Open Access Article
Open Access ArticlePhospholipase D encapsulated into metal-surfactant nanocapsules for enhancing biocatalysis in a two-phase system
Hui Liab,
Yang Pangab,
Xin Wangab,
Xun Caoab,
Xun Heab,
Kequan Chen *ab,
Ganlu Li*ab,
Pingkai Ouyangab and
Weiming Tanc
*ab,
Ganlu Li*ab,
Pingkai Ouyangab and
Weiming Tanc
aCollege of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing 210000, China. E-mail: kqchen@njtech.edu.cn; tkscb@163.com
bState Key Laboratory of Materials-Oriented Chemical Engineering, Nanjing, 210000, China
cNational Engineering Research Center for Coatings, CNOOC Changzhou Paint and Coatings Industry Research Institute Co., Ltd, Changzhou 213016, P. R. China
First published on 25th February 2019
Abstract
Methods for enhancing enzyme activities in two-phase systems are getting more attention. Phospholipase D (PLD) was successfully encapsulated into metal-surfactant nanocapsules (MSNCs) using a one-pot self-assembly technique in an aqueous solution. The highest yield for the production of high-value phosphatidylserine (PS) from low-value phosphatidylcholine (PC) in the two-phase system was achieved by encapsulating PLD into MSNCs formed from Ca2+ which gave an enzyme activity that was 133.6% of that of free PLD. The PLD@MSNC transformed the two-phase system into an emulsion phase system and improved the organic solvent tolerance, pH and thermal stabilities as well as the storage stability and reusability of the enzyme. Under optimal conditions, PLD@MSNC generated 91.9% PS over 8 h in the two-phase system, while free PLD generated only 77.5%.
1. Introduction
Biocatalytic reactions of enzymes are mostly conducted in aqueous media in the laboratory and in industrial applications. However, there are still many enzymatic catalytic reactions that need to be carried out in organic-aqueous two-phase systems.1,2 Two-phase systems increase the solubility of hydrophobic substrates and products and thus allow a possible shift of the reaction equilibrium.3 Moreover, two-phase systems can reduce the difficulty of product separation because the catalyst and products are present in distinct liquid phases.4 For these reasons, the development of enzymatic catalysis in two-phase systems is of great interest for fine chemical and pharmaceutical processes.5 However, unspecific binding of organic solvents to enzymes can promote unfolding or aggregation of the enzymes, which leads to irreversible inactivation. Organic solvents can also bind non-covalently to the substrate binding site or the substrate entrance channel of the enzyme, resulting in competitive inhibition.6,7 Moreover, free enzymes are difficult to recycle, resulting in low efficiency and high cost.8,9 Nanoscale immobilization of an enzyme can increase its stability in organic solvents while retaining a high enzyme activity.10,11 However, metal organic frameworks and nanoflowers have very strong rigid structures which has limited the structural variability of enzymes.12Bile salts, a class of surfactant molecules, are chiral building blocks that can be used in unidirectional self-aggregation reactions with divalent metal ions, leading to supramolecular nanocomposites.13,14 The carboxyl group of bile salts and divalent metal ions form metal coordination bonds in an aqueous solution. Metal-surfactant nanocapsules have flexible structures, which alleviate restrictions on enzyme structures. Surfactants can also enhance interfacial catalytic activity by interacting with protein secondary structure elements, thereby increasing the stability of the enzyme.15–17 Cao et al. reported that sodium deoxycholate (NaDC) with Co2+ can be used to immobilize lipase and enhance the enzyme activity in highly polar solvents.18 Bile salts can also transform a two-phase reaction system into an emulsifying reaction system, enhancing the mass transfer and solubility of substrates and products. Phospholipase D (PLD) is often used to catalyze the transphosphatidylation of low-value phosphatidylcholine (PC) to high-value phosphatidylserine (PS) at two-phase interfaces.19 PS is a membrane phospholipid component with broad applications in the pharmaceutical and functional food industries.20 However, because naturally-occurring PS is limited, the need for biosynthesis of PS is significant. Many researchers have attempted to reduce the cost of PS biosynthesis by improving the efficiency of PLD utilization.21,22
In this study, a two-phase system was used to biosynthesize PS from PC. PLD was encapsulated into metal-surfactant nanocapsules (MSNCs) and the biocatalytic activities for the conversion of PC to PS by both free PLD and encapsulated PLD (PLD@MSNC) at the two-phase interfaces were measured. PLD@MSNC transformed the two-phase reaction system of free PLD into an emulsion system of immobilized PLD, thus increasing the efficiency of transphosphatidylation. This approach is a promising way to improve the efficiency of enzyme utilization, prolong the lifetime of enzymes, and so reduce biocatalytic cost.
2. Materials and methods
2.1. Materials
Sodium deoxycholate (NaDC), L-serine, phosphatidylserine (PS) and phosphatidylcholine (PC) were purchased from Aladdin Ind. Co., Ltd. (China). Phospholipase D (PLD, EC 3.1.4.4) was prepared by our laboratory. CoCl2·6H2O, CaCl2, MgCl2·6H2O, NiCl2·6H2O, and ZnCl2 were purchased from Xilong Chemical Company (China). Dichloromethane, acetic acid and triethylamine were purchased from Sinopharm Chemical Reagent Co. (China). All chemicals used in this work were of analytical grade and were used without further purification.2.2. Preparation of phospholipase D encapsulated into metal-surfactant nanocapsules (PLD@MSNC)
In a typical experiment, free PLD (final concentration 0.6 mg mL−1) and sodium deoxycholate (NaDC, final concentration 10 mM) were added into a 50 mL conical flask in 10 mL deionized water. The mixture was stirred for 15 min at room temperature, and then 10 mL of 20 mM metal ion solution (Co2+, Ca2+, Mg2+, Ni2+, Zn2+) was added and the mixture was stirred again for 30 min at room temperature. After the reaction, the mixture was centrifuged for 15 min at 4 °C (7000g) to obtain supernatant and precipitate, and then the precipitate was washed two times with deionized water in order to remove excess material, and dried to a constant weight using a freezing vacuum (LGJ-10D, China) for 24 h. Finally, PLD-encapsulated metal-surfactant nanocapsules (PLD@MSNC) were obtained. The protein content of PLD was determined according to the Bradford method with bovine serum albumin as a standard.2.3. Measurements of free PLD and PLD@MSNC activity
The catalytic activities of free PLD and PLD@MSNC were assayed on the basis of the PLD-mediated transphosphatidylation of PC for the synthesis of PS, based on the method of Li et al. with modifications.22 Free PLD (0.3 mL; 0.85 mg protein, with and without 10 mM Co2+, Ca2+, Mg2+, Ni2+, or Zn2+) or PLD@MSNC (10 mg, equivalent protein) solution was added into 0.7 mL acetic acid-sodium acetate buffer (0.1 M, pH 5.5) containing 13 mM L-serine at 30 °C. The reaction was then initiated by adding 5 mL dichloromethane containing 0.22 mM PC at 30 °C, and incubated at 200 rpm and 30 °C for 120 min. The concentration of PS was detected by HPLC. One unit (U) of transphosphatidylation activity of PLD was defined as the amount of enzyme that produced 1 μmol of PS per minute under these assay conditions.2.4. Characterizations of PLD@MSNC
Scanning electron microscopy (SEM) was performed using a Hitachi S-3400 II (Japan) at an accelerating voltage of 3 kV. A surface area and pore size analyzer (Micromeritics 3Flex, USA) was employed to measure the surface areas and pore sizes of the different samples. The chemical functional groups were analyzed using a Nicolet IS5 FTIR spectrophotometer (USA). Thermogravimetric analysis (TGA) was performed using a TA Q500 (USA) in a nitrogen environment at a heating rate of 10 °C min−1 over the temperature range of 25–800 °C. The emulsifying properties of NaDC, MSNC and PLD@MSNC samples were measured using the volume of the aqueous phase, emulsion phase, and organic phase. All samples contained aqueous solutions comprising different concentrations of samples and equal volumes of lauric acid methyl ester; solutions were oscillated for 60 min to mix evenly, and then kept still for 60 min. The volume fractions of the aqueous phase, emulsion phase, and organic phase were determined for each sample.2.5. Effect of organic solvent, temperature and pH on the PLD-catalyzed synthesis of PS
Free PLD (0.85 mg protein) or PLD@MSNC (10 mg, equivalent protein) was incubated in 5 mL of a 50% solution of each examined organic solvent (ethyl acetate, cyclohexane, n-butanol, dichloromethane, chloroform, and methyl-tert-butyl ether) in the described reaction system to form PS. Free PLD or PLD@MSNC were incubated in the above reaction system to form PS at a range of temperatures (20–40 °C) and pH values (4.5–8.5) for 4 h.2.6. Kinetic study of the PLD-catalyzed synthesis of PS
Free PLD (0.85 mg protein) or PLD@MSNC (10 mg, equivalent protein) was incubated in the two-phase system or in acetic acid-sodium acetate buffer (0.1 M, pH 5.5) to synthesize PS at PC concentrations of 10 or 30 g L−1 for 24 h. The reaction mixtures were sampled every 4 h. Samples (10 μL) were taken from each reaction system and analyzed using HPLC.2.7. Storage stability of PLD@MSNC
Free PLD and PLD@MSNC were stored at 4 °C and at room temperature for 28 consecutive days. Every 4 days, samples of the free PLD and PLD@MSNC were incubated in the described reaction system to synthesize PS over a 4 h period, with a PC concentration of 10 g L−1.2.8. Reusability of PLD@MSNC
PLD@MSNC was incubated in the described reaction system to synthesize PS over a 4 h period, with a PC concentration of 10 g L−1. PLD@MSNC was recovered through centrifugation and washed and dried after each PS synthesis reaction. The recovered PLD@MSNC was used for the next batch of PS synthesis. This procedure was repeated eight times to determine the reusability of PLD@MSNC for PS synthesis reaction in a two-phase system.2.9. High-performance liquid chromatography (HPLC)
Samples were analyzed using an Agilent 1260 HPLC (USA) equipped with a Chromachem evaporative light scattering detector (ELSD). The samples from the two-phase system were centrifuged five times at 7000g for 15 min, and the organic phase was retained each time. The extracts were pooled for analysis by HPLC. The samples in aqueous solution were extracted with dichloromethane, centrifuged at 7000g for 15 min, and the organic phase was retained. This was repeated 5 times and the organic extracts were analyzed by HPLC. HPLC separation was performed on a ZORBAX Rx-SIL silica gel column (5 μm, 4.6 × 250 mm, Agilent). Mobile phase A contained 85% methanol, 14.5% water, 0.45% acetic acid, and 0.05% triethylamine; mobile phase B contained 20% n-hexane, 48% isopropanol, and 32% mobile phase A. The flow rate was 1.0 mL min−1. The elution conditions were as follows: initially 2% A; 5 min, 10% A; 9 min, 30% A; 14 min, 10% A; 17 min, 2% A. The column temperature, nebulizing temperature, and evaporating temperature were controlled at 38 °C, 72 °C, and 72 °C, respectively, and nitrogen was used as the nebulizing gas. The nitrogen gas flow rate was 2.0 SLM. Each phospholipid was determined from the retention time using calibration solutions of phospholipids, and the concentrations of the phospholipids in the samples were calculated from the peak areas by integration.3. Results and discussion
3.1. Synthesis and structural characterization of PLD@MSNC
NaDC is a food-grade surfactant23 that can self-assemble into a nanocomposite in the presence of divalent metal ions in aqueous solutions to encapsulate protein. Metal ions have important effects on enzyme activity.24,25 In this study, Co2+, Ca2+, Mg2+, Ni2+, and Zn2+ were tested for their ability to form nanocomposites with NaDC and to encapsulate PLD in aqueous solution (Fig. 1). It was found that Co2+, Ca2+, Mg2+, and Ni2+ promoted the activity of both free PLD and encapsulated PLD in MSNCs formed from Co2+, Ca2+, Mg2+, Ni2+, while Zn2+ displayed elevated activity for the encapsulated enzyme. This is because the metal ions acted as an activator and could enhance the activity of the enzyme26 while NaDC as the surfactant could also promote enzyme activity.27 Encapsulated PLD in MSNCs formed from Ca2+ displayed the highest specific enzyme activity of 10.51 U g−1 protein with PC as a substrate for the biosynthesis of PS, displaying 133.6% of the enzyme activity of free PLD (7.87 U g−1 protein); this was higher than the activities of PLD@MSNC prepared using other divalent metal ions. The amount of encapsulated PLD in PLD@MSNC was 85 mg g−1. Khatoon et al. reported that Ca2+ promotes transphosphatidylation of PLD and that the addition of NaDC can enhance transphosphatidylation in two-phase systems.1 Therefore, Ca2+ has two functions: one is to promote enzyme activity, the other is as a binding factor to form the nanocapsules.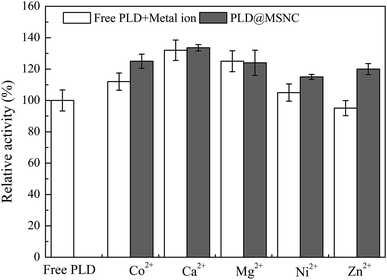 | ||
| Fig. 1 Effects of metal ions (Co2+, Ca2+, Mg2+, Ni2+, and Zn2+) on the activities of free PLD and PLD@MSNC. | ||
Morphologies of MSNC and PLD@MSNC (Fig. 2) were determined using scanning electron microscopy (SEM). The MSNCs exhibited a typical rod-like morphology with an average length of ∼1 μm and a diameter of ∼200 nm (Fig. 2a). However, supplementation with PLD changed the morphology to spherical structures of ∼200 nm in diameter or short rod structures (Fig. 2b). Because of the hydrophobic interactions of the surfactants, PLD@MSNC formed aggregates in aqueous media, but organic solvents weakened the hydrophobic forces of the surfactants and increased the dispersion of PLD@MSNC in the two-phase system (Fig. 2c), which contributed to the increase of catalytic activity. PLD encapsulation in MSNC therefore appeared to block the structural, unidirectional extension of MSNC by self-aggregation in the self-assembly process. It is possible that the PLD harbors electric charges on its surface which interact with Ca2+ to inhibit the axial growth of MSNC and convert MSNC from the typical rod-like form to a spherical form.28
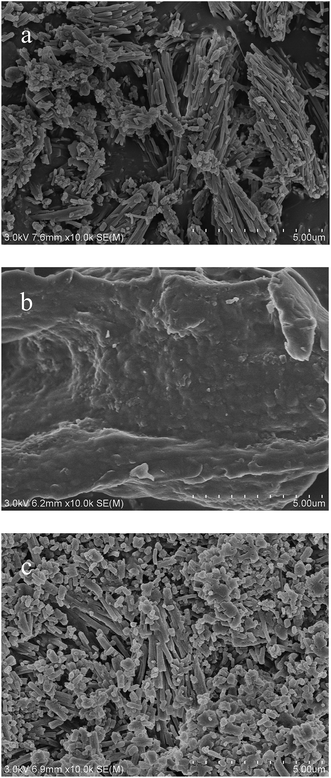 | ||
| Fig. 2 (a) Scanning electron microscopy (SEM) image of MSNCs. (b) SEM image of PLD@MSNC. (c) SEM image of PLD@MSNC in the two-phase reaction system. | ||
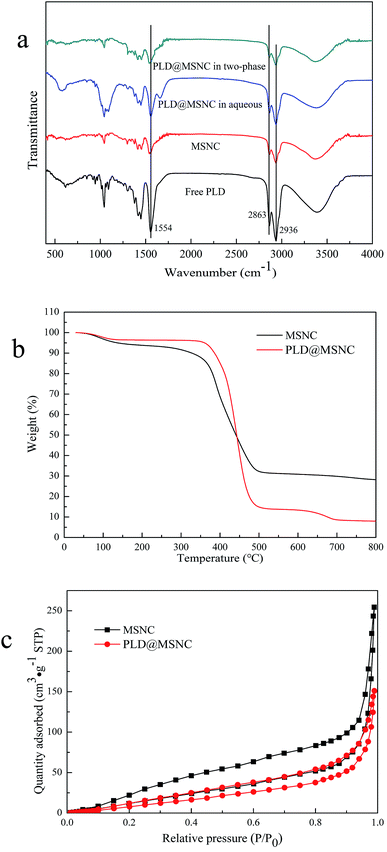
Fourier-transform infrared spectroscopy (FTIR) (Fig. 3a) confirmed the formation of complexes and the presence of protein in the nanocapsules. We observed a strong band for free PLD at 1554 cm−1 representing NH bending and CN stretching in the amide-II region; and bands at around 2863 and 2936 cm−1 which could be attributed to CH bonds. MSNCs without enzyme encapsulation had no characteristic bands of PLD. The characteristic bands of the PLD@MSNC at 1554, 2863, and 2936 cm−1 demonstrated the presence of PLD in the nanocapsules.28 The FTIR spectrum of PLD@MSNC in the two-phase reaction system was basically consistent with that of PLD@MSNC in the aqueous phase, which showed that the structure of PLD@MSNC was still intact in the two-phase reaction system. Thermogravimetric analysis (TGA) was conducted in a nitrogen atmosphere, as shown in Fig. 3b. PLD@MSNC and MSNCs exhibited different weight loss curves; PLD@MSNC endured 20% greater weight loss than MSNCs. TGA confirmed that the protein had been successfully wrapped in the nanocapsules. Nitrogen adsorption–desorption isotherms of MSNCs and PLD@MSNC were of type IV with H3 hysteresis, typical of porous materials (Fig. 3c). The H3 type hysteresis indicated that slit aperture openings were present in MSNCs and PLD@MSNC. The introduction of PLD into the MSNCs decreased the pore diameter from 9.607 to 8.741 nm, the pore volume from 0.3932 to 0.2334 cm3 g−1, and the surface area from 116.2 to 107.1 m2 g−1 (Table 1). The pores of MSNCs occupied by PLD contributed to the decreased BET surface area of PLD@MSNC.29 PLD changed the structural features of MSNCs, so that they became more conducive to mass transfer of substrates and products and thereby facilitated enzyme catalysis.30
| Samples | SBET (m2 g−1) | Pore volume (cm3 g−1) | Pore diameter (nm) |
|---|---|---|---|
| a SBET stands for BET surface area. | |||
| MSNC | 116.2 | 0.3932 | 9.607 |
| PLD@MSNC | 107.1 | 0.2334 | 8.741 |
The emulsifying properties of the different concentrations of NaDC, MSNCs, and PLD@MSNC in the two-phase system are shown in Table 2. MSNCs and PLD@MSNC had similar emulsifying properties, derived from that of NaDC. In the two-phase interface of an organic-aqueous system, organic molecules can deactivate free enzyme, while the surfactant forms a protective layer on the surface of the enzyme, preventing organic solvent from approaching the encapsulated enzyme.31 MSNCs not only provided protection to encapsulated PLD, but also facilitated the entry of substrates into PLD@MSNC.
| Samples | Concentration (g L−1) | 0 | 10 | 20 | 25 | 50 |
|---|---|---|---|---|---|---|
| NaDC | Aqueous phase (%) | 50 | 2.5 | 1.25 | 1.1 | 0 |
| Emulsion phase (%) | 0 | 60 | 63.75 | 65 | 68.75 | |
| Organic phase (%) | 50 | 37.5 | 35 | 33.9 | 31.25 | |
| MSNC | Aqueous phase (%) | 50 | 37.5 | 36.25 | 35 | 6.25 |
| Emulsion phase (%) | 0 | 13.75 | 18.75 | 21.25 | 55 | |
| Organic phase (%) | 50 | 48.75 | 45 | 43.75 | 38.75 | |
| PLD@MSNC | Aqueous phase (%) | 50 | 49 | 40 | 37.5 | 25 |
| Emulsion phase (%) | 0 | 1.25 | 12.5 | 18.5 | 35.25 | |
| Organic phase (%) | 50 | 49.75 | 47.5 | 44 | 39.75 |
3.2. Enzyme characteristics of PLD@MSNC
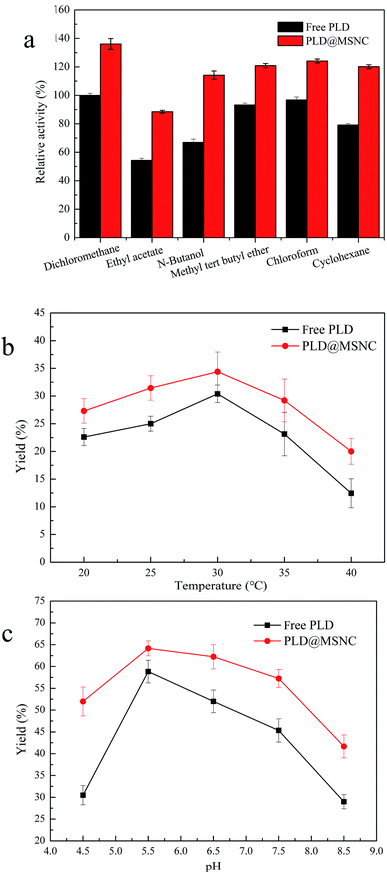
PLD@MSNC showed significantly better activity and organic solvent tolerance to all six solvents tested, compared with free PLD (Fig. 4a). The highest activity of PLD@MSNC was measured in dichloromethane. Organic solvents can damage the native structure of the enzyme, strip away protein-bound water, and rapidly deactivate the enzyme.6,7 PLD encapsulated in the MSNCs maintained its catalytic conformation and exhibited more structural rigidity due to its encapsulation, resulting in its greatly enhanced organic solvent tolerance. The temperature and pH characteristics of free PLD and PLD@MSNC were determined and the results are shown in Fig. 4b and c, respectively. The optimal temperature was 30 °C for the activities of free PLD and PLD@MSNC (Fig. 4b). However, PLD@MSNC exhibited higher activity over a broader temperature profile than did free PLD, and produced higher yields of PS across the temperature range from 20 °C to 40 °C. PLD encapsulation into the MSNCs enhanced thermal stability because it restricted changes to the protein configuration upon heating. PLD@MSNC and free PLD displayed maximum activity at pH 5.5 (Fig. 4c). However, PLD@MSNC displayed higher alkali and acid resistance over a broader pH range compared with free PLD, presumably due to the microenvironment and protective effect of the MSNCs.32 Nano-scale enzyme immobilization can maintain the structural stability and rigidity of the enzyme and endow it with superactivity.33 NaDC may change the spatial structure of PLD, while promoting an enzyme conformation that is more conducive to biocatalytic reactions.Under the optimal conditions, the effect of different concentrations of PC on PS biosynthesis and accumulation using free PLD or PLD@MSNC in the two-phase system or the aqueous system was determined (Fig. 5). Similar patterns were obtained for PS synthesis using free PLD and PLD@MSNC since both formed PS through the transphosphatidylation reaction. However, PLD@MSNC was more effective than free PLD, generating nearly 91.9% PS in 8 h at the concentration of 10 g L−1 PC, while free PLD only generated 77.5% PS. PLD@MSNC generated nearly 82.8% PS in 24 h at the concentration of 30 g L−1 PC, while free PLD only generated 60.3% PS. In addition, when PLD@MSNC was used in the aqueous system, it generated nearly 71.1% PS in 24 h at the concentration of 30 g L−1 PC which was a lower amount than that generated by the PLD@MSNC used in the two-phase system. This may be because substrates and products display better mass transfer in the two-phase reaction system. The by-product choline from the transphosphatidylation reaction could easily accumulate in proximity to free PLD resulting in its lower conversion of PC to PS.34 It is noteworthy that the relatively high yield generated by PLD@MSNC may be because PC (as a surfactant) can easily enter PLD@MSNC due to the interfacial and hydrophobic properties of PLD@MSNC. In summary, the PLD@MSNC can improve the yield of PS in three ways: (1) the surfactant increases enzyme stability and activity; (2) calcium ions promote enzyme activity; and (3) the emulsifying phase promotes mass transfer.
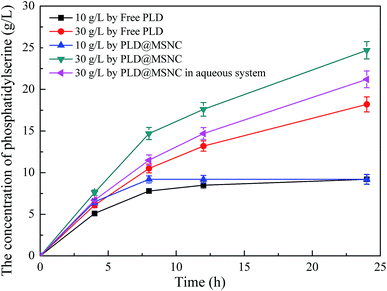 | ||
| Fig. 5 Time-dependence of biosynthesis and accumulation of PS using different concentrations of PC with free PLD or PLD@MSNC in the two-phase system and the aqueous system. | ||
3.3. Storage stability and reusability of PLD@MSNC
Free PLD and PLD@MSNC were stored at 4 °C and at room temperature for 28 days and their relative catalytic activities were detected every 4 days. The results are shown in Fig. 6. The PLD@MSNC exhibited long-term stability at 4 °C and room temperature, retaining 92.8% and 86.1%, respectively, of its initial activity after 28 days of storage. The relative enzyme activities of free PLD were 81.8% and 70.0%, respectively, of the initial activity in the same storage conditions. This drop of relative enzyme activity is probably due to the gradual inactivation of the free PLD and PLD@MSNC during storage.35 In general, in terms of storage stability, the PLD@MSNC was significantly superior to the free PLD, which increases its potential for industrial application.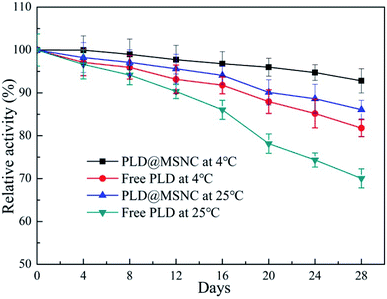 | ||
| Fig. 6 Storage stability of free PLD and PLD@MSNC at 4 °C and at room temperature over 28 consecutive days. | ||
In practical terms, the reusability of an enzyme is one of the essential factors in lowering the cost of biocatalysis. Compared with immobilized enzymes, free enzymes with high biocatalytic activity are more fragile and thus very difficult to recycle and reuse. The reusability of an enzyme increases its economic sustainability. Building on the experimental results above, the operational stability and reusability of PLD@MSNC in the two-phase system were investigated (Fig. 7a). PLD@MSNC displayed excellent operational stability in the two-phase system, and retained 54.3% of its initial activity after being used for eight cycles, highlighting the improved cost-effectiveness of the enzyme. After each cycle, the leakage of PLD from PLD@MSNC was analyzed (Fig. 7b). It was found that a small amount of PLD leaked from the PLD@MSNC after each cycle. However, the cumulative PLD loss after 8 cycles was 23.9%, which was lower than the total enzyme activity loss. Enzyme leakage, mechanical damage and organic solvent deactivation in the two-phase reaction conditions may all result in loss of activity.36 In the two-phase system, aggregation of free PLD might occur due to the use of the organic hydrophobic medium; the enzyme is insoluble in organic hydrophobic media, leading to a decreased activity and poor stability of free PLD.37 PLD@MSNC protects PLD from aggregation in organic solvents, and has a better dispersion in the two-phase interface. PLD@MSNC in the two-phase system has high stability and enzymatic catalytic activity, as demonstrated over the 32 h reaction time used in the recycling experiment.
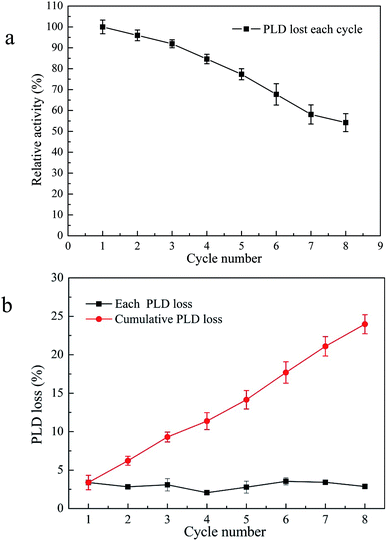 | ||
| Fig. 7 (a) Reusability of PLD@MSNC over eight successive cycles of biosynthesis of PS from PC in the two-phase system. (b) PLD loss from PLD@MSNC over eight successive cycles in the two-phase system. | ||
3.4. Comparison of the results from this work to other data
Many papers have reported on free PLD-mediated transphosphatidylation for the synthesis of PS from PC. However, immobilized PLD-mediated transphosphatidylation for the synthesis of PS from PC has rarely been reported. Li et al. reported recently an in situ cross-linking method to immobilize PLD on a ZnO nanowire/macroporous SiO2 composite support and catalyze the conversion of PC to PS in a biphasic system.35 The immobilized PLD exhibited a higher conversion efficiency than free PLD. However, according to the enzyme activity calculation method in this paper, the enzyme activity of the immobilized PLD was 2.9 U g−1 protein, which is lower than that reported in our work.4. Conclusions
In conclusion, we designed a novel approach to develop highly effective biocatalysis in a two-phase system, in which enzymes were encapsulated into MSNCs in a simple and effective way using one-pot self-assembly in an aqueous solution. This method provides a good foundation for applications of a heterogeneous reaction system. This novel method can be used further for other interfacial enzymes and the discovery of new interfacial catalytic reactions, increasing the catalytic range of enzymes and the available methods for nanoscale immobilization.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (21706126, 21606127, 21576134, 21390200), the National Key Research and Development Program (2016YFA0204300), the Open Funding Project of the State Key Laboratory of Bioreactor Engineering and the Jiangsu Synergetic Innovation Center for Advanced Bio-Manufacture (XTE1853).Notes and references
- H. Khatoon, J. Mansfeld and R. Ulbrich-Hofmann, J. Am. Oil Chem. Soc., 2016, 93, 365–372 CrossRef CAS.
- J. Scillipoti, C. Nioi, A. Marty, S. Camy and J. S. Condoret, Biochem. Eng. J., 2017, 117, 162–171 CrossRef CAS.
- A. Arca-Ramos, G. Eibes, M. T. Moreira, G. Feijoo and J. M. Lema, Process Biochem., 2012, 47, 1115–1121 CrossRef CAS.
- A. Bastida, R. M. Blanco, S. G. Zárate, E. García-Junceda and J. M. Guisán, Biocatal. Biotransform., 2018, 36, 371–378 CrossRef.
- X. Zhong, J. Qian, M. Liu and L. Ma, Eng. Life Sci., 2013, 13, 563–571 CrossRef CAS.
- M. Lotti, J. Pleiss, F. Valero and P. Ferrer, Biotechnol. J., 2018, 13, 1700155 CrossRef PubMed.
- A. A. Pollardo, H. Lee, D. Lee, S. Kim and J. Kim, J. Cleaner Prod., 2018, 185, 382–388 CrossRef CAS.
- E. Duboue-Dijon, E. Pluhařová, D. Domin, K. Sen, A. C. Fogarty, N. Chéron and D. Laage, J. Phys. Chem. B, 2017, 121, 7027–7041 CrossRef CAS PubMed.
- A. Toplak, T. Nuijens, P. J. L. M. Quaedflieg, B. Wu and D. B. Janssen, Enzyme Microb. Technol., 2015, 73–74, 20–28 CrossRef CAS PubMed.
- J. Ge, J. Lei and R. N. Zare, Nat. Nanotechnol., 2012, 7, 428–432 CrossRef CAS PubMed.
- M. Yan, J. Ge, Z. Liu and P. Ouyang, J. Am. Chem. Soc., 2006, 128, 11008–11009 CrossRef CAS PubMed.
- C. Doonan, R. Riccò, K. Liang, D. Bradshaw and P. Falcaro, Acc. Chem. Res., 2017, 50, 1423–1432 CrossRef CAS PubMed.
- A. Chakrabarty, U. Maitra and A. D. Das, J. Mater. Chem., 2012, 22, 18268–18274 RSC.
- N. F. Goldshleger, A. S. Lobach, V. E. Baulin and A. Y. Tsivadze, Russ. Chem. Rev., 2017, 86, 269–297 CrossRef CAS.
- L. Fan, P. Xie, Y. Wang, Z. Huang and J. Zhou, J. Agric. Food Chem., 2018, 66, 238–246 CrossRef CAS PubMed.
- A. Patra, N. Samanta, D. K. Das and R. K. Mitra, J. Phys. Chem. B, 2017, 121, 1457–1465 CrossRef CAS PubMed.
- N. Haque and N. P. Prabhu, Biochem. Biophys. Res. Commun., 2018, 499, 907–912 CrossRef CAS PubMed.
- X. Cao, Y. Ni, A. L. Zhang, S. Xu, K. Q. Chen and P. K. Ouyang, Chem. Commun., 2017, 53, 3134–3137 RSC.
- W. Qin, C. Wu, W. Song, X. Chen, J. Liu, Q. Luo and L. Liu, Process Biochem., 2018, 66, 146–149 CrossRef CAS.
- A. Kato-Kataoka, M. Sakai, R. Ebina, C. Nonaka, T. Asano and T. Miyamori, J. Clin. Biochem. Nutr., 2010, 47, 246–255 CrossRef CAS PubMed.
- B. Li, J. Wang, X. Zhang, B. Zhao and L. Niu, J. Agric. Food Chem., 2016, 64, 7555–7560 CrossRef CAS PubMed.
- B. Li, J. Wang, X. Zhang and B. Zhao, Ind. Eng. Chem. Res., 2016, 55, 10555–10565 CrossRef CAS.
- A. Pinsolle, P. Roy, C. Bure, A. Thienpont and M. Cansell, Colloids Surf., B, 2013, 106, 191–197 CrossRef CAS PubMed.
- X. Liu, Y. Chen and C. A. Fierke, J. Am. Chem. Soc., 2017, 139, 17457–17463 CrossRef CAS PubMed.
- M. J. Knape, M. Ballez, N. C. Burghardt, B. Zimmermann, D. Bertinetti, A. P. Kornev and F. W. Herberg, Metallomics, 2017, 9, 1576–1584 RSC.
- Z. Li, H. Xia, S. Li, J. Pang, W. Zhu and Y. Jiang, Nanoscale, 2017, 9, 15298–15302 RSC.
- S. S. Magalhaes, L. Alves, M. Sebastiao, B. Medronho, Z. L. Almeida, T. Q. Faria, R. M. M. Brito, M. J. Moreno and F. E. Antunes, Biotechnol. Prog., 2016, 32, 1276–1282 CrossRef CAS PubMed.
- J. Zeng, X. Gao, Z. Dai, B. Tang and X. Tang, Appl. Environ. Microbiol., 2014, 80, 2763–2772 CrossRef PubMed.
- H. Xia, X. Zhong, Z. Li and Y. Jiang, J. Colloid Interface Sci., 2019, 533, 1–8 CrossRef CAS PubMed.
- L. Zheng, Y. Zheng, C. Chen, Y. Zhan, X. Lin, Q. Zheng, K. Wei and J. Zhu, Inorg. Chem., 2009, 48, 1819–1825 CrossRef CAS PubMed.
- F. J. Lech, M. B. J. Meinders, P. A. Wierenga and H. Gruppen, Colloids Surf., A, 2015, 473, 18–23 CrossRef CAS.
- S. Cao, D. Yue, X. Li, T. J. Smith, N. Li, M. Zong, H. Wu, Y. Ma and W. Lou, ACS Sustainable Chem. Eng., 2016, 4, 3586–3595 CrossRef CAS.
- X. Ji, Z. Su, C. Liu, P. Wang and S. Zhang, Biochem. Eng. J., 2017, 121, 147–155 CrossRef CAS.
- J. O. Rich and Y. L. Khmelnitsky, Biotechnol. Bioeng., 2001, 72, 374–377 CrossRef CAS PubMed.
- S. Li, Y. Li, N. Long, F. Jiang and R. Zhang, Int. J. Biol. Macromol., 2018, 117, 1188–1194 CrossRef CAS PubMed.
- B. Li, D. Duan, J. Wang, H. Li, X. Zhang and B. Zhao, J. Biotechnol., 2018, 281, 67–73 CrossRef CAS PubMed.
- R. C. Rodrigues, C. Ortiz, Á. Berenguer-Murcia, R. Torres and R. Fernández-Lafuente, Chem. Soc. Rev., 2013, 42, 6290–6307 RSC.
| This journal is © The Royal Society of Chemistry 2019 |