Open Access Article
Open Access ArticleEffect of electron beam irradiation on the structural characteristics and functional properties of rice proteins
Xinxia Zhangabc,
Li Wang*abc,
Zhengxing Chen abc,
Yongfu Liab,
Xiaohu Luoabc and
Yanan Liabc
abc,
Yongfu Liab,
Xiaohu Luoabc and
Yanan Liabc
aState Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi, 214122, China. E-mail: wangli0318@jiangnan.edu.cn; zxchen_2008@126.com; Fax: +86-510-8519-7856; Tel: +86-510-8519-7856
bNational Engineering Laboratory for Cereal Fermentation Technology, Jiangnan University, Wuxi, 214122, China
cJiangsu Provincial Research Center for Bioactive Product Processing Technology, Jiangnan University, Lihu Road 1800, Wuxi, 214122, China
First published on 1st May 2019
Abstract
A study of the structural and functional changes of rice proteins (RPs) induced by electron beam irradiation (EBI) at 5 kGy, 10 kGy, 20 kGy, and 30 kGy was performed. The microcosmic surface structures of the RPs were changed and fragmented due to irradiation damage occurring on the RP surfaces. The changes in the UV visible spectra, intrinsic fluorescence spectra, surface hydrophobicity and SH and SS group contents indicated that the RPs unfolded after EBI treatment. In addition, the degree of conformational change was increased with increasing EBI treatment doses. FTIR analysis showed that the secondary structure redistributed, showing decreases in α-helices and concomitant increases in β-sheets, β-turns and random coils. The functional properties, emulsifying abilities, water adsorption capacities and oil adsorption capacities of the irradiated RPs improved dose-dependently, with maximums occurring at 30 kGy. The foaming properties were also enhanced by EBI; however, this effect was not dose-dependent. In contrast, all of the samples irradiated by electron beams presented lower emulsion stability than the control (0 kGy). These results provide a theoretical basis for the application of EBI in improving protein properties in the future.
1. Introduction
Rice proteins (RPs) are rice byproducts produced during the starch extraction process.1,2 RPs exhibit hypoallergenic activity3 and high protein efficiency ratios,4 which makes them good protein sources for human nutrition and even as suitable ingredients for infant food formulations.5 RPs are also attracting much interest in the food industry because they are plentiful and readily available.6,7 In addition, the hydrolysates of rice protein have crucial biological activities, including anti-oxidative,8 antiatherogenic9 and ACE-inhibitory properties.10 However, the utility of RDPs in the food industry is limited by their low digestibility and select functional properties.11 To obtain ideal properties, many technologies have been applied to improve the properties of the proteins, such as enzymatic hydrolysis8 and physical12 or chemical13 methods. Food irradiation is an emerging physical method used in the food industry to maintain nutritional properties and modify food properties.In recent years, studies of the application of irradiation technology on food protein modification have gradually increased and have primarily focused on physiochemical and functional properties. Shawrang et al. demonstrated that the emulsifying properties of protein in soybean meal were effectively improved by γ-irradiation. This improvement may be attributable to protein unfolding and scission induced by γ-irradiation. Such changes in protein structure could expose more hydrophobic amino acids, which have a close relationship with protein functional properties.14 It has also been found that 2.98 kGy irradiation had enormous effects on the foaming properties of egg white protein, mainly because of conformational conversion and increased surface hydrophobicity induced by γ-irradiation.15 In addition, the viscosity of irradiated liquid egg whites decreased significantly with increases in the irradiation dose level. This may be due to the chain scission of proteins induced by the irradiation. Scission of proteins reduces the number of peptide linkages and thereby decreases the viscosity.16
There are many categories of irradiation technologies, one of which is electron beam irradiation (EBI). Compared to the other irradiation methods, EBI is highly attractive in terms of great effectiveness, flexibility, cold process use and environmental friendliness.17 Several research groups have found that the use of EBI could modify the functional properties of food proteins.2,18 The application of EBI has also been reported by Wang et al., showing that the antioxidant activity of pea protein hydrolysates was effectively improved.2 Moreover, Akbarian et al. reported that EBI technology was applied during research into the digestion properties of soybeans.19
Rice protein has abundant nutritional value. However, the effect of EBI on rice protein has not yet been studied. To date, there is not a sufficient understanding of the structural characteristics and functional properties of rice protein irradiated by EBI. This study concentrated on elucidating the effects of EBI on rice protein. Therefore, the changes in the structural and functional properties of RPs induced by EBI have been evaluated. The results of this study are expected to provide a theoretical basis for the development of the electron beam irradiation technique and provide technical support for the further improvement of rice protein.
2. Materials and methods
2.1 Materials
Rice protein (82.9% protein, 7.9% water, 1.35% ash, 6.25% lipid, 1.60% sugar) was provided by Shanyuan Biotechnology Co. Ltd. (Wuxi, China), and 5,5′-dithio-bis-2-nitrobenzoic acid (DTNB) and 1-anilino-8-naphthalene-sulfonate (ANS) were purchased from Sigma (St. Louis, MO, USA). All chemicals and reagents were analytical grade or higher.2.2 Electron beam treatment
Electron beam irradiation was conducted by the AIBANG EB-Tech Co., Ltd. (Wuxi, China) using a high-energy linear accelerator. 100 g of rice protein was sealed in each sterile food-grade polyethylene plastic bag at a final thickness of 1.00 cm. Then, the rice protein was irradiated with varying doses (5 kGy, 10 kGy, 20 kGy, or 30 kGy) by electron beam irradiation at 5.0 MeV. The rice protein was irradiated in powder form. All irradiations were performed at room temperature. After irradiation, samples were stored at −20 °C until further use.2.3 Scanning electron microscopy (SEM)
The protein was adhered to a circular aluminum specimen stub by an adhesive tape and coated with gold-palladium for 90 s under the condition that the current was 15 mA. Then, the microcosmic surface structures of the sample granules were photographed under a potential of 5 kV with a scanning electron microscope FEI Quanta™ 20 0 (FEI Co., Hillsboro, USA). The surface structures of the samples were observed at 2400× magnification.2.4 UV visible spectrum
Protein solutions were prepared in phosphate buffer (10 mM, pH 8.0) and stirred for 20 min, then centrifuged at 5000 rpm for 10 min. The protein concentration in the supernatant was measured according to the Bradford method.20 Before being used, the sample solution was diluted to concentrations of 2 mg mL−1. The UV visible spectra of the sample solutions were recorded at 200–400 nm with a Varian Cary 100 UV-vis spectrophotometer (Varian Inc., Palo Alto, USA) at 25 °C. The scanning speed was 60 nm min−1, and the silt was 2.0 nm. For the blank, the phosphate buffer (0.01 M, pH 8.0) was used.2.5 Fourier transform infrared spectroscopy (FTIR) analysis
FTIR spectroscopy was recorded using a Nicolet iS10 FTIR spectrometer (ThermoFisher Scientific, Marietta, OH, USA). Protein powder (2 mg) was mixed with KBr, grounded, and then pressed into a pellet. Spectra were measured in a wavenumber range of 4000–400 cm−1 at a 2 cm−1 resolution. Omnic V8.1 (ThermoFisher Scientific, USA) and PeakFit 4.12 (SeaSolve Software Inc., USA) were used for determination of α-helices, β-sheets, turns and unordered structure percentages in the protein mixtures.2.6 Intrinsic fluorescence spectroscopy (IFS)
Protein solutions were prepared in phosphate buffer (10 mM, pH 7.0) and stirred for 20 min, then centrifuged at 5000 rpm for 10 min. The protein concentration in the supernatant was measured according to the Bradford method.20 Before being used, the sample solution was diluted to concentrations of 0.04 mg mL−1. The fluorescence intensity of the samples was performed with an F-7000 fluorescence spectrophotometer (Hitachi Co., Tokyo, Japan) at room temperature. Intrinsic spectra were recorded between 300 and 500 nm with an excitation wavelength of 295 nm (slit = 2.5 nm) at 10 nm s−1 of scanning speed.2.7 Surface hydrophobicity
The surface hydrophobicity (H0) of each sample was measured following the method described by Kato and Nakai21 using 1-anilino-8-naphthalene sulfonate (ANS). Protein solutions were prepared in phosphate buffer (10 mM, pH 7.0) and stirred for 20 min, then centrifuged at 5000 rpm for 10 min. The protein concentration in the supernatant was measured according to the Bradford method.20 Before being used, the sample solution was diluted to concentrations of 0.2 mg mL−1 to 0.8 mg mL−1. Then, 1 mL sample solution was mixed with 12.5 μL ANS (8 mM in 10 mM phosphate buffer, pH 7.0). The fluorescence intensity was measured at wavelengths of 365 nm (excitation) and 484 nm (emission) using an F-7000 fluorescence spectrophotometer (Hitachi Co., Tokyo, Japan). The surface hydrophobicity index was defined as the initial slope of the plot of fluorescence intensity as a function of protein concentration.2.8 Sulfhydryl and disulfide bond content
The exposed sulfhydryl group (SHE), total sulfhydryl group (SHT), and disulfide bond (S–S) content of the protein samples were determined using Ellman assays. For SHE content determination, 1.5 mg mL−1 protein samples were prepared with Tris-Gly buffer (0.1 mol L−1, pH 8.0). Then, 50 μL Ellman’ s reagent (DTNB in Tris-Gly buffer, 4 mg mL−1) was added to 1 mL protein solution. After binding for 5 min at room temperature, the mixture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 15 min. Absorbance of the supernatant at was then measured 412 nm.
000×g for 15 min. Absorbance of the supernatant at was then measured 412 nm.
2.9 X-ray photoelectron spectroscopy (XPS) measurements
X-ray photoelectron spectroscopy measurements were carried out with AXIS Supra by Kratos Analytical Inc. using monochromatized Al Kα radiation (hν = 1486.6 eV, 225 W) as X-ray source with a base pressure of 10−9 Torr. Survey scan spectra were acquired using a pass energy of 160 eV and a 1 eV step size. Narrow region scans were acquired using a pass energy of 40 eV and a 0.1 eV step size. The hybrid lens mode was used in both cases. The analyzed area of all XPS spectra was 300 × 700 μm2. A charge neutralizer was used throughout as the samples were mounted such that they were electrically isolated from the sample bar. All spectrums were calibrated by C 1s (284.8 eV).2.10 Emulsifying properties
Emulsifying capacity (EC) and emulsifying stability (ES) were determined using the turbidimetric method.22 A 1% RDP solution was prepared in phosphate buffer (10 mM, pH 7.0). Then, 15 mL sample solution was mixed with 5 mL of peanut oil and homogenized at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 1 min at room temperature with a homogenizer (T18BS25, IKA, Germany). A 50 μL aliquot of the emulsion sample was taken from the bottom of the test tube at 0 and 20 min and immediately dispersed into 5 mL of 0.1% SDS solution. The absorbance of the emulsion was measured at 500 nm with a spectrophotometer (UV-1800, Shimadzu, Japan). The EC was defined as the absorbance that was measured immediately after emulsion formation (A0). The ES was determined as follows:
000 rpm for 1 min at room temperature with a homogenizer (T18BS25, IKA, Germany). A 50 μL aliquot of the emulsion sample was taken from the bottom of the test tube at 0 and 20 min and immediately dispersed into 5 mL of 0.1% SDS solution. The absorbance of the emulsion was measured at 500 nm with a spectrophotometer (UV-1800, Shimadzu, Japan). The EC was defined as the absorbance that was measured immediately after emulsion formation (A0). The ES was determined as follows:| ES (%) = (A0 − A10)/A0 × 100 |
2.11 Foaming properties
Foaming capacity (FC) and foaming stability (FS) were determined following the method described by Liu et al.23 with slight modifications. A volume of 50 mL of 0.5% sample solution was added into a 250 mL cylinder and homogenized (T18BS25, IKA, Germany) at a speed of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 1 min at room temperature. The foam volumes were measured immediately (V0) and after standing for 60 min (V60). FC was expressed as V0. The ES was determined as follows:
000 rpm for 1 min at room temperature. The foam volumes were measured immediately (V0) and after standing for 60 min (V60). FC was expressed as V0. The ES was determined as follows:| FS (%) = V60/V0 × 100 |
2.12 Water absorption capacity (WAC) and oil absorption capacity (OAC)
The sample (0.1 g) was suspended in 1 mL of distilled water or peanut oil, vortexed intermittently and centrifuged at 1500 g for 15 min. Then, the aqueous supernatant or clear oil obtained after centrifuging was decanted. WAC and OAC were calculated as gram of water or oil absorbed per gram of sample, respectively.2.13 Statistical analysis
IMB SPSS statistics 2.0 software was used to analyze the data. The differences between the mean values of the samples were determined using the least significant difference (LSD) test at a significance level of 0.05.3. Results and discussion
3.1 Effect of EBI pretreatment on the microcosmic surface structure of RPs
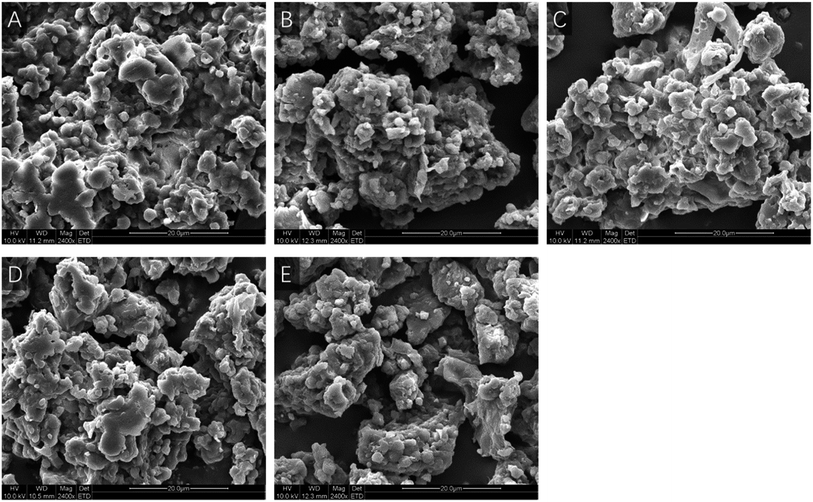
Fig. 1 shows the scanning electron microscope microphotographs of rice protein irradiated at different doses. As shown in Fig. 1A, nonirradiated RPs appeared to have globular aggregate structures, which were tight and rough. Moreover, the overall structures were intact. When RPs were subjected to a 5 kGy irradiation dose (Fig. 1B), tight aggregation was broken down and the proportion of small sized granules was increased; however, the surface structure remained compact compared with nonirradiated RPs. As shown in Fig. 1C, the structural integrity of RPs irradiated at 10 kGy decreased and fragments and cracks emerged compared with the nonirradiated RPs and the RPs irradiated at 5 kGy. As the irradiation dose continues to rise, the surface structure became loose and a large number of smaller fragments are observed (Fig. 1D and E). This type of breakage could be ascribed to the disintegration of rice protein caused by the highly energetic electron beams. The results revealed that the microcosmic surface structures of the RPs were obviously destroyed by the irradiation and that the degree of damage was enhanced by increasing the intensity of the treatment dose. Similar results were previously reported by Zhao et al. They observed that puncture pores appeared on the surface structure of the protein and the number of small fragments increased as the irradiation dose increased. Deformation of the protein has also been reported by Malik et al. in that gamma irradiation induced cracks on the surface of sunflower protein isolates. However, Wang et al. found that there were no obvious differences between the control and irradiated samples in terms of the microstructures of the proteins, indicating there was minimal degradation or aggregation of protein molecules in their solid form. Currently, two types of radiation damage to proteins have been observed: fragmentation and aggregation. As shown in Fig. 1, a broken protein surface and stratified structure do exist. Thus, this indicates that fragmentation resulting from irradiation damage occurred on the RPs surface, increased the proportion of small sized granules and enlarged the specific surface area, which may further influence the proteins' performance.3.2 Effect of EBI pretreatment on the conformation of RPs
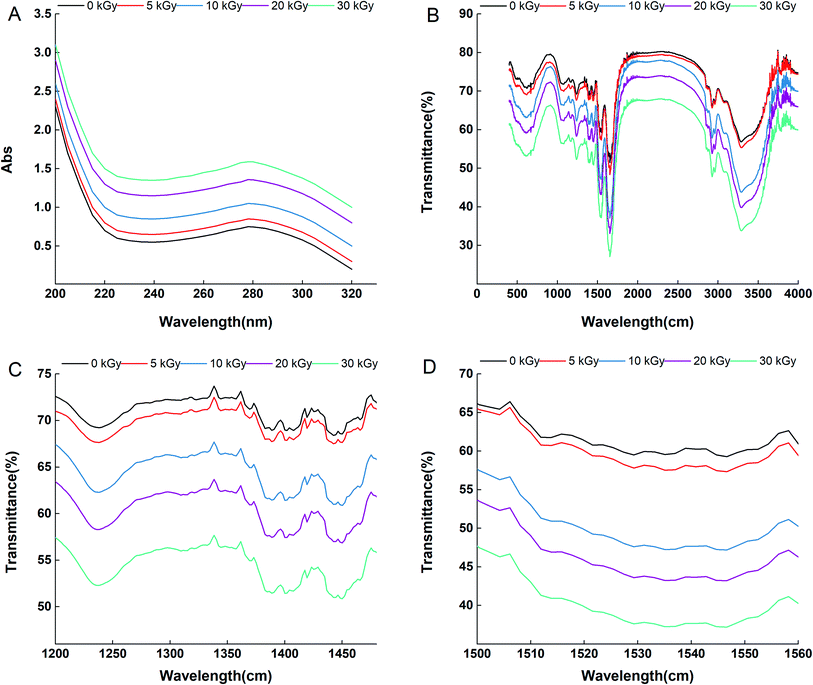
Fig. 2D shows the amide II band (1560–1500 cm−1) of rice protein irradiated by 0, 5, 10, 20 and 30 kGy. The amide II infrared absorption peak intensity can reflect the conformational changes of the tertiary structure of a protein. Tang et al. observed that the amide II peak intensity of soybean protein isolate increased in response to high pressure treatment, and the phenomenon was attributed to the unfolding of the protein tertiary structure.28 In this study, the peak intensity in the amide II region of the irradiated samples increased significantly with the increase in irradiation dose, indicating that the expansion degree of the RPs tertiary structure increased with the increase in the EBI treatment dose. This result is consistent with that determined by the UV visible spectra. However, the wavenumbers of most peaks in the amide II area were almost unaffected by EBI treatment, which indicates that the maintenance of the tertiary structure of RPs is mainly related to hydrophobic interactions and has nothing to do with hydrogen bonds within molecules.
The amide I band (1700–1600 cm−1) mainly comprises the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of the polypeptide chain; therefore, it can be used to discern α-helices, β-sheets, β-turns, and random coils through proper fitting to this area.29 In this study, FTIR analysis of the amide I band demonstrated that the secondary structural components changed considerably after EBI treatment (Table 1). Moreover, the changes were proportional to the irradiation dose. Compared to the nonirradiated group, EBI resulted in a maximum reduction of α-helices by 64.09% and an increase in β-sheets, β-turns and random coils by 16.44%, 8.03% and 28.07%, respectively. These results are consistent with those of Malik et al., who reported that gamma irradiation resulted in a decrease in α-helix structure and an increase in β-sheet, β-turn and random coil content compared to nonirradiated sun flower protein isolates.30 The α-helix structure is susceptible to conformational changes, and a reduction in its content might be related to partial unfolding of the protein, which results in exposure of buried groups.31,32 An increase in β-sheets and random coils may lead to loosening of the protein molecule while an increase in β-turns may promote protein aggregation.33 Based on these findings, we suggest that EBI changed the conformation of the proteins and caused unfolding of the RPs.
O stretching vibration of the polypeptide chain; therefore, it can be used to discern α-helices, β-sheets, β-turns, and random coils through proper fitting to this area.29 In this study, FTIR analysis of the amide I band demonstrated that the secondary structural components changed considerably after EBI treatment (Table 1). Moreover, the changes were proportional to the irradiation dose. Compared to the nonirradiated group, EBI resulted in a maximum reduction of α-helices by 64.09% and an increase in β-sheets, β-turns and random coils by 16.44%, 8.03% and 28.07%, respectively. These results are consistent with those of Malik et al., who reported that gamma irradiation resulted in a decrease in α-helix structure and an increase in β-sheet, β-turn and random coil content compared to nonirradiated sun flower protein isolates.30 The α-helix structure is susceptible to conformational changes, and a reduction in its content might be related to partial unfolding of the protein, which results in exposure of buried groups.31,32 An increase in β-sheets and random coils may lead to loosening of the protein molecule while an increase in β-turns may promote protein aggregation.33 Based on these findings, we suggest that EBI changed the conformation of the proteins and caused unfolding of the RPs.
| Irradiation does | |||||
|---|---|---|---|---|---|
| 0 kGy | 5 kGy | 10 kGy | 20 kGy | 30 kGy | |
| α-Helix | 19.80 ± 0.37 | 18.33 ± 0.34 | 14.81 ± 0.17 | 9.09 ± 0.51 | 7.11 ± 0.96 |
| β-Sheet | 31.03 ± 0.20 | 32.07 ± 0.19 | 32.89 ± 0.19 | 35.61 ± 0.12 | 36.13 ± 0.23 |
| β-Turn | 29.90 ± 0.34 | 30.30 ± 0.47 | 31.80 ± 0.40 | 31.90 ± 0.36 | 32.30 ± 0.21 |
| Random roils | 19.01 ± 0.15 | 19.30 ± 0.27 | 20.01 ± 0.23 | 23.40 ± 0.18 | 24.46 ± 0.33 |
| Treatment | SH and S–S contents (μmol g−1 protein) | |||
|---|---|---|---|---|
| H0 | SHE | SHT | S–S | |
| 0 kGy | 97.33 ± 1.15 | 7.63 ± 0.13 | 37.98 ± 0.41 | 41.75 ± 0.36 |
| 5 kGy | 112.27 ± 2.23 | 8.09 ± 0.19 | 32.88 ± 0.62 | 44.11 ± 0.62 |
| 10 kGy | 117.67 ± 3.15 | 8.87 ± 0.12 | 28.12 ± 0.54 | 52.73 ± 0.52 |
| 20 kGy | 178.00 ± 2.62 | 10.25 ± 0.24 | 25.16 ± 0.23 | 55.96 ± 0.24 |
| 30 kGy | 189.01 ± 2.00 | 11.36 ± 0.15 | 23.77 ± 0.47 | 58.44 ± 0.71 |
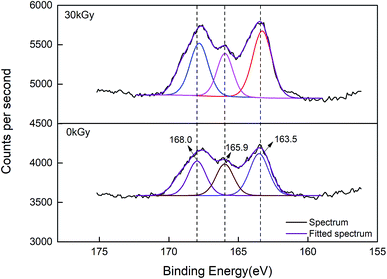
On the other hand, X-ray photoelectron spectroscopy (XPS) analysis was conducted to further confirm the surface chemical environment changes of the EBI treated rice protein.42–44 Fig. 4 shows the S 2p3/2 XPS spectra of rice protein irradiated by 0 kGy and 30 kGy. The S 2p3/2 peaks located at 163.5 and 168.0 eV suggested the present of –SH and S–S.45 Compared with the untreated rice protein, the S 2p3/2 peaks for the EBI treated rice protein (30 kGy) are located at 163.3 eV and 167.9 eV, being shifted slightly by about 0.1–0.2 eV toward lower binding energies owing to the –SH/S–S exchange reaction.
The results of the sulfhydryl group analysis and XPS analysis combined with those of the UV visible spectra, FTIR analysis, surface hydrophobicity and internal fluorescence determination indicated that electron beam irradiation will lead to protein denaturation and open the compact structure. As a consequence, the protein structure becomes more flexible, and many groups buried inside the molecules are exposed. These changes in protein structure may affect the functional properties and biological activities of the proteins.
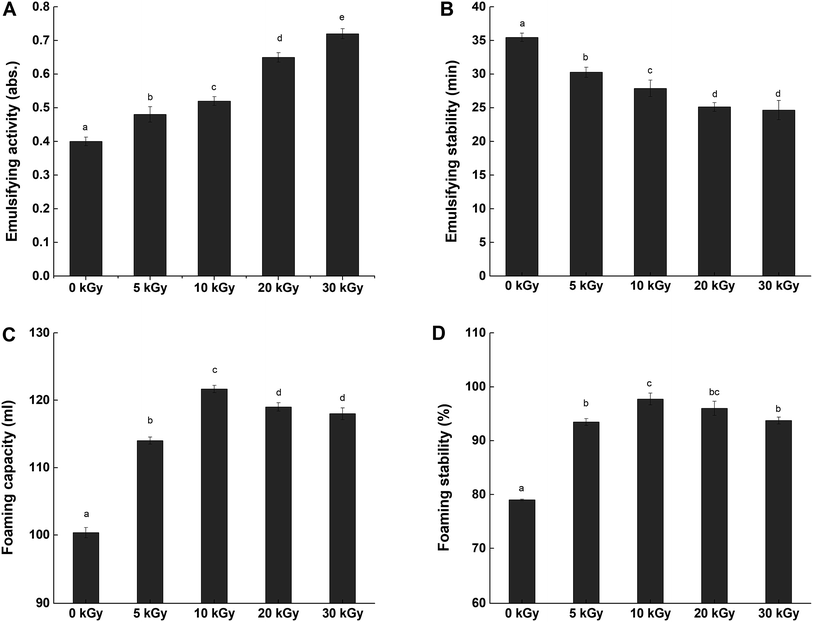
3.3 Effect of EBI pretreatment on the functional properties of RPs
In contrast, all of the samples irradiated by electron beams presented with lower ESs than the control (0 kGy). Meanwhile, the loss of stability became poorer as the irradiation dose increased. This result indicates that the emulsions formed by the irradiated proteins were vulnerable to environmental conditions. The decrease of emulsion stability is possibly due to the higher surface hydrophobicity, which would be expected to result in the flocculation of oil droplets.48
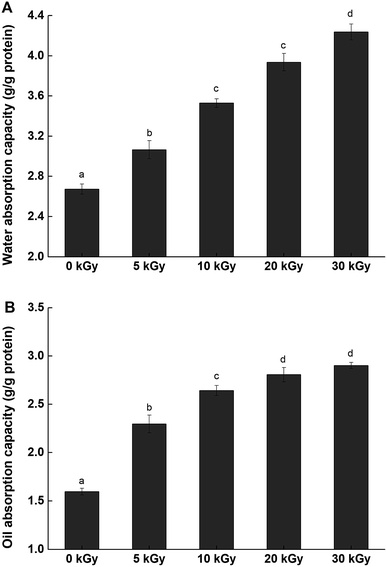 | ||
| Fig. 6 Water adsorption capacity (A) and oil adsorption capacity (B) of rice protein irradiated by 0, 5, 10, 20 and 30 kGy. The values reported represent the mean ± SD (n = 3). | ||
Oil absorption capacity (OAC) also plays an important role in enhancing mouth feel and maintaining flavor.57 The OACs of native and irradiated proteins are presented in Fig. 6. The OAC values of irradiated RPs presented marked increases when compared with the unmodified sample. As the irradiation dose increased, the OAC values increased up to a maximum value (2.81 ± 0.07 g g−1) at 30 kGy, which was an increase of 81.25%. There are many factors that can affect the OAC, such as surface hydrophobicity, physical entrapment of oil and macromolecule size.58 The increase in the OAC can be attributed to the partial unfolding of proteins in response to irradiation, causing more nonpolar protein residues inside the molecules to be exposed, which enhanced the interactions between the protein and the oil.
4. Conclusion
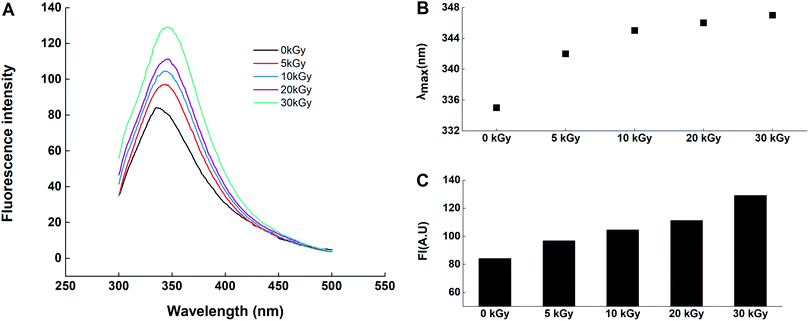
Rice proteins showed considerable changes after electron beam irradiation in terms of structure. The microcosmic surface structure of the RPs was changed and fragmentation occurred on the RPs surface. The microenvironment of tryptophan became more exposed, as shown by the UV visible spectrum and the endogenous fluorescence spectrum. Redistribution of the secondary structure was observed with a decrease in α-helices and concomitant increases in β-sheets, β-turns and random coils. The molecular mechanism of the responses of RPs pretreated by electron beams was also investigated, including the exposure of hydrophobic groups and the increase in SHE content. In terms of functional properties, the emulsifying ability, water adsorption capacity and oil adsorption capacity of the irradiated rice protein was improved dose-dependently, with a maximum occurring at 30 kGy. The foaming properties were also enhanced by electron beam irradiation; however, the effect was not dose-dependent. In contrast, all of the samples irradiated by electron beams presented lower emulsion stabilities than the control (0 kGy). Structural analysis demonstrated that EBI induces conformational changes and fragmentation of rice protein and enhances the release of many buried groups that are closely related to protein functions, thereby improving its functional properties. These results provide a theoretical basis for the application of EBI in improving protein properties in the future.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support for this research was provided by National Natural Science Foundation of China (No. 31471616), National Key R&D Program of China (2017YFD0401200), National Top Youth Talent for Grain Industry of China (LQ2016301), Special Scientific Research Project in Grain Public Welfare Industry (201513004) and National First-class Discipline Program of Food Science and Technology (JUFSTR20180203).References
- F. F. Shih, Mol. Nutr. Food Res., 2003, 47, 420–424 Search PubMed.
- L. Wang, X. Zhang, F. Liu, Y. Ding, R. Wang, X. Luo, Y. Li and Z. Chen, Innovative Food Science & Emerging Technologies, 2017, p. 41 Search PubMed.
- M. Shibasaki, S. Suzuki, H. Nemoto and T. Kuroume, J. Allergy Clin. Immunol., 1979, 64, 259–265 CrossRef CAS PubMed.
- M. Wang, N. S. Hettiarachchy, M. Qi, W. Burks and T. Siebenmorgen, J. Agric. Food Chem., 1999, 47, 411 CrossRef CAS PubMed.
- W. W. Koo and J. B. Lasekan, Minerva Pediatr., 2007, 59, 35–41 CAS.
- A. Fiocchi, M. Travaini, E. D'Auria, G. Banderali, L. Bernardo and E. Riva, Clin. Exp. Allergy, 2003, 33, 1576–1580 CrossRef CAS PubMed.
- Y. J. Chen, Y. Y. Chen, C. T. Wu, C. C. Yu and H. F. Liao, J. Cereal Sci., 2010, 51, 189–197 CrossRef CAS.
- X. Zhang, L. Wang, R. Wang, X. Luo, Y. Li and Z. Chen, Food Funct., 2016, 7, 1429–1437 RSC.
- R. L. Burris, C. H. Xie, P. Thampi, X. L. Wu, S. B. Melnyk and S. Nagarajan, Atherosclerosis, 2010, 212, 107–115 CrossRef CAS PubMed.
- J. Chen, S. Liu, R. Ye, G. Cai, B. Ji and Y. Wu, J. Funct. Foods, 2013, 5, 1684–1692 CrossRef CAS.
- I. Paraman, N. S. Hettiarachchy and C. Schaefer, Cereal Chem., 2008, 85, 76–81 CrossRef CAS.
- T. Wang, H. Zhang, L. Wang, R. Wang and Z. Chen, Food Chem., 2015, 178, 82–88 CrossRef CAS PubMed.
- X. H. Li, Y. L. Liu, C. P. Yi, Y. H. Cheng, S. M. Zhou and Y. F. Hua, J. Chin. Cereals Oils Assoc., 2010, 51, 7–12 CAS.
- A. A. Sadeghi, G. Raisali and M. Moradi, Anim. Feed Sci. Technol., 2007, 134, 140–151 CrossRef.
- C. Y. Ma, M. R. Sahasrabudhe, L. M. Poste, V. R. Harwalkar, J. R. Chambers and K. P. J. O'Hara, Can. Inst. Food Sci. Technol. J., 1990, 23, 226–232 CrossRef.
- H. P. Song, B. Kim, J. H. Choe, S. Jung, K. S. Kim, D. H. Kim and C. Jo, Radiat. Phys. Chem., 2009, 78, 217–221 CrossRef CAS.
- P. Supriya, K. R. Sridhar, S. Nareshkumar and S. Ganesh, Food Bioprocess Technol., 2012, 5, 1049–1060 CrossRef CAS.
- L. Wang, Y. Ding, X. Zhang, Y. Li, R. Wang, X. Luo, Y. Li, J. Li and Z. Chen, J. Food Eng., 2017, 202, 9–17 CrossRef CAS.
- A. Akbarian, M. Khorvash, G. R. Ghorbani, E. Ghasemi, M. Dehghan-Banadaky, P. Shawrang and M. H. Ghaffari, Livest. Sci., 2014, 168, 45–52 CrossRef.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS.
- A. Kato and S. Nakai, Biochim. Biophys. Acta, 1980, 624, 13–20 CrossRef CAS.
- K. N. Pearce and J. E. Kinsella, J. Agric. Food Chem., 1978, 26, 716–723 CrossRef CAS.
- S. Liu, C. Elmer, N. H. Low and M. T. Nickerson, Food Res. Int., 2010, 43, 489–495 CrossRef CAS.
- M. H. Simonian, Curr. Protoc. Cell Biol., 2002, 15, A.3B.1–A.3B.7 CrossRef PubMed.
- L. Yongwoo and S. Kyung Bin, J. Biochem. Mol. Biol., 2002, 35, 590–594 Search PubMed.
- H. K. S. Mudasir Ahmad Malik and C. Singh Saini, Food Hydrocolloids, 2017, 72, 312–322 CrossRef.
- A. J. Peacock and A. Calhoun, Polym. Sci., 2006, 87, 171–181 Search PubMed.
- C. H. Tang and C. Y. Ma, LWT–Food Sci. Technol., 2009, 42, 606–611 CrossRef CAS.
- L. Gang, L. Ji, S. Ke, W. Su, C. Jiwang, L. Ying and H. Qingrong, J. Agric. Food Chem., 2009, 57, 4552–4558 CrossRef PubMed.
- M. A. Malik, H. K. Sharma and C. S. Saini, Food Hydrocolloids, 2017, 72, 312–322 CrossRef CAS.
- H. W. Yeom, Q. H. Zhang and C. P. Dunne, Food Chem., 1999, 67, 53–59 CrossRef CAS.
- K. M. Mingchun Lv, H. Zhang, D. Xu and W. Yang, Food Chem., 2018, 254, 64–69 CrossRef PubMed.
- J. Jian, H. Ma, W. Kai, E. G. A. Yagoub, J. Owusu, W. Qu, R. He, C. Zhou and X. Ye, Ultrason. Sonochem., 2015, 24, 55–64 CrossRef PubMed.
- M. Parisi, A. Mazzini, R. T. Sorbi, R. Ramoni, S. Grolli and R. Favilla, Biochim. Biophys. Acta, 2003, 1652, 115–125 CrossRef CAS.
- Z. Wei and R. Yang, Eur. Food Res. Technol., 2008, 228, 47–54 CrossRef.
- Y. Sun, J. Chen, S. Zhang, H. Li, L. Jing, L. Lu, H. Uluko, Y. Su, W. Cui and W. Ge, J. Food Eng., 2014, 124, 11–18 CrossRef CAS.
- S. Tang, N. S. Hettiarachchy, R. Horax and S. Eswaranandam, J. Food Sci., 2010, 68, 152–157 CrossRef.
- W. Tao, F. Liu, W. Ren, W. Li, Z. Hao and Z. Chen, Food Funct., 2015, 6, 423–430 RSC.
- A. C. Alting, M. Weijers, E. H. de Hoog, V. D. P. Am, M. A. Cohen Stuart, R. J. Hamer, C. G. de Kruif and R. W. Visschers, J. Agric. Food Chem., 2004, 52, 623–631 CrossRef CAS PubMed.
- J. Z. Chaoting Wen, J. Zhou, Y. Duan, H. Zhang and H. Ma, Ultrason. Sonochem., 2018, 49, 294–302 CrossRef PubMed.
- K. M. Mingchun Lv, H. Zhang, D. Xu and W. Yang, Food Chem., 2018, 254, 64–69 CrossRef PubMed.
- Y. Zhang and S. J. Park, J. Catal., 2018, 361, 238–247 CrossRef CAS.
- Y. Zhang and S. J. Park, J. Catal., 2017, 355, 1–10 CrossRef CAS.
- Y. Zhang and S. J. Park, Carbon, 2017, 122, 287–297 CrossRef CAS.
- P. S. Jin and Y. Zhang, J. Mater. Chem. A, 2018, 6, 20304–20312 RSC.
- E. Li Chan, S. Nakai and D. F. Wood, J. Food Sci., 2010, 49, 345–350 CrossRef.
- A. Venktesh and V. Prakash, J. Agric. Food Chem., 1993, 41, 99–108 Search PubMed.
- R. S. H. Lam and M. T. Nickerson, LWT–Food Sci. Technol., 2015, 60, 427–434 CrossRef CAS.
- P. J. Halling, Crit. Rev. Food Sci. Nutr., 1981, 15, 155–203 CrossRef CAS PubMed.
- B. Miroljub, C. Slavica, P. Mirjana, S. Sla Ana, P. E. Milica, M. E. Ognjen and R. Nikola, Int. J. Mol. Sci., 2011, 12, 8372–8387 CrossRef PubMed.
- S. Tang, N. S. Hettiarachchy, R. Horax and S. Eswaranandam, J. Food Sci., 2003, 68, 152–157 CrossRef CAS.
- T. Wang, H. Zhang, L. Wang, R. Wang and Z. Chen, Food Chem., 2015, 178, 82–88 CrossRef CAS PubMed.
- D. C. Clark, I. F. Kiss, P. J. Wilde and D. R. Wilson, Food Hydrocolloids, 1992, 5, 541–548 CrossRef CAS.
- U. Singh, J. Food Sci. Technol., 2001, 38, 191–199 CAS.
- G. K. Chandi and D. S. Sogi, J. Food Eng., 2007, 79, 592–597 CrossRef CAS.
- J. F. Zayas, Functionality of Proteins in Food, Springer, Berlin, Heidelberg, 1997 Search PubMed.
- J. E. Kinsella and N. Melachouris, Crit. Rev. Food Sci. Nutr., 1976, 7, 219–280 CrossRef CAS.
- W. Prinyawiwatkul, K. H. McWatters, L. R. Beuchat and R. D. Phillips, J. Agric. Food Chem., 1997, 45, 1891–1899 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2019 |