Open Access Article
Open Access ArticleSignificant role of Asn-247 and Arg-64 residues in close proximity of the active site in maintaining the catalytic function of CTX-M-15 type β-lactamase†
Lubna Maryam,
Shamsi Khalid,
Abid Ali and
Asad U. Khan *
*
Medical Microbiology and Molecular Biology Laboratory, Interdisciplinary Biotechnology Unit, Aligarh Muslim University, Aligarh, Uttar Pradesh 202 002, India. E-mail: asad.k@rediffmail.com; Fax: +91-571-2721776; Tel: +91-983-7021912
First published on 12th February 2019
Abstract
Members of Enterobacteriaceae cause antibiotic-resistant infections worldwide. One such marker, CTX-M-15, expressed by Enterobacteriaceae produces β-lactamases, which hydrolyze the cephalosporin group of antibiotics, such as cefotaxime, used in the treatment of both Gram-positive and negative bacterial infections. Amino acid residues present in close proximity of the active site might also play a major role in the structure and function of CTX-M-15, hence the objective of this study was to investigate the significance of two amino acid residues, Asn-247 and Arg-64, present near to the active site in the hydrolysis of cefotaxime. blaCTX-M-15, cloned from the E. cloacae strain, and using Polymerase Chain Reaction (PCR)-based site-directed mutagenesis, Asn247Val and Arg64Leu mutations were introduced. The minimum inhibitory concentrations of cefotaxime for the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutants were reduced by 512 and 128 fold, respectively. Proteins/enzymes of wild-type CTX-M-15, CTX-M-15 (N247V) and CTX-M-15 (R64L) mutants were expressed and purified. Kinetic studies showed that the catalytic efficiencies of the N247V mutant and R64L mutant enzymes in the hydrolysis of cefotaxime were reduced by 89.66% and 71.11%, respectively. Circular dichroism spectroscopic studies showed considerable changes in the α-helical content of the mutant enzymes. A fluorescence study showed that N247V mutant-cefotaxime and R64L mutant-cefotaxime underwent complex formation with strong interactions. The study provides an understanding of the crucial role of the amino acid residues asparagine 247 and arginine 64 present in close proximity of the active site in the hydrolytic mechanism of CTX-M-15 type β-lactamases. Hence, Asn-247 and Arg-64 can be used as potential target sites for the design of inhibitory molecules against CTX-M-15-producing bacterial strains.
Introduction
Enterobacteriaceae belonging Gram-negative bacteria cause various infections in the human population worldwide, including infections of the respiratory tract, skin, joints, bone, soft tissues, urinary tract, central nervous system, eyes, etc. The current issues related to the emergence and spread of resistance to currently available antibiotics in Enterobacteriaceae are making the treatment of serious nosocomial infections a challenge.1 Emerging resistance in Enterobacteriaceae is thus a serious problem that requires immediate attention. The production of extended spectrum β-lactamases (ESBLs) in members of Enterobacteriaceae is one of the major concerns of infection control due to resistance. In response to environmental pressure, in time new variants are evolving via mutation to gain stability and fitness.2 The production of these resistance conferring enzyme (ESBL) producing Gram-negative bacteria is the reason for higher mortality and morbidity rates in the human population.3 Moreover, the diseases caused by such bacteria are difficult to identify by routine laboratory assays, leading to delayed diagnosis and therapy. Broad spectrum β-lactam antibiotics are used for the treatment of both Gram-positive and negative bacterial infections, and include penicillins, cephalosporins, monobactams and carbapenems.4 These antibiotics have a β-lactam ring in their molecular structure. The primary mechanism of the development of resistance to β-lactam antibiotics is the cleavage of the β-lactam ring by β-lactamase enzymes through the action of hydrolysis. ESBLs belong to class A β-lactamases, which are also defined as plasmid-mediated enzymes that play a role in the hydrolysis of oxyimino-cephalosporins.5 One of the most common ESBLs is CTX-M-types.6 In the evolution of the resistance mechanism, CTX-M β-lactamases are considered as the base. Genetic incorporation of CTX-M via mobilization into various hierarchical structures and the selective force exerted by cefotaxime (a cephalosporin) has fuelled a mutational event leading to the diversification of various newer clusters of CTX-M and thereby the emergence of several variants in the population.7 It has been reported that around 20% of K. pneumoniae infections and 31% Enterobacter species infections in ICU departments in the United States are caused by strains that are resistant to third generation cephalosporins.1 CTX-M stands for cefotaxime hydrolyzing β-lactamase family and hence has the ability to hydrolyze the drug cefotaxime.8 However, majorly dispersed infections are caused by the CTX-M-15 genotype of the CTX-M family.7,9 In India, CTX-M-15 has been reported from six unrelated members of Enterobacteriaceae, one K. pneumonia, one E. aerogenes, and four E. coli strains, and is considered as one of the most widespread ESBLs.10 Its extensive dissemination has led to difficulty in the treatment of the community and hospital acquired infections.11,12 CTX-M-15 type ESBL plays a major role in the hydrolysis of cefotaxime.13 There have been several studies on the relevance of active site residues in the catalytic efficiency of CTX-M-15.13 However, residues present in close proximity of the active site might also play a crucial role in the structural configuration of the CTX-M-15 enzyme in maintaining its hydrolytic efficiency towards antibiotics.Hence, this study was designed to understand the role of the amino acid residues asparagine 247 and arginine 64 present in close proximity of the active site in the overall activity of CTX-M-15 by replacing them with valine and leucine, respectively, so that if the findings are significant, they could be used as potential target sites for the design of appropriate inhibitors.
Material
Strains for the cloning and expression of CTX-M-15
Enterobacter cloacae (EC-15) strain (Genebank accession no.: JN860195.1) for extraction of the CTX-M-15 gene,14 pQE-2 (cloning vector), competent E. coli DH5α cells and competent E. coli BL21 cells.Antibiotics/chemicals
The antibiotic cefotaxime was purchased from Sigma chemical co. (St. Louis, MO). The substrate nitrocefin, which was used in the enzyme kinetic studies, was purchased from Calbiochem (USA). Analytical grade reagents and chemicals were used for the study. Double distilled water was used to carry out all the experiments.Methodology
Site-directed mutagenesis and cloning
For the extraction of the plasmid harbouring CTX-M-15 gene from the Enterobacter cloacae strain, a Qiagen plasmid extraction kit was used. Using a forward primer (5′ ATATCATATGGTTAAAAAATCACTG 3′) with a NdeI restriction site and reverse primer (5′ ATATAAGCTTTTACAAACCGTCGGTGAC 3′) with a HindIII restriction site, blaCTX-M-15 was amplified. The Asn247Val (N247V) mutation in blaCTX-M-15 was introduced by Polymerase Chain Reaction (PCR)-based site-directed mutagenesis using a forward primer (5′ GGCACCACCGTAGATATCGCGGTG 3′) and reverse primer (5′ CACCGCGATATCTACGGTGGTGCC 3′) to obtain the blaCTX-M-15(N247V) gene. Similarly, the Arg64Leu (R64L) mutation in the CTX-M-15 gene was introduced using a forward primer (5′ ATACTTTATCTGGCTGATGAGCGC 3′) and reverse primer (5′ GCGCTCATCAGCCAGATAAAGTAT 3′) to obtain the blaCTX-M-15(R64L) gene.The conditions used for the PCR were denaturation at 95 °C for 30 seconds, annealing of the primers at 54 °C for 25 seconds and extension at 72 °C for 40 seconds. 35 cycles of this process were carried out.15 The promoter region was not present in the amplified gene, since vector pQE-2 in which we were going to ligate the genes, already has a promoter region. The amplified PCR products (blaCTX-M-15, blaCTX-M-15(N247V) and blaCTX-M-15(R64L)) and the high copy cloning vector pQE-2 were double digested using NdeI and HindIII restriction endonucleases. Then blaCTX-M-15, blaCTX-M-15(N247V) and blaCTX-M-15(R64L) were ligated to the pQE-2 vector. The ligated products were transformed into competent E. coli DH5α cells and BL21 cells using a heat shock method (keeping at 4 °C for 30 minutes, giving a heat shock at 42 °C for 90 seconds and then maintaining the temperature at 4 °C for 10 minutes). Transformants harbouring blaCTX-M-15, blaCTX-M-15(N247V) and blaCTX-M-15(R64L) were selected on Luria–Bertani agar plates containing 100 μg ml−1 of ampicillin as a selection marker. The clones were confirmed for the presence of CTX-M-15, CTX-M-15(N247V) and CTX-M-15 (R64L) genes by doing double restriction digestion of the transformed cells using NdeI and HindIII restriction enzymes and sequencing according to standard procedures.
Expression and purification of wild-type and mutant proteins/enzymes
A 10 ml overnight culture of transformed E. coli BL21 (DE3) cells harbouring blaCTX-M-15, blaCTX-M-15(N247V) and blaCTX-M-15(R64L) in a pQE-2 vector was grown in 100 μg ml−1 of ampicillin containing Luria–Bertani broth. This culture was used to inoculate 100 μg ml−1 of ampicillin containing 1 liter of Luria–Bertani culture and the bacteria were allowed to grow at 120 rpm at 37 °C, until 0.6–0.8 optical density was reached at a wavelength of 600 nm.16 For the expression of the CTX-M-15 protein and the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant proteins, the respective cultures of each that had reached an absorbance of 0.6–0.8 were cooled and induced using 0.2 mM IPTG (Isopropyl β-D-1-thiogalactopyranoside) as an inducer. The induced cultures were left for 12–16 h at 16 °C at 150 rpm. After that, the cells were collected by centrifugation and suspended in 20 ml of lysis buffer (containing 50 mM of sodium phosphate at pH 8.0, 300 mM of NaCl and 10 mM of imidazole) and β-mercaptoethanol (0.1%). Then, sonication was performed at 35% amplitude for 10 min, to rupture the cells. By centrifuging at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min, the supernatants containing protein were obtained from the rupture cells dissolved in lysis buffer. These supernatants were loaded onto a Ni-NTA column, which was used for the purification of the desired protein. Three Ni-NTA protein purification columns were pre-equilibrated with lysis buffer and the supernatants were then loaded onto it and allowed to bind. The columns were then washed with lysis buffer containing 50 mM of imidazole. To elute bound proteins, phosphate buffer saline (PBS) containing 50 mM of pH 8.0 sodium phosphate and 300 mM of NaCl along with 250 mM of imidazole was used. After dialysis in PBS, pure proteins/enzymes of CTX-M-15, CTX-M-15 (N247V) mutant and CTX-M-15 (R64L) mutant were obtained. Using the 25, 440 M−1 cm−1 molar extinction coefficient, the concentration of the proteins/enzymes were calculated at 280 nm.17
000 rpm for 30 min, the supernatants containing protein were obtained from the rupture cells dissolved in lysis buffer. These supernatants were loaded onto a Ni-NTA column, which was used for the purification of the desired protein. Three Ni-NTA protein purification columns were pre-equilibrated with lysis buffer and the supernatants were then loaded onto it and allowed to bind. The columns were then washed with lysis buffer containing 50 mM of imidazole. To elute bound proteins, phosphate buffer saline (PBS) containing 50 mM of pH 8.0 sodium phosphate and 300 mM of NaCl along with 250 mM of imidazole was used. After dialysis in PBS, pure proteins/enzymes of CTX-M-15, CTX-M-15 (N247V) mutant and CTX-M-15 (R64L) mutant were obtained. Using the 25, 440 M−1 cm−1 molar extinction coefficient, the concentration of the proteins/enzymes were calculated at 280 nm.17
Antimicrobial susceptibility testing
The minimum inhibitory concentration (MIC) of the antibiotic cefotaxime for wild-type and mutant cells was determined using a microdilution method according to guidelines laid out by the CLSI (Clinical Laboratory Standards Institute).18 The lowest concentrations of cefotaxime that completely inhibited visible growth of wild and mutant-carrying bacterial cultures were reported as MIC values.Steady-state enzyme kinetics study
To understand the effect on the CTX-M-15 β-lactamase enzyme upon insertion of the N247V and R64L mutations, an enzyme kinetics study was carried out. For the hydrolytic activities of CTX-M-15, and the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant enzymes towards a chromogenic cephalosporin β-lactamase substrate, nitrocefin was compared in order to understand the effect of the mutation on the hydrolytic efficiency of each enzyme. This was done by examining the rate of appearance and disappearance of nitrocefin.19 The experiment was performed using a spectrophotometer (UV-1800) purchased from Shimadzu International Co. Ltd., Kyoto, Japan. By varying the nitrocefin concentration from 0 to 750 μM, the activities of 6 nM of CTX-M-15, and the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant enzymes were screened. 20 μg ml−1 of BSA was added to 6 nM of the enzymes for dilution and to prevent denaturating prior to screening, since at this concentration of BSA the activities of the enzymes will not be influenced.20 Hydrolysis of nitrocefin was measured for 70 s at 486 nm and the observed absorbance was used to measure the concentration of nitrocefin by taking the molar extinction coefficient 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1.16 The kinetics were measured at 298 K in the presence of pH 7.4 sodium phosphate buffer (50 mM).
000 M−1 cm−1.16 The kinetics were measured at 298 K in the presence of pH 7.4 sodium phosphate buffer (50 mM).
Another enzyme kinetics study was carried out to identify the effect of mutations on the hydrolysis of cefotaxime by the CTX-M-15 enzyme. For that, the activities of 6 nM each of CTX-M-15, and the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant enzymes were monitored at 298 K by varying the concentration of cefotaxime from 0 to 850 μM in the presence of 50 mM of pH 7.4 sodium phosphate buffer. Hydrolysis of cefotaxime was measured for 70 s at 264 nm by taking the value of molar extinction coefficient −7250 M−1 cm−1.21
Using the following eqn (1) and (2), the kinetic parameters Km, kcat and kcat/Km were determined:
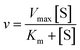 | (1) |
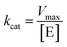 | (2) |
CD spectroscopic study
To gain insight into the conformation of the secondary structure of the CTX-M-15 enzyme upon insertion of the mutation, CD (circular dichroism) spectroscopy was carried out. The study was performed using a J-810 spectropolarimeter (PTC-423S/15) purchased from Jasco International Co. Ltd., Tokyo, Japan equipped with a water bath and temperature controller (Peltier type). Far UV-CD spectra of 5 μM of CTX-M-15, and the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant enzymes were recorded in a 0.1 cm cuvette at 298 K. The instrument was calibrated using (+)-10-camphor sulfonic acid and the scan speed and response time were set at 100 nm min−1 and 1 s, respectively. Each measurement was corrected using the appropriate blanks. Using the following eqn (3), the MRE (mean residual ellipticity) values were estimated in deg cm2 dmol−1 from the values of the observed ellipticity ([θ]obs) in mdeg.22
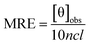 | (3) |
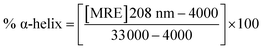 | (4) |
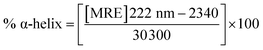 | (5) |
Studies based on fluorescence spectroscopy
To monitor the effect of the insertion of the mutation on the fluorescence intensity of the CTX-M-15 enzyme and to gain insight into the mechanism of interaction24,25 of cefotaxime with the CTX-M-15 enzyme, and the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant enzymes, fluorescence spectroscopic studies were carried out. The study was carried out using a spectrofluorometer (RF-5301 PC) purchased from Shimadzu Corporation, Kyoto, Japan, equipped with a thermostatically-controlled cell holder and a water bath to maintain a particular temperature. Fluorescence spectra of 2 μM of the CTX-M-15 enzyme, and the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant enzymes were recorded in the presence of 50 mM of pH 7.4 sodium phosphate buffer at 298 K. Also, to gain insight into the CTX-M-15-cefotaxime interaction, and the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant-cefotaxime interactions, another fluorescence spectroscopy study was carried out at 298 K for each enzyme. To a 450 μl sample of the enzymes at a concentration of 2 μM, in the presence of sodium phosphate buffer (50 mM, pH 7.4), 2 μM of cefotaxime was added successively until an 18 μM concentration was reached. The samples were excited at 295 nm and in fast scanning mode the fluorescence intensities were measured between 300-450 nm wavelengths.26 The settings of the excitation and emission slits were made at 5 nm. Corrections for the inner filter effect were made for all fluorescence intensities. To analyze the fluorescence intensity at the emission maxima, the following Stern–Volmer eqn (6) was used:25| F°/F = 1 + Ksv[Q] = 1 + Kqτ°[Q] | (6) |
| Kq = Ksv/τ° | (7) |
The other binding parameters, Ka (binding constant) and n (no. of binding sites), were deduced using the following modified Stern–Volmer eqn (8):27
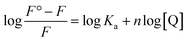 | (8) |
Results
The blaCTX-M-15, blaCTX-M-15(N247V) and blaCTX-M-15(R64L) were cloned and the transformants harbouring the genes were confirmed for the presence of genes by carrying out double restriction digestion using the NdeI and HindIII restriction enzymes. The agarose gel of the double digestion showed two bands in each lane, one of 4.8 kb in size corresponding to the pQE-2 vector and the other approximately 800 bp corresponding to the CTX-M-15, CTX-M-15 (N247V) and CTX-M-15 (R64L) genes (Fig. S1†).Purified proteins/enzymes of CTX-M-15, and the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutants were obtained in 97%, 94% and 93% purity, respectively, as seen in a single band of 31 kDa each by SDS-PAGE. The concentrations of the proteins/enzymes were calculated using the 25, 440 M−1 cm−1 molar extinction coefficient at 280 nm and were found to be 2.5 mg ml−1 for CTX-M-15, 2.2 mg ml−1 for CTX-M-15 (N247V) and 1.9 mg ml−1 for CTX-M-15 (R64L) (Fig. S2†).
The MIC of cefotaxime for the blaCTX-M-15 gene harbouring the DH5α cells was reported to be 512 μg ml−1,28 which is in a high resistance range, the MIC of cefotaxime for the blaCTX-M-15(N247V) gene harbouring DH5α cells was found to be 1 μg ml−1 and the MIC for blaCTX-M-15(R64L) harbouring DH5α cells was found to be ≤4 μg ml−1, values that are in the sensitive range (Table 1).
| Antimicrobial agent | DH5α (pQE-2-CTX-M-15) MIC (μg ml−1) | DH5α (pQE-2-CTX-M-15 (N247V)) MIC (μg ml−1) | DH5α (pQE-2-CTX-M-15 (R64L)) MIC (μg ml−1) | DH5α (pQE-2) MIC (μg ml−1) |
|---|---|---|---|---|
| CTX (cefotaxime) | 512 | 1 | ≤4 | ≤0.5 |
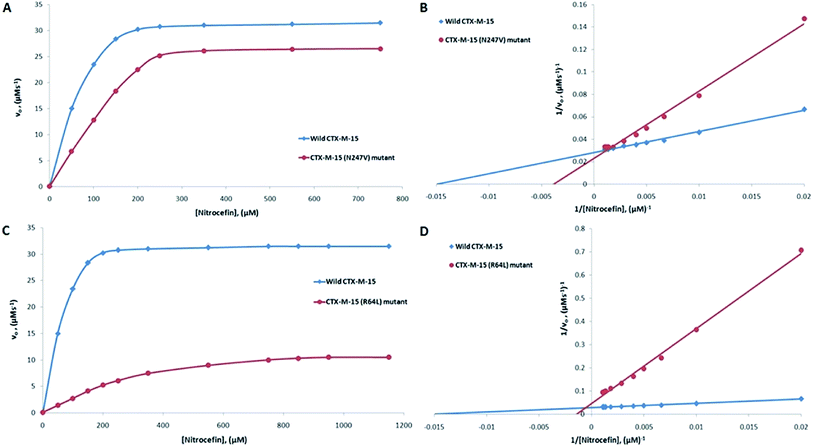
CTX-M-15 is a β-lactamase enzyme and nitrocefin (a chromogenic substrate) is used for the detection of β-lactamase activity. In order to monitor the effect on the enzyme activity and efficiency of CTX-M-15, and the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant β-lactamases to hydrolyze nitrocefin, an enzyme kinetics study was carried out. The Michaelis–Menten and Lineweaver–Burk plots of wild-type CTX-M-15, and the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant enzymes are shown in Fig. 1. The kinetic parameters kcat (catalytic activity), Km (Michaelis–Menten constant) and kcat/Km (catalytic efficiency) were calculated using the Lineweaver–Burk plot (Tables 2 and 3) and it was observed that Km increased from 66.703 to 241.830 μM, and kcat/Km decreased from 92.542 to 28.670 (μM−1 s−1) upon changing asparagine with valine at site 247 in the CTX-M-15 β-lactamase. In the same way, upon changing arginine with leucine at site 64 in the CTX-M-15 β-lactamase, Km increased from 66.703 to 655.504 μM, and kcat/Km decreased from 92.542 to 5.188 (μM−1 s−1).
| Antibiotics | Wild-type CTX-M-15 | CTX-M-15 (N247V) mutant | Residual (kcat/Km) of N247V (%) | ||||
|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | ||
| a Data are reported as an average of ±standard error from three independent experiments. | |||||||
| Nitrocefin | 66.70 | 6172.83 | 92.54 | 241.83 | 6933.30 | 28.67 | 69.01 |
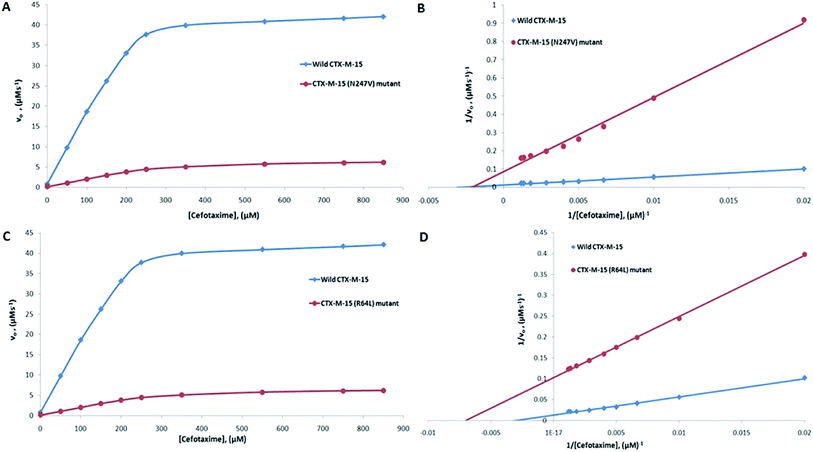
| Cefotaxime | 324.15 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 820.50 820.50 |
39.55 | 479.50 | 1960.60 | 4.09 | 89.66 |
| Antibiotics | Wild-type CTX-M-15 | CTX-M-15 (R64L) mutant | Residual (kcat/Km) of N247V (%) | ||||
|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | ||
| a Data are reported as an average of ±standard error from three independent experiments. | |||||||
| Nitrocefin | 66.70 | 6172.83 | 92.54 | 655.50 | 3401.30 | 5.19 | 94.39 |
| Cefotaxime | 324.15 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 820.50 820.50 |
39.55 | 143.03 | 1633.83 | 11.42 | 71.11 |
The major concern of CTX-M-15 carrying bacterial cells is its ability to hydrolyze the antibiotic cefotaxime. So, in order to understand the effect of the mutations on the hydrolytic ability of the CTX-M-15 enzyme to hydrolyze cefotaxime, another enzyme kinetics study was carried out. The Michaelis–Menten plots and the corresponding Lineweaver–Burk plots are shown in Fig. 2. The Lineweaver–Burk plots were used to deduce the kinetic parameters (Tables 2 and 3). It was observed that Km increased from 324.153 to 479.500 μM, and kcat/Km decreased from 39.550 to 4.088 (μM−1 s−1) upon insertion of the N247V mutation in CTX-M-15 β-lactamase. Similarly, upon insertion of the R64L mutation in CTX-M-15 β-lactamase, Km was observed to decrease from 324.153 to 143.025 μM, and kcat/Km from 39.550 to 11.423 (μM−1 s−1).
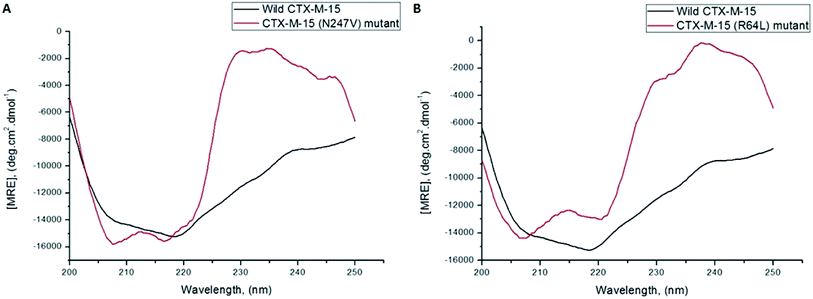
A CD study was carried to determine the secondary structure of the wild CTX-M-15 enzyme and to monitor any conformational changes that might have occurred upon insertion of the mutation into the CTX-M-15 enzyme. Hence, CD spectra of the CTX-M-15 enzyme, and the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant enzymes were recorded in the far UV-CD range of 200–250 nm at 298 K (Fig. 3). Using eqn (3), the MRE was calculated at both 208 and 222 nm using values of the observed ellipticity, and using eqn (4) and (5), the % α-helical content of CTX-M-15, CTX-M-15 (N247V), and CTX-M-15 (R64L) at 208 and 222 nm were calculated using the MRE values (Table 4). When the spectrum of the wild CTX-M-15 was recorded, the MRE values were found to be −14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 196.3 and −14
196.3 and −14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 237.2 deg cm2 dmol−1 at 208 and 222 nm, respectively, and the percentages of the α-helical content present at 208 and 222 nm were 35.16% and 39.26%, respectively. When the spectrum of the CTX-M-15 (N247V) mutant was recorded, the MRE values had changed to −16
237.2 deg cm2 dmol−1 at 208 and 222 nm, respectively, and the percentages of the α-helical content present at 208 and 222 nm were 35.16% and 39.26%, respectively. When the spectrum of the CTX-M-15 (N247V) mutant was recorded, the MRE values had changed to −16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 473.2 and −14
473.2 and −14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 681.5 deg cm2 dmol−1 at 208 and 222 nm, respectively and the percentages of the α-helical content present at 208 and 222 nm had changed to 43.01% and 40.73%, respectively. Similarly, when the spectrum of CTX-M-15 (R64L) was recorded, the MRE values had changed to −14
681.5 deg cm2 dmol−1 at 208 and 222 nm, respectively and the percentages of the α-helical content present at 208 and 222 nm had changed to 43.01% and 40.73%, respectively. Similarly, when the spectrum of CTX-M-15 (R64L) was recorded, the MRE values had changed to −14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 941.7 and −13
941.7 and −13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 242.5 deg cm2 dmol−1 at 208 and 222 nm, respectively and the percentages of the α-helical content present at 208 and 222 nm had changed to 37.73% and 35.98%, respectively.
242.5 deg cm2 dmol−1 at 208 and 222 nm, respectively and the percentages of the α-helical content present at 208 and 222 nm had changed to 37.73% and 35.98%, respectively.
| MRE208 (deg cm2 dmol−1) | % α-helix at 208 | MRE222 (deg cm2 dmol−1) | % α-helix at 222 | |
|---|---|---|---|---|
| Wild CTX-M-15 | −14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 196.3 ± 140 196.3 ± 140 |
35.16 ± 1.5 | −14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 237.2 ± 90 237.2 ± 90 |
39.26 ± 1.8 |
| CTX-M-15 (N247V) mutant | −16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 473.2 ± 98 473.2 ± 98 |
43.01 ± 1.3 | −14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 681.5 ± 120 681.5 ± 120 |
40.73 ± 1.4 |
| CTX-M-15 (R64L) mutant | −14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 941.7 ± 76 941.7 ± 76 |
37.73 ± 1.0 | −13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 242.5 ± 117 242.5 ± 117 |
35.98 ± 2.0 |
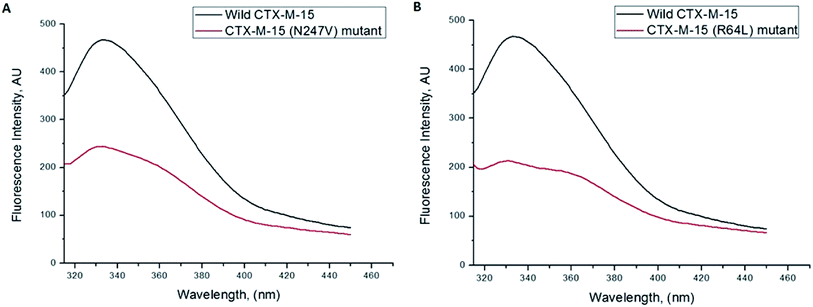
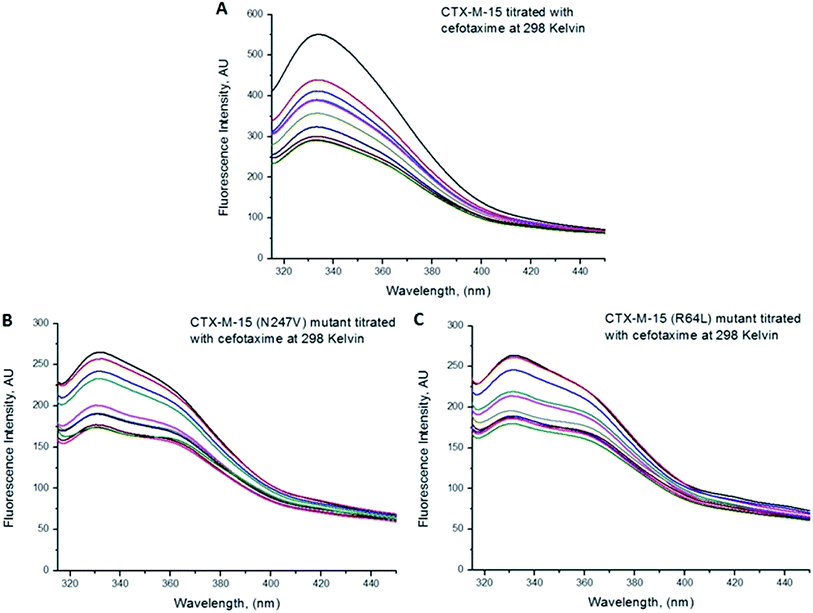
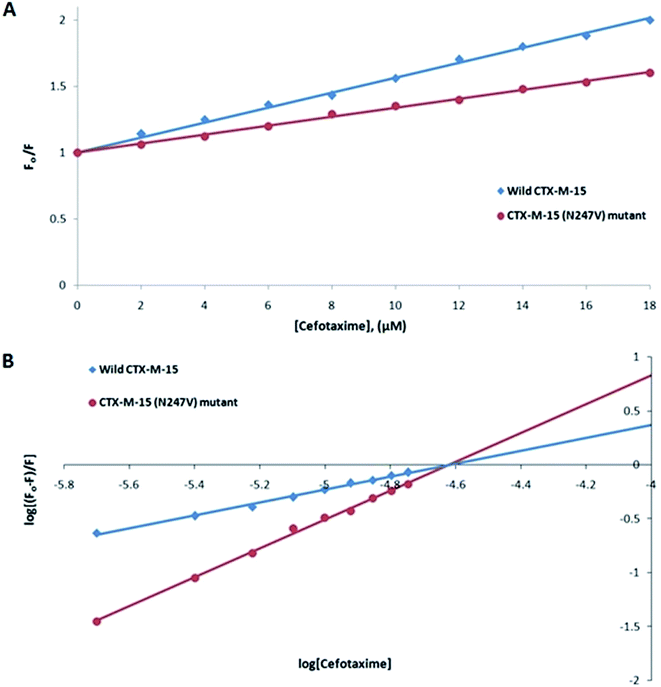
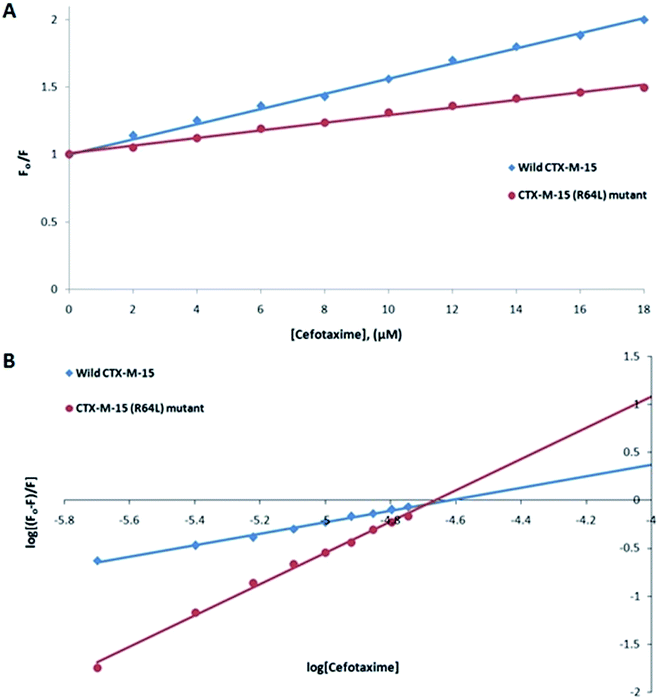
Fluorescence spectroscopic studies were carried out to understand the protein–ligand/enzyme–ligand interactions.29 To monitor the effect of the mutations on the fluorescence intensity of the CTX-M-15 enzyme, fluorescence spectra were recorded. The fluorescence spectra of the CTX-M-15, CTX-M-15 (N247V), and CTX-M-15 (R64L) were recorded at 298 K (Fig. 4). A considerable decrease in the fluorescence intensity curves was observed in the case of the mutant enzymes. Moreover, since the study was focused on the effect of the mutation on the cefotaxime binding and its hydrolysis, to understand the interaction of the antibiotic cefotaxime with CTX-M-15, CTX-M-15 (N247V) and CTX-M-15 (R64L), fluorescence spectroscopy was carried out. A progressive decrease in the fluorescence intensity curves of the wild enzyme and mutant enzymes was observed at 298 K due to a quenching phenomenon (Fig. 5). Using the Stern–Volmer eqn (6) and (7) and modified Stern–Volmer eqn (8), the Stern–Volmer and modified Stern–Volmer plots were determined for CTX-M-15 and the CTX-M-15 (N247V) mutant enzyme (Fig. 6) and for CTX-M-15 and the CTX-M-15 (R64L) mutant enzyme (Fig. 7). Also using eqn (6)–(8), binding parameters were calculated (Table 5). The Ksv (Stern–Volmer constant) values were found to be in the order of 104 M−1. The Kq (quencher rate coefficient) values were determined to be in the order of 1013 M−1 s−1 for the CTX-M-15 enzyme and in the order of 1012 M−1 s−1 for the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant enzymes. The Ka (binding constant) values were in the order of 102 M−1, 107 M−1 and 106 M−1 for CTX-M-15, CTX-M-15 (N247V), and CTX-M-15 (R64L), respectively (Table 5). The number of binding sites (n) available for the binding of cefotaxime on CTX-M-15 and CTX-M-15 (N247V) was 1, whereas there were 2 on CTX-M-15 (R64L).
| Ksv (M−1) | Kq (M−1 s−1) | Ka (M−1) | n | R2 | |
|---|---|---|---|---|---|
| Wild-type CTX-M-15 at 298 K | 5.5 × 104 | 1.28 × 1013 | 7.43 × 102 | 0.618 | 0.994 |
| CTX-M-15 (N247V) mutant at 298 K | 3.3 × 104 | 7.66 × 1012 | 1.51 × 106 | 1.336 | 0.997 |
| CTX-M-15 (R64L) mutant at 298 K | 2.8 × 104 | 6.50 × 1012 | 7.05 × 107 | 1.685 | 0.995 |
Discussion
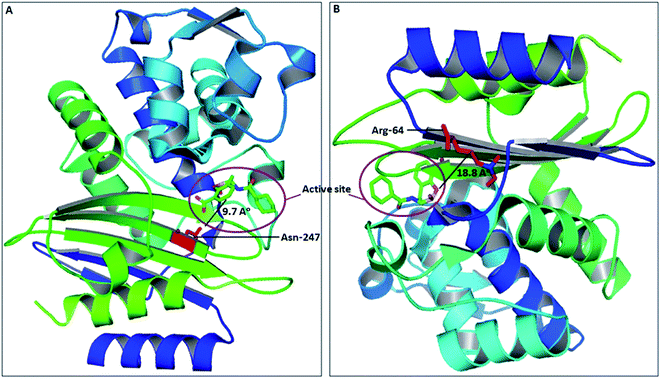
The spread of CTX-M-15 and its variants is one of the major concerns in infection control, caused by multi-drug resistant strains.30 Due to continuous evolution, newer variants of CTX-M-15 are continuously arising, thus making the situation much worse. Several studies have shown the significance of active site residues of CTX-M-15, such as Ser-70, Thr-235, Ser-130, Asn-104, Arg-274, Ser-237, etc., in substrate binding and catalysis.13,16 However, other residues near to the active site might also contribute significantly in determining the broad specificity of the CTX-M-15 enzyme, as observed after closely examining the CTX-M-15 structure. Also, in the course of CTX-M-15 variant evolution, the residues present near to the active site play a major role. Therefore, a detailed study is needed to be carried out to understand the importance of these residues in the structure and function of the CTX-M-15 enzyme that lead to evolution. In this study, we explored the significance of two residues, Asn-247 and Arg-64, present near to the active site, in the structure and function of CTX-M-15 (PDB-ID: 5T66) by replacing them with Val-247 and Leu-64, respectively. Asn-247 and Arg-64 are located in close proximity of the active site at 9.7 Å and 18.8 Å distance from the active site, respectively, as shown in Fig. 8.To evaluate the effect of the mutation on the resistance pattern, the MICs of E. coli DH5α cells harbouring blaCTX-M-15, blaCTX-M-15(N247V) and blaCTX-M-15(R64L) were determined. The E. coli DH5α cells harbouring blaCTX-M-15 showed strong resistance to cefotaxime with a MIC value of 512 μg ml−1. On replacing Asn-247 with Val-247, the MIC of cefotaxime was reduced by 512 fold. Similarly, on replacing Arg-64 with Leu-64 in blaCTX-M-15, the MIC of cefotaxime was reduced by 128 fold.
Using purified proteins/enzymes, steady-state enzyme kinetics studies were performed. First of all, the enzyme kinetics were investigated by varying the concentration of nitrocefin to monitor the effect on the enzyme upon mutation. Nitrocefin is a chromogenic cephalosporin substrate, which exhibits a hydrolytic profile of a characteristic β-lactamase enzyme. The β-lactam ring of nitrocefin is hydrolyzed by a β-lactamase enzyme, producing a shift in the UV absorption, which is detected as a measure of the β-lactamase activity. The Km value of the N247V mutant was increased considerably, which means that the affinity of the N247V mutant towards the substrate was reduced considerably. Similarly, the Km value of the R64L mutant was increased much more than that of the N247V mutant, indicating much more of a decrease in the affinity of the R64L mutant towards the substrate. The overall catalytic efficiency of the CTX-M-15 enzyme on changing Asn-247 to Val-247 towards the hydrolysis of nitrocefin was decreased by 69.01%. And, the efficiency of the enzyme on changing Arg-64 to Leu-64 towards the hydrolysis of nitrocefin was decreased by 94.39%. CTX-M-15 is characterized by its ability to hydrolyze the antibiotic cefotaxime. So, to gain an insight into the effect of the mutation on the cefotaxime hydrolytic ability of the enzyme, another enzyme kinetics study was carried out. An increase in Km and a significant decrease in kcat was observed for the N247V mutant enzyme suggested a decrease in the affinity of the enzyme towards cefotaxime and a decrease in the catalytic activity of the enzyme towards the hydrolysis of cefotaxime. The catalytic efficiency (kcat/Km) of the N247V mutant enzyme towards the hydrolysis of cefotaxime was decreased by 89.66%. Similarly, a significant decrease in the Km and kcat values was observed for the R64L mutant enzyme, suggesting an increase in the affinity and a decrease in the catalytic activity of the enzyme. Overall, the catalytic efficiency (kcat/Km) of the R64L mutant enzyme towards the hydrolysis of cefotaxime was decreased by 71.11%.
The kinetics data, together with the MIC values, clearly showed that the Asn247Val and Arg64Leu mutants reduced the ability of the CTX-M-15 enzyme to hydrolyze cefotaxime.
CD studies help to determine the secondary and tertiary structures of the protein, as well as to determine intermediate states, such as molten globules, during the alteration in the conformation of a particular protein/enzyme.31 The fraction of protein/enzyme molecules present in various conformations of the secondary structure, such as α-helices, β-sheets, beta-turn, random coil, etc., can also be estimated. Hence, to investigate the effect of the mutation on the overall structure and stability of the CTX-M-15 enzyme, CD spectroscopic studies were performed. The CD spectrum of wild CTX-M-15 displayed a characteristic α-helical protein curve with minima at 208 nm and 222 nm, resembling the previously reported far UV-CD spectrum of CTX-M-1.32 On mutating Asn-247 to Val-247 in CTX-M-15, a distorted far-UV CD spectrum was observed with increased α-helical content and MRE values at 208 and 222 nm. On mutating Arg-64 to Leu-64 in CTX-M-15, an altered far-UV CD spectrum was observed with increased α-helical content and a MRE value at 208 nm and decreased α-helical content and MRE value at 222 nm.
Fluorescence spectroscopic studies are carried out to understand the interaction of a particular drug with protein molecules. When an electron from a higher energy level returns back to a lower energy level, fluorescence is observed, referred to as photon emission.33 Fluorescence phenomena involve the absorption of energy in energy levels of the electron and emission at a longer wavelength. Fluorescence spectroscopic studies measure the decrease in the fluorescence intensity upon drug binding, known as quenching phenomena. Quenching is either static or dynamic, with static quenching forming the ground-state complex and in dynamic quenching, the quencher interacts with the molecules indirectly. Quenching is observed due to the transfer of energy or due to molecular collisions, rearrangement and interaction in the excited state.33 This technique also helps in the determination of the conformational alterations of proteins/enzymes. Changing Asn-247 with Val-247 and Arg-64 with Leu-64 in the CTX-M-15 enzyme led to a significant decrease in the fluorescence intensity peak, suggesting an alteration in the conformation of the CTX-M-15 enzyme upon insertion of the mutation. It was observed that upon binding of cefotaxime with the CTX-M-15 enzyme and mutant enzymes, the fluorescence intensity curves decreased progressively, indicating conformational changes occurring in CTX-M-15 and the mutants upon binding of cefotaxime. Ksv values in the order of 104 M−1 signified significant interaction of cefotaxime with the enzymes. Kq values in the order of 1013 M−1 s−1 for the wild CTX-M-15 enzyme and 1012 M−1 s−1 for the mutant enzymes signified that all of the cefotaxime–enzyme interactions undergo complex formation, since these values exceeded the maximum value of the dynamic quenching constant (1010 M−1 s−1). A binding constant in the order of 102 M−1 for CTX-M-15 indicates medium to strong interaction with cefotaxime, and in the order of 107 M−1 and 106 M−1 for the CTX-M-15 (N247V) and CTX-M-15 (R64L) mutant enzymes indicates very strong interactions of these mutants with cefotaxime, respectively. The changing of Arg-64 with Leu-64 induces conformational changes in the CTX-M-15 enzyme meaning that cefotaxime can bind at two sites.
Conclusion
It can be concluded that Asn-247 and Arg-64 play a significant role in maintaining the overall stability and proper orientation of the active site of the CTX-M-15 enzyme. Hence, these amino acid residues could be targeted for the development of novel inhibitors in the future. The study shows that the non-active site amino acid residues play a very crucial role in modulating the sensitivity of the active site and maintaining the structure and function of the β-lactamase enzyme. The development of inhibitors against them can also be an approach to fight infections against β-lactamase-producing strains.Funding
The work was supported by the Department of Biotechnology, Government of India, grant no. BT/PR8281/BID/7/448/2013.Author contributions
LM performed all of the experiments, did the calculations and graph plotting and wrote the manuscript; SK helped in the various experiments, AA helped in the protein purification, AUK designed the study, provided guidance and checked the manuscript.Conflicts of interest
There are no conflicts to declare.Abbreviations
| CD | Circular dichroism |
| MIC | Minimum inhibitory concentration |
Acknowledgements
The author acknowledges the support of the Interdisciplinary Biotechnology Unit, Aligarh Muslim University under DST-PURSE Phase II.References
- D. L. Paterson, Resistance in Gram-negative bacteria: Enterobacteriaceae, Am. J. Infect. Control, 2006, 34(5), S20–S28 CrossRef PubMed.
- A. U. Khan, L. Maryam and R. Zarrilli, Structure, genetics and worldwide spread of New Delhi metallo-β-lactamase (NDM): a threat to public health, BMC Microbiol., 2017, 17(1), 101, DOI:10.1186/s12866-017-1012-8.
- R. H. P. Dhillon and J. Clark, ESBLs: a clear and present danger?, Crit. Care Res. Prac., 2012, 625170 Search PubMed.
- K. B. Holten and E. M. Onusko, Appropriate prescribing of oral beta-lactam antibiotics, Am. Fam. Physician, 2000, 62, 611–620 CAS.
- P. A. Bradford, Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat, Clin. Microbiol. Rev., 2001, 14(4), 933–951 CrossRef CAS PubMed.
- M. E. Rupp and P. D. Fey, Extended spectrum β-lactamase (ESBL)-producing Enterobacteriaceae, Drugs, 2003, 63(4), 353–365 CrossRef CAS PubMed.
- R. Cantón, J. M. González-Alba and J. C. Galán, CTX-M enzymes: origin and diffusion, Front. Microbiol., 2012, 3, 110 Search PubMed.
- A. Bauernfeind, M. Holley, R. Jungwirth, P. Mangold, T. Röhnisch, S. Schweighart and M. Goldberg, A new plasmidic cefotaximase from patients infected with Salmonella typhimurium, Infection, 1992, 20(3), 158–163 CrossRef CAS PubMed.
- L. Poirel, R. A. Bonnin and P. Nordmann, Genetic support and diversity of acquired extended-spectrum β-lactamases in Gram-negative rods, Infect., Genet. Evol., 2012, 12(5), 883–893, DOI:10.1016/j.meegid.2012.02.008.
- A. Karim, L. Poirel, S. Nagarajan and P. Nordmann, Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence IS Ecp1, FEMS Microbiol. Lett., 2001, 201(2), 237–241 CAS.
- S. Shakil and A. U. Khan, Infected foot ulcers in male and female diabetic patients: a clinico-bioinformative study, Ann. Clin. Microbiol. Antimicrob., 2010, 9(1), 2, DOI:10.1186/1476-0711-9-2.
- K. Bush, Bench-to-bedside review: the role of β-lactamases in antibiotic-resistant Gram-negative infections, Crit. Care, 2010, 14(3), 224, DOI:10.1186/cc8892.
- S. D. Lahiri, S. Mangani, T. Durand-Reville, M. Benvenuti, F. De Luca, G. Sanyal and J. D. Docquier, Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-lactamases, Antimicrob. Agents Chemother., 2013 DOI:10.1128/AAC.02247-12.
- L. F. Chen, J. T. Freeman, B. Nicholson, A. Keiger, S. Lancaster, M. Joyce and A. L. Cromer, Widespread dissemination of CTX-M-15 genotype extended-spectrum-β-lactamase-producing Enterobacteriaceae among patients presenting to community hospitals in the southeastern United States, Antimicrob. Agents Chemother., 2014, 58(2), 1200–1202, DOI:10.1128/AAC.01099-13.
- L. Maryam and A. U. Khan, A Mechanism of Synergistic Effect of Streptomycin and Cefotaxime on CTX-M-15 Type β-lactamase Producing Strain of E. cloacae: A First Report, Front. Microbiol., 2016, 7, 2007, DOI:10.3389/fmicb.2016.02007.
- L. Maryam and A. U. Khan, Structural insight into mode of binding of Meropenem to CTX-M-15 type β-lactamase, Int. J. Biol. Macromol., 2017, 96, 78–86, DOI:10.1016/j.ijbiomac.2016.12.032.
- L. Maryam and A. U. Khan, Synergistic effect of doripenem and cefotaxime to inhibit CTX-M-15 type β-lactamases: Biophysical and microbiological views, Front. Pharmacol., 2017, 8, 449, DOI:10.3389/fphar.2017.00449.
- P. A. Wayne and Clinical and Laboratory Standards Institute, Performance standards for antimicrobial susceptibility testing: 21st informational supplement, 2011, M100-S2 Search PubMed.
- C. H. O'Callaghan, A. Morris, S. M. Kirby and A. H. Shingler, Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate, Antimicrob. Agents Chemother., 1972, 1(4), 283–288 CrossRef.
- M. Galleni, N. Franceschini, B. Quinting, L. Fattorini, G. Orefici, A. Oratore and G. Amicosante, Use of the chromosomal class A beta-lactamase of Mycobacterium fortuitum D316 to study potentially poor substrates and inhibitory beta-lactam compounds, Antimicrob. Agents Chemother., 1994, 38(7), 1608–1614 CrossRef CAS PubMed.
- A. U. Khan and M. T. Rehman, Role of Non-Active-Site Residue Trp-93 in the Function and Stability of New Delhi Metallo-β-Lactamase 1, Antimicrob. Agents Chemother., 2016, 60(1), 356–360, DOI:10.1128/AAC.01194-15.
- M. Ishtikhar, S. S. Usmani, N. Gull, G. Badr, M. H. Mahmoud and R. H. Khan, Inhibitory effect of copper nanoparticles on rosin modified surfactant induced aggregation of lysozyme, Int. J. Biol. Macromol., 2015, 78, 379–388, DOI:10.1016/j.ijbiomac.2015.03.069.
- Y. H. Chen, J. T. Yang and H. M. Martinez, Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion, Biochemistry, 1972, 11(22), 4120–4131 CrossRef CAS.
- M. R. Eftink and C. A. Ghiron, Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies, Biochemistry, 1976, 15(3), 672–680 CrossRef CAS PubMed.
- J. R. Lakowicz, Principles of frequency-domain fluorescence spectroscopy and applications to cell membranes, in Fluorescence Studies on Biological Membranes, 1988, Springer, Boston, MA, pp. 89–126 Search PubMed.
- L. Maryam and A. U. Khan, Combination of aztreonam and cefotaxime against CTX-M-15 type β-lactamases: A mechanism based effective therapeutic approach, Int. J. Biol. Macromol., 2018, 116, 1186–1195, DOI:10.1016/j.ijbiomac.2018.05.153.
- J. Kang, Y. Liu, M. X. Xie, S. Li, M. Jiang and Y. D. Wang, Interactions of human serum albumin with chlorogenic acid and ferulic acid, Biochim. Biophys. Acta, 2004, 1674(2), 205–214 CrossRef CAS PubMed.
- M. Faheem, M. T. Rehman, M. Danishuddin and A. U. Khan, Biochemical characterization of CTX-M-15 from Enterobacter cloacae and designing a novel non-β-lactam-β-lactamase inhibitor, PLoS One, 2013, 8(2), e56926, DOI:10.1371/journal.pone.0056926.
- L. Maryam, A. Sharma, M. W. Azam, S. N. Khan and A. U. Khan, Understanding the mode of binding mechanism of doripenem to human serum albumin: spectroscopic and molecular docking approaches, J. Mol. Recognit., 2018, e2710, DOI:10.1002/jmr.2710.
- R. Bonnet, Growing group of extended-spectrum β-lactamases: the CTX-M enzymes, Antimicrob. Agents Chemother., 2004, 48(1), 1–14 CrossRef CAS PubMed.
- M. F. Alam, A. A. Laskar, L. Maryam and H. Younus, Activation of human salivary aldehyde dehydrogenase by sulforaphane: mechanism and significance, PLoS One, 2016, 11(12), e0168463, DOI:10.1371/journal.pone.0168463.
- F. J. Pérez-Llarena, F. Kerff, O. Abián, S. Mallo, M. C. Fernández, M. Galleni and G. Bou, Distant and new mutations in CTX-M-1 β-lactamase affect cefotaxime hydrolysis, Antimicrob. Agents Chemother., 2011 DOI:10.1128/AAC.00298-11.
- A. Ali, Danishuddin, L. Maryam, G. Srivastava, A. Sharma and A. U. Khan, Designing of inhibitors against CTX-M-15 type β-lactamase: potential drug candidate against β-lactamases-producing multi-drug-resistant bacteria, J. Biomol. Struct. Dyn., 2018, 36(7), 1806–1821, DOI:10.1080/07391102.2017.1335434.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra10313e |
| This journal is © The Royal Society of Chemistry 2019 |