology in combating multidrug resistance. Nanoparticles are seen to have ability to cross and disrupt the membrane. Thus loading drugs on the nanoparticle surface may increase the efficiency of biocidal motion by additional effect than disrupting the membrane. Thus it can have the future option to decode bacterial lipopolysaccharide Achilles. Toxicity of silver nanoparticles is also a major issue. Although biomolecular capping in green synthesis process reduces the toxicity, but it is imperative to develop more efficient capping and reducing agents for particular uses. The current trend of research is mainly carried out using the whole plant extracts as reducing and capping agents. This serves as a drawback regarding not clearly understanding the role of isolated pure bio molecules in synthesis and activity of nanoparticles. Synthesis using pure compound in an optimized condition may additionally give solution regarding the poly disparity of green nanoparticle synthesis. Thus studies using pure compounds would be carried out for better understanding the science behind the green synthesis. In near future new opportunities may also come with new problem solutions. From the previous discussion it can be concluded that this is the beginning of green nanoparticle synthesis. Green synthesis may serve many medicinal applications in near future.Conflicts of interest
There are no conflicts to declare.Acknowledgements
O. B. and M. D. Y. wish to thank Konya Food and Agriculture University (KFAU) and KFAU Research and Development Center for Diagnostic Kits (KIT-ARGEM) for their support.References
- A. Roy, O. L. Franco and S. M. Mandal, Curr. Protein Pept. Sci., 2013, 14, 580–587 CrossRef CAS.
- S. M. Mandal, A. Roy, A. K. Ghosh, T. K. Hazra, A. Basak and O. L. Franco, Front. Pharmacol., 2014, 5, 105 Search PubMed.
- A. Roy, K. Khanra, A. Mishra and N. Bhattacharyya, Int. J. Adv. Res., 2013, 1, 193–198 Search PubMed.
- S. M. Mandal, A. Roy, D. Mahata, L. Migliolo, D. O. Nolasco and O. L. Franco, J. Antibiot., 2014, 67, 771–775 CrossRef CAS PubMed.
- I. K. Sen, A. K. Mandal, R. Chakraborty, B. Behera, K. K. Yadav, T. K. Maiti and S. S. Islam, Carbohydr. Polym., 2014, 101, 188–195 CrossRef CAS PubMed.
- D. M. Livermore, M. Blaser, O. Carrs, G. Cassell, N. Fishman, R. Guidos, S. Levy, J. Powers, R. Norrby, G. Tillotson, R. Davies, S. Projan, M. Dawson, D. Monnet, M. Keogh-Brown, K. Hand, S. Garner, D. Findlay, C. Morel, R. Wise, R. Bax, F. Burke, I. Chopra, L. Czaplewski, R. Finch, D. Livermore, L. J. V. Piddock and T. White, J. Antimicrob. Chemother., 2011, 66, 1941–1944 CrossRef CAS PubMed.
- M. K. Rai, S. D. Deshmukh, A. P. Ingle and A. K. Gade, J. Appl. Microbiol., 2012, 112, 841–852 CrossRef CAS PubMed.
- J. A. Lemire, J. J. Harrison and R. J. Turner, Nat. Rev. Microbiol., 2013, 11, 371–384 CrossRef CAS PubMed.
- J. W. Alexander, Surg. Infect., 2009, 10, 289–292 CrossRef PubMed.
- G. Borkow and J. Gabbay, Curr. Chem. Biol., 2009, 3, 272–278 CAS.
- L.-J. Ming, Med. Res. Rev., 2003, 23, 697–762 CrossRef CAS PubMed.
- Z. H. Chohan, C. T. Supuran and A. Scozzafava, J. Enzyme Inhib. Med. Chem., 2005, 20, 303–307 CrossRef CAS PubMed.
- S. Moharrer, B. Mohammadi, R. A. Gharamohammadi, Yargoli and M. Yargoli, Indian J. Sci. Technol., 2012, 5, 2443–2444 CAS.
- H.-J. Lee, J. Y. Song and B. S. Kim, J. Chem. Technol. Biotechnol., 2013, 88, 1971–1977 CAS.
- S. Azizi, M. B. Ahmad, F. Namvar and R. Mohamad, Mater. Lett., 2014, 116, 275–277 CrossRef CAS.
- P. S. Schabes-Retchkiman, G. Canizal, R. Herrera-Becerra, C. Zorrilla, H. B. Liu and J. A. Ascencio, Opt. Mater., 2006, 29, 95–99 CrossRef CAS.
- R. Raliya and J. C. Tarafdar, Agric. Res., 2013, 2, 48–57 CrossRef CAS.
- H. Gu, P. L. Ho, E. Tong, L. Wang and B. Xu, Nano Lett., 2003, 3, 1261–1263 CrossRef CAS.
- J. R. Morones-Ramirez, J. A. Winkler, C. S. Spina and J. J. Collins, Sci. Transl. Med., 2013, 5, 190ra81 Search PubMed.
- M. Zhou, M. Lin, L. Chen, Y. Wang, X. Guo, L. Peng, X. Guo and W. Ding, Chem. Commun., 2015, 51, 5116–5119 RSC.
- M. Rai, A. Yadav and A. Gade, Biotechnol. Adv., 2009, 27, 76–83 CrossRef CAS PubMed.
- S. Gurunathan, J. W. Han, D.-N. Kwon and J.-H. Kim, Nanoscale Res. Lett., 2014, 9, 373 CrossRef PubMed.
- P. Gong, H. Li, X. He, K. Wang, J. Hu, W. Tan, S. Zhang and X. Yang, Nanotechnology, 2007, 18, 285604 CrossRef.
- P. Jain and T. Pradeep, Biotechnol. Bioeng., 2005, 90, 59–63 CrossRef CAS PubMed.
- I. Sawada, R. Fachrul, T. Ito, Y. Ohmukai, T. Maruyama and H. Matsuyama, J. Membr. Sci., 2012, 387–388, 1–6 CrossRef CAS.
- I. Sawada, R. Fachrul, T. Ito, Y. Ohmukai, T. Maruyama and H. Matsuyama, J. Membr. Sci., 2012, 387–388, 1–6 CrossRef CAS.
- A. Das, A. Kumar, N. B. Patil, C. Viswanathan and D. Ghosh, Carbohydr. Polym., 2015, 130, 254–261 CrossRef CAS PubMed.
- M. Shah, D. Fawcett, S. Sharma, S. Tripathy, G. Poinern, M. Shah, D. Fawcett, S. Sharma, S. K. Tripathy and G. E. J. Poinern, Materials, 2015, 8, 7278–7308 CrossRef CAS.
- S. F. Adil, M. E. Assal, M. Khan, A. Al-Warthan, M. R. H. Siddiqui and L. M. Liz-Marzán, Dalton Trans., 2015, 44, 9709–9717 RSC.
- J. Virkutyte and R. S. Varma, Sustainable Preparation of Metal Nanoparticles, Royal Society of Chemistry, Cambridge, 2012 Search PubMed.
- P. Christian, F. Von der Kammer, M. Baalousha and T. Hofmann, Ecotoxicology, 2008, 17, 326–343 CrossRef CAS PubMed.
- A. K. Mittal, Y. Chisti and U. C. Banerjee, Biotechnol. Adv., 2013, 31, 346–356 CrossRef CAS PubMed.
- A. D. Dwivedi and K. Gopal, Colloids Surf., A, 2010, 369, 27–33 CrossRef CAS.
- A. Rai, A. Prabhune and C. C. Perry, J. Mater. Chem., 2010, 20, 6789 RSC.
- M. Demurtas and C. C. Perry, Gold Bull., 2013, 47, 103–107 CrossRef.
- S. Francis, S. Joseph, E. P. Koshy and B. Mathew, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 795–804 CrossRef CAS.
- M. A. Asghar, E. Zahir, S. M. Shahid, M. N. Khan, M. A. Asghar, J. Iqbal and G. Walker, Lebensm.-Wiss. -Technol., 2018, 90, 98–107 CrossRef CAS.
- B. Ajitha, Y. A. K. Reddy, H.-J. Jeon and C. W. Ahn, Adv. Powder Technol., 2018, 29, 86–93 CrossRef CAS.
- M. Baghayeri, B. Mahdavi, Z. Hosseinpor-Mohsen Abadi and S. Farhadi, Appl. Organomet. Chem., 2018, 32, e4057 CrossRef.
- L. Wang, C. Hu and L. Shao, Int. J. Nanomed., 2017, 12, 1227–1249 CrossRef CAS PubMed.
- Y. He, F. Wei, Z. Ma, H. Zhang, Q. Yang, B. Yao, Z. Huang, J. Li, C. Zeng and Q. Zhang, RSC Adv., 2017, 7, 39842–39851 RSC.
- Y. He, X. Li, Y. Zheng, Z. Wang, Z. Ma, Q. Yang, B. Yao, Y. Zhao and H. Zhang, New J. Chem., 2018, 42, 2882–2888 RSC.
- S. Hamedi, S. A. Shojaosadati and A. Mohammadi, J. Photochem. Photobiol., B, 2017, 167, 36–44 CrossRef CAS.
- Y. K. Mohanta, S. K. Panda, R. Jayabalan, N. Sharma, A. K. Bastia and T. K. Mohanta, Front. Mol. Biosci., 2017, 4, 14 Search PubMed.
- N. Jain, A. Bhargava, M. Rathi, R. V. Dilip and J. Panwar, PLoS One, 2015, 10, e0134337 CrossRef PubMed.
- M. N. Gallucci, J. C. Fraire, A. P. V. Ferreyra Maillard, P. L. Páez, I. M. Aiassa Martínez, E. V. Pannunzio Miner, E. A. Coronado and P. R. Dalmasso, Mater. Lett., 2017, 197, 98–101 CrossRef CAS.
- A. R. Allafchian, S. Z. Mirahmadi-Zare, S. A. H. Jalali, S. S. Hashemi and M. R. Vahabi, J. Nanostruct. Chem., 2016, 6, 129–135 CrossRef CAS.
- A. Ravichandran, P. Subramanian, V. Manoharan, T. Muthu, R. Periyannan, M. Thangapandi, K. Ponnuchamy, B. Pandi and P. N. Marimuthu, J. Photochem. Photobiol., B, 2018, 185, 117–125 CrossRef CAS PubMed.
- K. Anandalakshmi, J. Venugobal and V. Ramasamy, Appl. Nanosci., 2016, 6, 399–408 CrossRef CAS.
- P. S. Ramesh, T. Kokila and D. Geetha, Spectrochim. Acta, Part A, 2015, 142, 339–343 CrossRef CAS PubMed.
- K. Ali, B. Ahmed, S. Dwivedi, Q. Saquib, A. A. Al-Khedhairy and J. Musarrat, PLoS One, 2015, 10, e0131178 CrossRef PubMed.
- H. M. M. Ibrahim, J. Radiat. Res. Appl. Sci., 2015, 8, 265–275 CrossRef.
- N. Muniyappan and N. S. Nagarajan, Process Biochem., 2014, 49, 1054–1061 CrossRef CAS.
- P. Kumar, S. Senthamil Selvi and M. Govindaraju, Appl. Nanosci., 2013, 3, 495–500 CrossRef CAS.
- K. Paulkumar, G. Gnanajobitha, M. Vanaja, S. Rajeshkumar, C. Malarkodi, K. Pandian and G. Annadurai, Sci. World J., 2014, 2014, 829894 Search PubMed.
- P. Prakash, P. Gnanaprakasam, R. Emmanuel, S. Arokiyaraj and M. Saravanan, Colloids Surf., B, 2013, 108, 255–259 CrossRef CAS PubMed.
- F. T. Minhas, G. Arslan, I. H. Gubbuk, C. Akkoz, B. Y. Ozturk, B. Asıkkutlu, U. Arslan and M. Ersoz, Int. J. Biol. Macromol., 2018, 107, 157–165 CrossRef CAS PubMed.
- E. F. Aboelfetoh, R. A. El-Shenody and M. M. Ghobara, Environ. Monit. Assess., 2017, 189, 349 CrossRef PubMed.
- V. S. Ramkumar, A. Pugazhendhi, K. Gopalakrishnan, P. Sivagurunathan, G. D. Saratale, T. N. B. Dung and E. Kannapiran, Biotechnol. Rep., 2017, 14, 1–7 CrossRef PubMed.
- Z. Salari, F. Danafar, S. Dabaghi and S. A. Ataei, J. Saudi Chem. Soc., 2016, 20, 459–464 CrossRef CAS.
- T. Kathiraven, A. Sundaramanickam, N. Shanmugam and T. Balasubramanian, Appl. Nanosci., 2015, 5, 499–504 CrossRef CAS.
- N. Aziz, M. Faraz, R. Pandey, M. Shakir, T. Fatma, A. Varma, I. Barman and R. Prasad, Langmuir, 2015, 31, 11605–11612 CrossRef CAS PubMed.
- M. Yousefzadi, Z. Rahimi and V. Ghafori, Mater. Lett., 2014, 137, 1–4 CrossRef CAS.
- S. P. Sivagnanam, A. T. Getachew, J. H. Choi, Y. B. Park, H. C. Woo and B. S. Chun, Green Process. Synth., 2016, 6, 147–160 Search PubMed.
- D. Inbakandan, R. Venkatesan and S. Ajmal Khan, Colloids Surf., B, 2010, 81, 634–639 CrossRef CAS PubMed.
- T. Kathiraven, A. Sundaramanickam, N. Shanmugam and T. Balasubramanian, Appl. Nanosci., 2015, 5, 499–504 CrossRef CAS.
- H. M. El-Rafie, M. H. El-Rafie and M. K. Zahran, Carbohydr. Polym., 2013, 96, 403–410 CrossRef CAS PubMed.
- P. Kumar, S. S. Selvi, A. L. Prabha, K. P. Kumar, R. S. Ganeshkumar and M. Govindaraju, Nano Biomed. Eng., 2013, 52, 562–566 Search PubMed.
- A. Pugazhendhi, D. Prabakar, J. M. Jacob, I. Karuppusamy and R. G. Saratale, Microb. Pathog., 2018, 114, 41–45 CrossRef CAS PubMed.
- S. Murugesan, S. Bhuvaneswari and V. Sivamurugan, Int. J. Pharm. Pharm. Sci., 2017, 9, 192–197 CrossRef CAS.
- A. P. de Aragão, T. M. de Oliveira, P. V. Quelemes, M. L. G. Perfeito, M. C. Araújo, J. de A. S. Santiago, V. S. Cardoso, P. Quaresma, J. R. de Souza de Almeida Leite and D. A. da Silva, Arabian J. Chem. DOI:10.1016/J.ARABJC.2016.04.014.
- F. WA, A. E. BEE, S. WF and F. WA, J. Nanomed. Nanotechnol., 2016, 07, 1–4 Search PubMed.
- V. Lavakumar, K. Masilamani, V. Ravichandiran, N. Venkateshan, D. V. R. Saigopal, C. K. Ashok Kumar and C. Sowmya, Chem. Cent. J., 2015, 9, 42 CrossRef CAS PubMed.
- H. M. El-Rafie, M. H. El-Rafie and M. K. Zahran, Carbohydr. Polym., 2013, 96, 403–410 CrossRef CAS PubMed.
- V. Venkatpurwar and V. Pokharkar, Mater. Lett., 2011, 65, 999–1002 CrossRef CAS.
- A. S. Sonker, Richa, J. Pathak, Rajneesh, V. K. Kannaujiya and R. P. Sinha, Can. J. Biotechnol., 2017, 1, 26–37 CrossRef.
- N. O. San Keskin, N. Koçberber Kılıç, G. Dönmez and T. Tekinay, J. Nano Res., 2016, 40, 120–127 Search PubMed.
- V. Patel, D. Berthold, P. Puranik and M. Gantar, Biotechnol. Rep., 2015, 5, 112–119 CrossRef PubMed.
- G. Singh, P. K. Babele, S. K. Shahi, R. P. Sinha, M. B. Tyagi and A. Kumar, J. Microbiol. Biotechnol., 2014, 24, 1354–1367 CrossRef CAS PubMed.
- F. M. Morsy, N. A. Nafady, M. H. Abd-Alla and D. A. Elhady, Univers. J. Microbiol. Res., 2014, 2, 36–43 CAS.
- S. Neethu, S. J. Midhun, E. K. Radhakrishnan and M. Jyothis, Microb. Pathog., 2018, 116, 263–272 CrossRef CAS.
- T. Singh, K. Jyoti, A. Patnaik, A. Singh, R. Chauhan and S. S. Chandel, J. Genet. Eng. Biotechnol., 2017, 15, 31–39 CrossRef.
- L. Ma, W. Su, J.-X. Liu, X.-X. Zeng, Z. Huang, W. Li, Z.-C. Liu and J.-X. Tang, Mater. Sci. Eng., C, 2017, 77, 963–971 CrossRef CAS.
- R. Al-Bahrani, J. Raman, H. Lakshmanan, A. A. Hassan and V. Sabaratnam, Mater. Lett., 2017, 186, 21–25 CrossRef CAS.
- M. Eugenio, N. Müller, S. Frasés, R. Almeida-Paes, L. M. T. R. Lima, L. Lemgruber, M. Farina, W. de Souza and C. Sant'Anna, RSC Adv., 2016, 6, 9893–9904 RSC.
- J. G. Fernández, M. A. Fernández-Baldo, E. Berni, G. Camí, N. Durán, J. Raba and M. I. Sanz, Process Biochem., 2016, 51, 1306–1313 CrossRef.
- B. Xue, D. He, S. Gao, D. Wang, K. Yokoyama and L. Wang, Int. J. Nanomed., 2016, 11, 1899–1906 CAS.
- S. K. Singdevsachan, U. K. Parida, H. Bae, Y. K. Mohanta, T. K. Mohanta and S. K. Panda, IET Nanobiotechnol., 2016, 10, 184–189 CrossRef PubMed.
- F. Niknejad, M. Nabili, R. Daie Ghazvini and M. Moazeni, Curr. Med. Mycol., 2015, 1, 17–24 CAS.
- P. Ramalingmam, S. Muthukrishnan and P. Thangaraj, Journal of Nanoscience and Nanoengineering, 2015, 1, 241–247 Search PubMed.
- D. Singh, V. Rathod, S. Ninganagouda, J. Herimath and P. Kulkarni, J. Pharm. Res., 2013, 7, 448–453 CAS.
- M. Saravanan, S. Arokiyaraj, T. Lakshmi and A. Pugazhendhi, Microb. Pathog., 2018, 117, 68–72 CrossRef CAS PubMed.
- L. Gan, S. Zhang, Y. Zhang, S. He and Y. Tian, Prep. Biochem. Biotechnol., 2018, 1–7 Search PubMed.
- M. A. Shaker and M. I. Shaaban, J. Taibah Univ. Med. Sci., 2017, 12, 291–297 Search PubMed.
- P. Singh, Y. J. Kim, C. Wang, R. Mathiyalagan and D. C. Yang, Artif. Cells, Nanomed., Biotechnol., 2016, 44, 1569–1575 CrossRef CAS PubMed.
- P. S. Nayak, M. Arakha, A. Kumar, S. Asthana, B. C. Mallick and S. Jha, RSC Adv., 2016, 6, 8232–8242 RSC.
- J. H. Jo, P. Singh, Y. J. Kim, C. Wang, R. Mathiyalagan, C.-G. Jin and D. C. Yang, Artif. Cells, Nanomed., Biotechnol., 2016, 44, 1576–1581 CrossRef CAS.
- R. Thomas, A. Janardhanan, R. T. Varghese, E. V Soniya, J. Mathew and E. K. Radhakrishnan, Braz. J. Microbiol., 2014, 45, 1221–1227 CrossRef CAS PubMed.
- C. Malarkodi, S. Rajeshkumar, K. Paulkumar, M. Vanaja, G. D. G. Jobitha and G. Annadurai, Drug Invent. Today, 2013, 5, 119–125 CrossRef CAS.
- M. S.-V. Nélly, E. D. C. Nelson, d. A. S. Taciana, V. Q. Patrick, R. d. A. Alyne, C. M. d. M. Ana, L. d. S. C. Ana, P. F. L. João, B. A. Ricardo, A. d. S. Durcilene, R. d. S. d. A. L. José and F. S. T. Maria, Afr. J. Biotechnol., 2017, 16, 2072–2082 CrossRef.
- B. Buszewski, V. Railean-Plugaru, P. Pomastowski, K. Rafińska, M. Szultka-Mlynska, P. Golinska, M. Wypij, D. Laskowski and H. Dahm, J. Microbiol., Immunol. Infect., 2018, 51, 45–54 CrossRef CAS PubMed.
- H. M. Abd-Elnaby, G. M. Abo-Elala, U. M. Abdel-Raouf and M. M. Hamed, Egypt. J. Aquat. Res., 2016, 42, 301–312 CrossRef.
- P. Sanjenbam, J. V. Gopal and K. Kannabiran, J. Mycol. Med., 2014, 24, 211–219 CrossRef CAS PubMed.
- P. Manivasagan, J. Venkatesan, K. Senthilkumar, K. Sivakumar and S.-K. Kim, BioMed Res. Int., 2013, 2013, 287638 Search PubMed.
- R. S. Prakasham, S. K. Buddana, S. K. Yannam and G. S. Guntuku, J. Microbiol. Biotechnol., 2012, 22, 614–621 CrossRef CAS.
- K. Khanra, A. Roy and N. Bhattacharyya, J. Nanosci. Nanotechnol., 2013, 1, 1–6 Search PubMed.
- S. Ahmed, M. Ahmad, B. L. Swami and S. Ikram, J. Adv. Res., 2016, 7, 17–28 CrossRef CAS PubMed.
- M. Rai, A. Yadav and A. Gade, Crit. Rev. Biotechnol., 2008, 28, 277–284 CrossRef CAS.
- P. Mohanpuria, N. K. Rana and S. K. Yadav, J. Nanopart. Res., 2008, 10, 507–517 CrossRef CAS.
- N. Roy, A. Gaur, A. Jain, S. Bhattacharya and V. Rani, Environ. Toxicol. Pharmacol., 2013, 36, 807–812 CrossRef CAS PubMed.
- V. L. Das, R. Thomas, R. T. Varghese, E. V Soniya, J. Mathew and E. K. Radhakrishnan, 3 Biotech, 2014, 4, 121–126 CrossRef PubMed.
- K. Kalimuthu, R. Suresh Babu, D. Venkataraman, M. Bilal and S. Gurunathan, Colloids Surf., B, 2008, 65, 150–153 CrossRef CAS PubMed.
- S. R. Kumar Pandian, V. Deepak, K. Kalishwaralal, P. Viswanathan and S. Gurunathan, Braz. J. Microbiol., 2010, 41, 805–809 CrossRef PubMed.
- S. Anil Kumar, M. K. Abyaneh, S. W. Gosavi, S. K. Kulkarni, R. Pasricha, A. Ahmad and M. I. Khan, Biotechnol. Lett., 2007, 29, 439–445 CrossRef CAS PubMed.
- R. M. Slawson, M. I. Van Dyke, H. Lee and J. T. Trevors, Plasmid, 1992, 27, 72–79 CrossRef CAS PubMed.
- K. B. Narayanan and H. H. Park, Eur. J. Plant Pathol., 2014, 140, 185–192 CrossRef CAS.
- P. Mukherjee, A. Ahmad, D. Mandal, S. Senapati, S. R. Sainkar, M. I. Khan, R. Parishcha, P. V. Ajaykumar, M. Alam, R. Kumar and M. Sastry, Nano Lett., 2001, 1, 515–519 CrossRef CAS.
- A. M. Fayaz, K. Balaji, M. Girilal, R. Yadav, P. T. Kalaichelvan and R. Venketesan, Nanomedicine, 2010, 6, 103–109 CrossRef CAS PubMed.
- M. Rai and N. E. Duran, Metal nanoparticles in microbiology, Springer, 2011 Search PubMed.
- K. B. Narayanan and N. Sakthivel, J. Hazard. Mater., 2011, 189, 519–525 CrossRef CAS PubMed.
- A. Ahmad, S. Senapati, M. I. Khan, R. Kumar, R. Ramani, V. Srinivas and M. Sastry, Nanotechnology, 2003, 14, 824–828 CrossRef CAS.
- D. Inbakandan, R. Venkatesan and S. Ajmal Khan, Colloids Surf., B, 2010, 81, 634–639 CrossRef CAS PubMed.
- S. Iravani, Green Chem., 2011, 13, 2638 RSC.
- A. K. Jha, K. Prasad, K. Prasad and A. R. Kulkarni, Colloids Surf., B, 2009, 73, 219–223 CrossRef CAS PubMed.
- Y. Park, Y. N. Hong, A. Weyers, Y. S. Kim and R. J. Linhardt, IET Nanobiotechnol., 2011, 5, 69–78 CrossRef CAS PubMed.
- M. H. El-Rafie, M. E. El-Naggar, M. A. Ramadan, M. M. G. Fouda, S. S. Al-Deyab and A. Hebeish, Carbohydr. Polym., 2011, 86, 630–635 CrossRef CAS.
- X. Gao, L. Wei, H. Yan and B. Xu, Mater. Lett., 2011, 65, 2963–2965 CrossRef CAS.
- M. M. Kemp, A. Kumar, S. Mousa, E. Dyskin, M. Yalcin, P. Ajayan, R. J. Linhardt and S. A. Mousa, Nanotechnology, 2009, 20, 455104 CrossRef PubMed.
- J. Koller, Drugs Exp. Clin. Res., 2004, 30, 183–190 CAS.
- I. K. Sen, A. K. Mandal, S. Chakraborti, B. Dey, R. Chakraborty and S. S. Islam, Int. J. Biol. Macromol., 2013, 62, 439–449 CrossRef CAS PubMed.
- D. K. Manna, A. K. Mandal, I. K. Sen, P. K. Maji, S. Chakraborti, R. Chakraborty and S. S. Islam, Int. J. Biol. Macromol., 2015, 80, 455–459 CrossRef CAS PubMed.
- Z. Lu, K. Rong, J. Li, H. Yang and R. Chen, J. Mater. Sci.: Mater. Med., 2013, 24, 1465–1471 CrossRef CAS PubMed.
- S. Pal, Y. K. Tak and J. M. Song, Appl. Environ. Microbiol., 2007, 73, 1712–1720 CrossRef CAS PubMed.
- M. A. Raza, Z. Kanwal, A. Rauf, A. N. Sabri, S. Riaz and S. Naseem, Nanomaterials, 2016, 6, 74 CrossRef PubMed.
- M. J. Yacamán, J. A. Ascencio, H. B. Liu and J. Gardea-Torresdey, J. Vac. Sci. Technol., B: Microelectron. Nanometer Struct., 2001, 19, 1091 CrossRef.
- A. Panáček, L. Kvítek, R. Prucek, M. Kolář, R. Večeřová, N. Pizúrová, V. K. Sharma, T. Nevěčná and R. Zbořil, J. Phys. Chem. B, 2006, 110, 16248–16253 CrossRef PubMed.
- J. Li, K. Rong, H. Zhao, F. Li, Z. Lu and R. Chen, J. Nanosci. Nanotechnol., 2013, 13, 6806–6813 CrossRef PubMed.
- A. Ivask, I. Kurvet, K. Kasemets, I. Blinova, V. Aruoja, S. Suppi, H. Vija, A. Käkinen, T. Titma, M. Heinlaan, M. Visnapuu, D. Koller, V. Kisand and A. Kahru, PLoS One, 2014, 9, e102108 CrossRef PubMed.
- M. R. El-Zahry, A. Mahmoud, I. H. Refaat, H. A. Mohamed, H. Bohlmann and B. Lendl, Talanta, 2015, 138, 183–189 CrossRef CAS PubMed.
- A. Alshareef, K. Laird and R. B. M. Cross, Appl. Surf. Sci., 2017, 424, 310–315 CrossRef CAS.
- P. Van Dong, C. H. Ha, L. T. Binh and J. Kasbohm, Int. Nano Lett., 2012, 2, 9 CrossRef.
- A. Ahmad, Y. Wei, F. Syed, K. Tahir, A. U. Rehman, A. Khan, S. Ullah and Q. Yuan, Microb. Pathog., 2017, 102, 133–142 CrossRef CAS PubMed.
- S. Pal, Y. K. Tak and J. M. Song, Appl. Environ. Microbiol., 2007, 73, 1712–1720 CrossRef CAS PubMed.
- S. Chen and D. L. Carroll, Nano Lett., 2002, 2, 1003–1007 CrossRef CAS.
- L. Actis, A. Srinivasan, J. L. Lopez-Ribot, A. K. Ramasubramanian and J. L. Ong, J. Mater. Sci.: Mater. Med., 2015, 26, 215 CrossRef PubMed.
- A. Parveen, M. S. Yalagatti, V. Abbaraju and R. Deshpande, J. Drug Delivery, 2018, 2018, 1–10 CrossRef PubMed.
- P. Van Viet, T. T. Sang, N. H. N. Bich and C. M. Thi, J. Photochem. Photobiol., B, 2018, 182, 108–114 CrossRef PubMed.
- J. S. Kim, E. Kuk, K. N. Yu, J.-H. Kim, S. J. Park, H. J. Lee, S. H. Kim, Y. K. Park, Y. H. Park, C.-Y. Hwang, Y.-K. Kim, Y.-S. Lee, D. H. Jeong and M.-H. Cho, Nanomedicine, 2007, 3, 95–101 CrossRef CAS PubMed.
- E. Pazos-Ortiz, J. H. Roque-Ruiz, E. A. Hinojos-Márquez, J. López-Esparza, A. Donohué-Cornejo, J. C. Cuevas-González, L. F. Espinosa-Cristóbal and S. Y. Reyes-López, J. Nanomater., 2017, 2017, 1–9 CrossRef.
- D. Mandal, S. Kumar Dash, B. Das, S. Chattopadhyay, T. Ghosh, D. Das and S. Roy, Biomed. Pharmacother., 2016, 83, 548–558 CrossRef CAS PubMed.
- K. Shameli, M. Bin Ahmad, E. A. Jaffar Al-Mulla, N. A. Ibrahim, P. Shabanzadeh, A. Rustaiyan, Y. Abdollahi, S. Bagheri, S. Abdolmohammadi, M. S. Usman and M. Zidan, Molecules, 2012, 17, 8506–8517 CrossRef CAS PubMed.
- P. Phanjom and G. Ahmed, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2017, 8, 045016 Search PubMed.
- A. M. El Badawy, R. G. Silva, B. Morris, K. G. Scheckel, M. T. Suidan and T. M. Tolaymat, Environ. Sci. Technol., 2011, 45, 283–287 CrossRef CAS PubMed.
- S. Ashraf, N. Akhtar, M. Ghauri, M. Rajoka, Z. M. Khalid and I. Hussain, Nanoscale Res. Lett., 2012, 7, 267 CrossRef PubMed.
- K. Chitra and G. Annadurai, BioMed Res. Int., 2014, 2014, 1–6 CrossRef PubMed.
- H. Mu, J. Tang, Q. Liu, C. Sun, T. Wang and J. Duan, Sci. Rep., 2016, 6, 18877 CrossRef CAS PubMed.
- T. S. Peretyazhko, Q. Zhang and V. L. Colvin, Environ. Sci. Technol., 2014, 48, 11954–11961 CrossRef CAS PubMed.
- A. P. V. Ferreyra Maillard, P. R. Dalmasso, B. A. López de Mishima and A. Hollmann, Colloids Surf., B, 2018, 171, 320–326 CrossRef CAS PubMed.
- T. C. Dakal, A. Kumar, R. S. Majumdar and V. Yadav, Front. Microbiol., 2016, 7, 1831 Search PubMed.
- N. Durán, M. Durán, M. B. de Jesus, A. B. Seabra, W. J. Fávaro and G. Nakazato, Nanomedicine, 2016, 12, 789–799 CrossRef PubMed.
- Y. N. Slavin, J. Asnis, U. O. Häfeli and H. Bach, J. Nanobiotechnol., 2017, 15, 65 CrossRef PubMed.
- M. T. Madigan, J. M. Martinko, K. Bender, D. P. Buckley and D. A. Stahl, Brock Biology of Microorganisms Plus Masteringmicrobiology With Etext, Benjamin-Cummings Pub Co, 2014 Search PubMed.
- T. J. Silhavy, D. Kahne and S. Walker, Cold Spring Harbor Perspect. Biol., 2010, 2, a000414 Search PubMed.
- V. Gopinath, S. Priyadarshini, M. F. Loke, J. Arunkumar, E. Marsili, D. MubarakAli, P. Velusamy and J. Vadivelu, Arabian J. Chem., 2017, 10, 1107–1117 CrossRef CAS.
- Q. Feng, J. Wu and G. Chen, J. Biomed. Mater. Res., 2000, 52, 662–668 CrossRef CAS PubMed.
- T. J. Beveridge, J. Bacteriol., 1999, 181, 4725–4733 CAS.
- A. Abbaszadegan, Y. Ghahramani, A. Gholami, B. Hemmateenejad, S. Dorostkar, M. Nabavizadeh and H. Sharghi, J. Nanomater., 2015, 2015, 1–8 CrossRef.
- B. Ramalingam, T. Parandhaman and S. K. Das, ACS Appl. Mater. Interfaces, 2016, 8, 4963–4976 CrossRef CAS PubMed.
- E. Z. Gomaa, J. Gen. Appl. Microbiol., 2017, 63, 36–43 CrossRef CAS PubMed.
- B. Das, S. Dash, D. Mandal, J. Adhikary, S. Chattopadhyay, S. Tripathy, A. Dey, S. Manna, S. Dey, D. Das and S. Roy, BLDE Univ. J. Health Sci., 2016, 1, 89 CrossRef.
- C. Krishnaraj, E. G. Jagan, S. Rajasekar, P. Selvakumar, P. T. Kalaichelvan and N. Mohan, Colloids Surf., B, 2010, 76, 50–56 CrossRef CAS PubMed.
- T. A. Abalkhil, S. A. Alharbi, S. H. Salmen and M. Wainwright, Biotechnol. Biotechnol. Equip., 2017, 31, 411–417 CrossRef CAS.
- Y.-G. Yuan, Q.-L. Peng and S. Gurunathan, Int. J. Mol. Sci., 2017, 18, 569 CrossRef PubMed.
- M. A. Ansari and M. A. Alzohairy, Evid.-Based Complementary Altern. Med., 2018, 2018, 1–9 CrossRef PubMed.
- J. S. McQuillan, H. Groenaga Infante, E. Stokes and A. M. Shaw, Nanotoxicology, 2012, 6, 857–866 CrossRef CAS PubMed.
- S. Ninganagouda, V. Rathod, D. Singh, J. Hiremath, A. K. Singh, J. Mathew and M. ul-Haq, BioMed Res. Int., 2014, 2014, 753419 Search PubMed.
- B. Chopade, S. Ghosh, M. Patil, R. Ahire, S. Kitture, K. Jabgunde, S. S. Kale, J. Pardesi, D. D. Cameotra, A. Bellare and B. A. Dhavale, Int. J. Nanomed., 2012, 7, 483 CrossRef PubMed.
- S. Y. Liau, D. C. Read, W. J. Pugh, J. R. Furr and A. D. Russell, Lett. Appl. Microbiol., 1997, 25, 279–283 CrossRef CAS PubMed.
- K. B. Holt and A. J. Bard, Biochemistry, 2005, 44, 13214–13223 CrossRef CAS PubMed.
- H. S. Toh, C. Batchelor-McAuley, K. Tschulik and R. G. Compton, Sci. China: Chem., 2014, 57, 1199–1210 CrossRef CAS.
- C.-N. Lok, C.-M. Ho, R. Chen, Q.-Y. He, W.-Y. Yu, H. Sun, P. K.-H. Tam, J.-F. Chiu and C.-M. Che, J. Proteome Res., 2006, 5, 916–924 CrossRef CAS PubMed.
- J. S. McQuillan and A. M. Shaw, Nanotoxicology, 2014, 8, 177–184 CrossRef CAS PubMed.
- S. K. Gogoi, P. Gopinath, A. Paul, A. Ramesh, S. S. Ghosh and A. Chattopadhyay, Langmuir, 2006, 22, 9322–9328 CrossRef CAS PubMed.
- Y.-H. Hsueh, K.-S. Lin, W.-J. Ke, C.-T. Hsieh, C.-L. Chiang, D.-Y. Tzou and S.-T. Liu, PLoS One, 2015, 10, e0144306 CrossRef PubMed.
- O. Bondarenko, A. Ivask, A. Käkinen, I. Kurvet and A. Kahru, PLoS One, 2013, 8, e64060 CrossRef CAS PubMed.
- S. Vishnupriya, K. Chaudhari, R. Jagannathan and T. Pradeep, Part. Part. Syst. Charact., 2013, 30, 1056–1062 CrossRef CAS.
- M. Rahban, A. Divsalar, A. A. Saboury and A. Golestani, J. Phys. Chem. C, 2010, 114, 5798–5803 CrossRef CAS.
- S. Pramanik, S. Chatterjee, A. Saha, P. S. Devi and G. Suresh Kumar, J. Phys. Chem. B, 2016, 120, 5313–5324 CrossRef CAS PubMed.
- S. Chowdhury, A. Basu and S. Kundu, Nanoscale Res. Lett., 2014, 9, 365 CrossRef PubMed.
- M. A. Radzig, V. A. Nadtochenko, O. A. Koksharova, J. Kiwi, V. A. Lipasova and I. A. Khmel, Colloids Surf., B, 2013, 102, 300–306 CrossRef CAS PubMed.
- P. Dutta, A. Harrison, S. Sabbani, R. S. Munson, P. K. Dutta and W. J. Waldman, Int. J. Nanomed., 2011, 6, 1833 CrossRef PubMed.
- K. Siriwardana, A. Wang, M. Gadogbe, W. E. Collier, N. C. Fitzkee and D. Zhang, J. Phys. Chem. C, 2015, 119, 2910–2916 CrossRef CAS PubMed.
- K. Poole, Efflux-mediated antimicrobial resistance, 2005, vol. 56 Search PubMed.
- X. Yan, B. He, L. Liu, G. Qu, J. Shi, L. Hu and G. Jiang, Metallomics, 2018, 10, 557–564 RSC.
- N. S. Wigginton, A. de Titta, F. Piccapietra, J. Dobias, V. J. Nesatyy, M. J. F. Suter and R. Bernier-Latmani, Environ. Sci. Technol., 2010, 44, 2163–2168 CrossRef CAS PubMed.
- Z. Yuan, J. Li, L. Cui, B. Xu, H. Zhang and C.-P. Yu, Chemosphere, 2013, 90, 1404–1411 CrossRef CAS PubMed.
- T. S. Radniecki, D. P. Stankus, A. Neigh, J. A. Nason and L. Semprini, Chemosphere, 2011, 85, 43–49 CrossRef CAS PubMed.
- M. Yamanaka, K. Hara and J. Kudo, Appl. Environ. Microbiol, 2005, 71, 7589–7593 CrossRef CAS PubMed.
- M. Schieber and N. S. Chandel, Curr. Biol., 2014, 24, R453–R462 CrossRef CAS PubMed.
- S. K. Verma, E. Jha, B. Sahoo, P. K. Panda, A. Thirumurugan, S. K. S. Parashar and M. Suar, RSC Adv., 2017, 7, 40034–40045 RSC.
- H. Fu, J. Yuan and H. Gao, Arch. Biochem. Biophys., 2015, 584, 28–35 CrossRef CAS PubMed.
- S. B. Farr and T. Kogoma, Microbiol. Rev., 1991, 55, 561–585 CAS.
- Y. Zhang, D. Meng, Z. Wang, H. Guo and Y. Wang, FEMS Microbiol. Lett., 2012, 334, 95–101 CrossRef CAS PubMed.
- W. Yang, C. Shen, Q. Ji, H. An, J. Wang, Q. Liu and Z. Zhang, Nanotechnology, 2009, 20, 085102 CrossRef PubMed.
- L. Zou, J. Wang, Y. Gao, X. Ren, M. E. Rottenberg, J. Lu and A. Holmgren, Sci. Rep., 2018, 8, 11131 CrossRef PubMed.
- B. Das, S. K. Dash, D. Mandal, T. Ghosh, S. Chattopadhyay, S. Tripathy, S. Das, S. K. Dey, D. Das and S. Roy, Arabian J. Chem., 2017, 10, 862–876 CrossRef CAS.
- S. Belluco, C. Losasso, I. Patuzzi, L. Rigo, D. Conficoni, F. Gallocchio, V. Cibin, P. Catellani, S. Segato and A. Ricci, Front. Microbiol., 2016, 7, 307 Search PubMed.
- K. Soo-Hwan, H. Seon Lee, D. Seon Ryu, S. Jae Choi and D. Seok Lee, Korean J. Microbiol. Biotechnol., 2011, 39, 77–85 Search PubMed.
- P. Korshed, L. Li, Z. Liu and T. Wang, PLoS One, 2016, 11, e0160078 CrossRef PubMed.
- M. A. Quinteros, V. Cano Aristizábal, P. R. Dalmasso, M. G. Paraje and P. L. Páez, Toxicol. In Vitro, 2016, 36, 216–223 CrossRef CAS PubMed.
- D. Soni, A. Bafana, D. Gandhi, S. Sivanesan and R. A. Pandey, Environ. Toxicol. Chem., 2014, 33, 2126–2132 CrossRef CAS PubMed.
- M. Y. Galperin, Environ. Microbiol., 2004, 6, 552–567 CrossRef CAS PubMed.
- A. Kobir, L. Shi, A. Boskovic, C. Grangeasse, D. Franjevic and I. Mijakovic, Biochim. Biophys. Acta, Gen. Subj., 2011, 1810, 989–994 CrossRef CAS PubMed.
- S. Shrivastava, T. Bera, A. Roy, G. Singh, P. Ramachandrarao and D. Dash, Nanotechnology, 2007, 18, 225103 CrossRef.
- A. Roy, S. L. Shrivastava, S. M. Mandal, S. K. Roy and S. K. Srivastava, Adv. Sci., Eng. Med., 2015, 7, 465–472 CrossRef CAS.
- M. Yamanaka, K. Hara and J. Kudo, Appl. Environ. Microbiol., 2005, 71, 7589–7593 CrossRef CAS PubMed.
- W. K. Jung, H. C. Koo, K. W. Kim, S. Shin, S. H. Kim and Y. H. Park, Appl. Environ. Microbiol., 2008, 74, 2171–2178 CrossRef CAS PubMed.
- D. P. Li, Z. Zheng, Y. Lei, F. L. Yang, S. X. Ge, Y. D. Zhang, B. J. Huang, Y. H. Gao, K. W. Wong and W. M. Lau, Chem.–Eur. J., 2011, 17, 7694–7700 CrossRef CAS PubMed.
- H. Dong, Q. Xiang, Y. Gu, Z. Wang, N. G. Paterson, P. J. Stansfeld, C. He, Y. Zhang, W. Wang and C. Dong, Nature, 2014, 511, 52–56 CrossRef CAS PubMed.
- L. Scorzoni, A. C. A. de Paula e Silva, C. M. Marcos, P. A. Assato, W. C. M. A. de Melo, H. C. de Oliveira, C. B. Costa-Orlandi, M. J. S. Mendes-Giannini and A. M. Fusco-Almeida, Front. Microbiol., 2017, 08, 36 Search PubMed.
- C. G. Kumar and Y. Poornachandra, Colloids Surf., B, 2015, 125, 110–119 CrossRef CAS PubMed.
- K. Tutaj, R. Szlazak, K. Szalapata, J. Starzyk, R. Luchowski, W. Grudzinski, M. Osinska-Jaroszuk, A. Jarosz-Wilkolazka, A. Szuster-Ciesielska and W. I. Gruszecki, Nanomedicine, 2016, 12, 1095–1103 CrossRef CAS PubMed.
- A. Ahmad, Y. Wei, F. Syed, K. Tahir, R. Taj, A. U. Khan, M. U. Hameed and Q. Yuan, Microb. Pathog., 2016, 99, 271–281 CrossRef CAS PubMed.
- K. Ishida, T. F. Cipriano, G. M. Rocha, G. Weissmüller, F. Gomes, K. Miranda and S. Rozental, Mem. Inst. Oswaldo Cruz, 2014, 109, 220–228 CrossRef PubMed.
- N. T. Khan and M. Mushtaq, Biol. Med., 2017, 09, 1–4 Search PubMed.
- M. Wypij, J. Czarnecka, H. Dahm, M. Rai and P. Golinska, J. Basic Microbiol., 2017, 57, 793–800 CrossRef CAS PubMed.
- A. H. A. Kelkawi, A. Abbasi Kajani and A.-K. Bordbar, IET Nanobiotechnol., 2017, 11, 370–376 CrossRef PubMed.
- M. Bhagat, R. Anand, R. Datt, V. Gupta and S. Arya, J. Inorg. Organomet. Polym. Mater., 2018, 1–9 CAS.
- Y.-K. Jo, B. H. Kim and G. Jung, Plant Dis., 2009, 93, 1037–1043 CrossRef CAS.
- S. M. Ouda, Res. J. Microbiol., 2014, 9, 34–42 CrossRef CAS.
- S. M. Ali, N. M. H. Yousef and N. A. Nafady, J. Nanomater., 2015, 2015, 1–10 Search PubMed.
- A. M. Elgorban, A. E.-R. M. El-Samawaty, M. A. Yassin, S. R. Sayed, S. F. Adil, K. M. Elhindi, M. Bakri and M. Khan, Biotechnol. Biotechnol. Equip., 2016, 30, 56–62 CrossRef CAS.
- H. J. C. de Vries, S. H. Reedijk and H. D. F. H. Schallig, Am. J. Clin. Dermatol., 2015, 16, 99–109 CrossRef PubMed.
- A. Ahmad, Y. Wei, F. Syed, S. Khan, G. M. Khan, K. Tahir, A. U. Khan, M. Raza, F. U. Khan and Q. Yuan, J. Photochem. Photobiol., B, 2016, 161, 17–24 CrossRef CAS PubMed.
- A. Allahverdiyev, E. Ş. Abamor, M. Bagirova, C. B. Ustundag, C. Kaya, F. Kaya and M. Rafailovich, Int. J. Nanomed., 2011, 6, 2705 CrossRef CAS PubMed.
- S. K. Kalangi, A. Dayakar, D. Gangappa, R. Sathyavathi, R. S. Maurya and D. Narayana Rao, Exp. Parasitol., 2016, 170, 184–192 CrossRef CAS PubMed.
- A. A. Zahir, I. S. Chauhan, A. Bagavan, C. Kamaraj, G. Elango, J. Shankar, N. Arjaria, S. M. Roopan, A. A. Rahuman and N. Singh, Antimicrob. Agents Chemother., 2015, 59, 4782–4799 CrossRef CAS PubMed.
- A. Ahmad, F. Syed, A. Shah, Z. Khan, K. Tahir, A. U. Khan and Q. Yuan, RSC Adv., 2015, 5, 73793–73806 RSC.
- L. C. S. Lopes, L. M. Brito, T. T. Bezerra, K. N. Gomes, F. A. D. A. Carvalho, M. H. Chaves and W. Cantanhêde, An. Acad. Bras. Cienc., 2018, 90, 2679–2689 Search PubMed.
- M. El-khadragy, E. Alolayan, D. Metwally, M. El-Din, S. Alobud, N. Alsultan, S. Alsaif, M. Awad and A. Abdel Moneim, Int. J. Environ. Res. Public Health, 2018, 15, 1037 CrossRef PubMed.
- M. Ovais, A. T. Khalil, A. Raza, N. U. Islam, M. Ayaz, M. Saravanan, M. Ali, I. Ahmad, M. Shahid and Z. K. Shinwari, Appl. Microbiol. Biotechnol., 2018, 102, 4393–4408 CrossRef CAS PubMed.
- J. R. Fanti, F. Tomiotto-Pellissier, M. M. Miranda-Sapla, A. H. D. Cataneo, C. G. T. de J. Andrade, C. Panis, J. H. da S. Rodrigues, P. F. Wowk, D. Kuczera, I. N. Costa, C. V. Nakamura, G. Nakazato, N. Durán, W. R. Pavanelli and I. Conchon-Costa, Acta Trop., 2018, 178, 46–54 CrossRef CAS PubMed.
- I. Ullah, G. Cosar, E. S. Abamor, M. Bagirova, Z. K. Shinwari and A. M. Allahverdiyev, 3 Biotech, 2018, 8, 98 CrossRef PubMed.
- J. S. Kim, T.-J. Yoon, K. N. Yu, B. G. Kim, S. J. Park, H. W. Kim, K. H. Lee, S. B. Park, J.-K. Lee and M. H. Cho, Toxicol. Sci., 2006, 89, 338–347 CrossRef CAS PubMed.
- D. E. Gorka, J. S. Osterberg, C. A. Gwin, B. P. Colman, J. N. Meyer, E. S. Bernhardt, C. K. Gunsch, R. T. DiGulio and J. Liu, Environ. Sci. Technol., 2015, 49, 10093–10098 CrossRef CAS PubMed.
- J.-J. Lin, W.-C. Lin, R.-X. Dong and S. Hsu, Nanotechnology, 2012, 23, 065102 CrossRef PubMed.
- K. C. Nguyen, V. L. Seligy, A. Massarsky, T. W. Moon, P. Rippstein, J. Tan and A. F. Tayabali, J. Phys.: Conf. Ser., 2013, 429, 012025 CrossRef.
- Y. Cheng, L. Yin, S. Lin, M. Wiesner, E. Bernhardt and J. Liu, J. Phys. Chem. C, 2011, 115, 4425–4432 CrossRef CAS.
- I. M. Pongrac, L. B. Ahmed, H. Mlinarić, D. D. Jurašin, I. Pavičić, A. M. Marjanović Čermak, M. Milić, S. Gajović and I. Vinković Vrček, J. Trace Elem. Med. Biol., 2018, 50, 684–692 CrossRef CAS PubMed.
- L. Brkić Ahmed, M. Milić, I. M. Pongrac, A. M. Marjanović, H. Mlinarić, I. Pavičić, S. Gajović and I. Vinković Vrček, Food Chem. Toxicol., 2017, 107, 349–361 CrossRef PubMed.
- M. Beg, A. Maji, A. K. Mandal, S. Das, M. N. Aktara, P. K. Jha and M. Hossain, J. Mol. Recognit., 2017, 30, e2565 CrossRef PubMed.
- B. Venkatesan, V. Subramanian, A. Tumala and E. Vellaichamy, Asian Pac. J. Trop. Med., 2014, 7, S294–S300 CrossRef CAS.
- K. Jadhav, S. Deore, D. Dhamecha, H. R. Rajeshwari, S. Jagwani, S. Jalalpure and R. Bohara, ACS Biomater. Sci. Eng., 2018, 4, 892–899 CrossRef CAS.
Footnotes |
| † Dedicated to Prof. Anil Bhuimali, Hon'ble Vice-Chancellor Raiganj University on the occasion of his 59th birthday. |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |





