Open Access Article
Open Access ArticlePreparation, identification and molecular docking study of novel osteoblast proliferation-promoting peptides from yak (Bos grunniens) bones†
Mengliang Ye a,
Wei Jiaab,
Chunhui Zhang
a,
Wei Jiaab,
Chunhui Zhang *a,
Qingshan Shena,
Lingyu Zhuac and
Lisha Wanga
*a,
Qingshan Shena,
Lingyu Zhuac and
Lisha Wanga
aInstitute of Food Science and Technology, Chinese Academy of Agricultural Sciences, No. 2 Yuan Ming Yuan West Road, Haidian District, Beijing 100193, China. E-mail: dr_zch@163.com; Fax: +86 10 6281 5950; Tel: +86 10 6281 9469
bInner Mongolia Tianqi Biotechnology Co., LTD, Chifeng 024000, China
cSchool of Tea and Food Science & Technology, Anhui Agricultural University, Hefei 230036, China
First published on 10th May 2019
Abstract
The aim of this study was to isolate and identify osteogenic bioactive peptides from yak bones collagen, while simultaneously investigating their underlying mechanisms for promoting osteoblast proliferation. Response surface methodology (RSM) was employed to investigate the effect of hydrolysis variables on the production of peptides with osteoblast proliferation-promoting activity (OPPA). The concentration of soluble peptides reached 0.5169 mg mL−1, which was well matched with the value (0.5189 mg mL−1) predicted by the model, with the following optimized conditions: hydrolysis time, 3.6 h; pH, 6.12; hydrolysis temperature, 54 °C; E/S (enzyme to substrate) of 5637 U g−1. Hydrolysates were then separated using an ultrafiltration membrane system, and the peptides (<3 kDa) possessed excellent OPPA with a dose–response relationship. A total of 59 novel peptides were identified by HPLC-MS/MS with Mascot analysis. GPSGPAGKDGRIGQPG (GP-16) and GDRGETGPAGPAGPIGPV (GD-18) were selected for docking to investigate the underlying mechanisms of interaction. The molecular docking study revealed that osteoblast proliferation stimulation activity of GP-16 and GD-18 was mainly attributed to the formation of very strong hydrogen bonds with the epidermal growth factor receptor (EGFR). These results indicate that such peptides are promising in the discovery of potential candidates for the future industrial production of functional peptides, which could be used in the mediated treatment of osteoporosis.
1 Introduction
As the world's population is transitioning gradually to an ageing stage, the incidence of osteoporosis increases annually. Osteoporosis, a major public health concern characterized by reduced bone mineral density and enhanced susceptibility to bone fractures, has affected more than 200 million people.1 Under normal physiological conditions, osteoblasts are responsible for new bone formation during bone development and remodeling.2 Currently, there are several pharmaceuticals (risedronate, teriparatide, alendronic acid and zoledronic acid) available to prevent bone loss in clinical practice.3–8 However, these treatments may cause several side effects including risk of esophagitis, nausea and abdominal pain.9,10 Therefore, seeking safer products that can promote bone formation and potentially reverse bone structural damage are attracting more attentions.The yak (Bos grunniens) is a unique kind of cattle that lives in a cold plateau region three kilometers (or more) above sea level without environmental pollution.11 Due to its special living environment, yak meat has been considered as a green food. However, yak bone, which is rich in collagen, is a major by-product of yak meat processing, has not been utilized effectively. Yak bone is one of the major components of Tibetan medicine (traditional Chinese medicine) and its function has mainly been associated with an improvement in bone health. However, the functional bioactive ingredients and the underlying mechanisms are still unclear. There are several published studies concerning the bioactive peptides (i.e., from collagen and lactoferrin) promoting the proliferation of osteoblasts and serving as bone growth factors, which counteract osteoporosis.12–14 Furthermore, bone collagen hydrolysates have had positive impact on osteogenesis in ovariectomized rats.15 Therefore, peptides from yak bone collagen are probably the key functional bioactive ingredients for bone health improvement.
Different enzymatic hydrolysates are characterized by different compositions and lengths of peptide chains.16 Therefore, the choice of enzyme determines the properties of the peptides, including the amino acids composition, flavor, and functionality. Previous studies have reported the functions of peptides, such as antioxidant activity, metal-chelation, radical-scavenging and antihypertensive activity.17–19 Nevertheless, there remains a little information about the isolation and identification of osteoblast proliferation-promoting bioactive peptides, especially those acquired by the targeted enzymatic hydrolysis of collagen from yak bones.
To better understand the osteogenic activities of hydrolysates from yak bones collagen, the peptides contributing to those activities should be investigated. The objective of the present study was to isolate and identify osteogenic bioactive peptides from yak bones collagen, while simultaneously investigating their underlying mechanisms of osteoblast proliferation-promoting activity (OPPA). Yak bones collagen was hydrolyzed using six commercial proteolytic enzymes, including trypsin, Alcalase, Flavourzyme, Protamex, Neutrase and papain. Hydrolysates were withdrawn during different hydrolysis phases and their degree of hydrolysis, and OPPA were determined. The effects of key processing variables (hydrolysis time, initial pH, temperature and enzyme to substrate (E/S) ratio) on the production of peptides were investigated using response surface methodology (RSM) in the present study. The bioactivity of peptides promoting MC3T3-E1 osteoblast proliferation was verified by the MTT method. Novel peptides with excellent OPPA were identified from hydrolysates by HPLC-MS/MS with Mascot analysis. Furthermore, the binding interaction of identified peptides within the active site of the epidermal growth factor receptor (EGFR) was determined using Discovery Studio 2018 software, which was recently reported to be the most popular docking program with high accuracy and versatility.12,21 This study was the first attempt to identify osteogenic bioactive peptides from the hydrolysate of yak bone collagen, which provides a feasible strategy for the preparation of peptides with OPPA and their potential applications in functional foods and pharmaceuticals.
2 Materials and methods
2.1 Materials and chemicals
Yak bones (leg bone) were obtained from the Tibet Academy of Agricultural and Animal Husbandry Sciences (Tibet, China). The bones were mechanically ground into granules with a diameter ranging from 3–5 mm and stored at −80 °C until analyzed. Pepsin (EC number 3.4.23.1, 1200 U mg−1), trypsin (EC number 3.4.21.4, 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 383 U mg−1), Alcalase (EC number 3.4.21.63, 221
383 U mg−1), Alcalase (EC number 3.4.21.63, 221![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 756 U mg−1), Flavourzyme (EC number 3.4.11.1, 15
756 U mg−1), Flavourzyme (EC number 3.4.11.1, 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 311 U mg−1), Protamex (56
311 U mg−1), Protamex (56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 084 U mg−1), Neutrase (EC number 3.4.24.28, 19
084 U mg−1), Neutrase (EC number 3.4.24.28, 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 267 U mg−1) and papain (EC number 3.4.22.2, 37
267 U mg−1) and papain (EC number 3.4.22.2, 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 018 U mg−1) were purchased from Solarbio (Beijing, China). Soluble peptides content was measured using a bicinchoninic acid (BCA) Protein Assay Kit from Beyotime (Shanghai, China). Trifluoroacetic acid (TFA), sodium dodecyl sulfate (SDS), and trinitrobenzene sulfonic acid (TNBS) were purchased from Sigma-Aldrich (Shanghai, China). HPLC grade acetonitrile (ACN) was purchased from Thermo Fisher Scientific (Waltham, USA). GP-16 and GD-18 (purity > 98%) was chemical synthesized and purified using high performance liquid chromatography (HPLC) by Beijing Protein Innovation (Beijing, China). All other chemicals and reagents were of analytical grade, unless otherwise stated.
018 U mg−1) were purchased from Solarbio (Beijing, China). Soluble peptides content was measured using a bicinchoninic acid (BCA) Protein Assay Kit from Beyotime (Shanghai, China). Trifluoroacetic acid (TFA), sodium dodecyl sulfate (SDS), and trinitrobenzene sulfonic acid (TNBS) were purchased from Sigma-Aldrich (Shanghai, China). HPLC grade acetonitrile (ACN) was purchased from Thermo Fisher Scientific (Waltham, USA). GP-16 and GD-18 (purity > 98%) was chemical synthesized and purified using high performance liquid chromatography (HPLC) by Beijing Protein Innovation (Beijing, China). All other chemicals and reagents were of analytical grade, unless otherwise stated.
2.2 Preparation of yak bone collagen hydrolysates
The collagen of yak bones was prepared according to the method of Kittiphattanabawon et al. with slight modification and then dispersed and homogenized with PBS solution (with pH values adjusted for different enzymes) at a concentration of 0.5% (w/w).22 Enzymes were added to the solution in a ratio of 5000 U g−1 (enzyme/protein) to initiate the hydrolysis. Aliquots were taken at 0, 0.25, 0.5, 1, 2, 3, 4, and 5 h. After proteolysis, the hydrolysates were heated to 95 °C for 10 min and centrifuged at 8000 × g for 20 min. The supernatant was lyophilized and stored at −20 °C for future analysis. The degree of hydrolysis (DH) indicating the ratio of hydrolyzed peptide bonds was assayed by the TNBS method.232.3 Determination of soluble peptides content
The content of soluble peptides (CSP) was determined using a BCA Protein Assay Reagent Kit following the manufacturer's instructions.24 Briefly, 0.25 mL hydrolysates from different treatments were homogenized with equal volumes of 10% (w/v) TCA in a 1 mL tube and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g at 4 °C for 20 min. The supernatant (50 μL) was pipetted into a 96-well plate, followed by the addition of 200 μL BCA working solution. The plates were incubated at room temperature for 2 h and then the absorbance values were measured at 562 nm with a microplate reader. The CSP was calculated from a standard curve (Y = 1.1842X + 0.1366, R2 = 0.997) by varying standard bovine serum albumin (BSA) concentrations (from 0.025 mg mL−1 to 0.5 mg mL−1) at an absorbance at 562 nm.
000 × g at 4 °C for 20 min. The supernatant (50 μL) was pipetted into a 96-well plate, followed by the addition of 200 μL BCA working solution. The plates were incubated at room temperature for 2 h and then the absorbance values were measured at 562 nm with a microplate reader. The CSP was calculated from a standard curve (Y = 1.1842X + 0.1366, R2 = 0.997) by varying standard bovine serum albumin (BSA) concentrations (from 0.025 mg mL−1 to 0.5 mg mL−1) at an absorbance at 562 nm.
2.4 Cell culture and MTT assays
The MC3T3-E1 osteoblast cell line was purchased from the Institute of Basic Medicine at the Chinese Academy of Medical Sciences (Beijing, China). The cells were cultured in α-MEM medium containing 10% foetal bovine serum and 1% penicillin–streptomycin under a humidified atmosphere of 5% CO2 and 95% air at 37 °C (Stericycle CO2 incubator, Thermo Fisher Scientific Inc., Waltham, MA). MC3T3-E1 cells were passaged at 90% confluence after digestion in 0.25% trypsin. The number of cells was counted by using a blood counting chamber and was adjusted to a cell density of 1.25 × 104 cells per mL. For the proliferation and survival assays, 200 μL of cell mediums were seeded into 96-well plates, and the assays were conducted three times at each concentration. The proliferation of osteoblasts was measured by the MTT method after incubating for 24, 48, 72 and 96 h.2.5 Experimental design-based on response surface methodology
Optimum enzymatic hydrolysis conditions for yak bone collagen peptides (YBCP) were investigated using RSM. The effect of changing a single factor on the CSP was analysed to determine the preliminary range of the hydrolysis variables of hydrolysis time, pH, temperature and E/S. Then, the hydrolysis parameters were optimized using RSM. The Design-Expert package (Version 8.0, Minneapolis, USA) was employed for regression analysis of the data. The independent variables and their levels were chosen based on the results of preliminary experiments. Soluble peptides content was the dependent variable.2.6 Fractionation of YBCP
The lyophilized YBCP was dissolved in distilled water and fractionated by ultrafiltration. The fractions were collected separately by their different molecular weights (MW) and lyophilized to evaluate their OPPA. The distribution profile of the MW of the fraction (<3 kDa) was estimated by size exclusion chromatography and methods described by Zhang et al. (2017).232.7 Peptides identification by HPLC-MS/MS
The active fraction was further analyzed by a reverse phase capillary column (Phenomenex, Aqua® 150 × 4.6 mm, 5 μm C18 125 Å, USA). The elution was conducted with a 65 min gradient from 1% to 35% (v/v) solvent B (0.1% formic acid in ACN) in solvent A (0.1% formic acid in water). Mass spectroscopy analysis was performed using a Thermo Q-Exactive high resolution mass spectrometer (Thermo Scientific, Waltham, MA, USA) in positive ion mode with a capillary temperature of 320 °C and a 2 kV spray voltage applied to the emitter. Peptide sequences were determined based on the MS/MS spectra and Swiss-Prot database search, which was performed using a Mascot 2.4 search engine (Matrix Science, Boston, MA, USA).2.8 Molecular docking
The structures of identified peptides were constructed using Chem Office 2014 software (Cambridge Soft Co., USA). Molecular docking of the estimated osteoblast proliferation-promoting bioactive peptides and epidermal growth factor receptor (EGFR) was carried out using Discovery Studio 2018 software (NeoTrident Technology Ltd., Beijing, China). The crystal structure of the EGFR-EGF protein-ligand complex (PDB ID: 1IVO) was derived from the RCSB Protein Data Bank (https://www.rcsb.org/structure/1ivo) and used as the template to compare the docking mode of peptides bound to EGFR.25 Before the docking, the complex crystal structure was constructed with the clean and prepare protein program to model missing loop regions, remove water, add hydrogen and complete other procedures as required. Additionally, the active site was set at the original position (EGF site) in the 1IVO complex for the docking of bioactive peptides and EGFR. The binding sites were created with the coordinates x, y, and z coordinates of 124.403, 75.629 and 51.305, respectively, and a radius of 22.248 Å. In this study, we compared three kinds of common used docking algorithm (CDOCKER, LibDock and LigandFit) to evaluate and select the most suitable docking algorithm. The EGF, the native ligand of 1IVO, was removed and docked again to the initial active site of EGFR using the above three kind of docking algorithms. The number of generated poses and the RMSD values (set initial EGF conformation as reference) were set as standard of validation. The CDOCKER algorithm (with 10 poses and the minimum RMSD value of 1.9480) was finally selected for further investigation (Table S1†). The potential mechanisms of action of certain bioactive peptides promoting osteoblast proliferation were conducted according to a similar molecular docking procedure reported by Shi et al. (2017).12 Molecular docking results were evaluated based on the CDOCKER energy scores, active site, and types of interaction with EGFR.2.9 Statistical analysis
Statistical analysis was conducted using SPSS software, version 20 (SPSS Inc., Chicago, IL, USA). Data were presented as means ± standard deviations (SD) (n = 3). One-way analysis of variance (ANOVA) followed by Duncan's multiple range test was performed to evaluate statistical significance. Statistical significance was set at P < 0.05.3 Results and discussion
3.1 DH and promotion of osteoblast proliferation of hydrolysates
The DH is defined as the percentage of the number of peptide bonds broken accounting for the total number of peptide bonds in the substrate. DH is associated with the chain length of the peptides, which was an indicator of the cleavage of peptide bonds.23,26 Moreover, the DH is proportionally associated with the size or structure of peptides, and thus affects the exposure of certain amino acids and biological activities.27 The hydrolysates of collagen from yak bones were obtained using enzymatic hydrolysis with trypsin, Alcalase, Flavourzyme, Protamex, Neutrase and papain. The results are shown in Fig. S1,† and Alcalase caused the most extensive hydrolysis. Protamex and Neutrase behaved very similarly in hydrolyzing the yak bone collagen with almost the same DH after 2 h of hydrolysis. In the present study, Alcalase and Flavourzyme hydrolysis were more effective than other treatments. Generally, the substrate type involved in the enzymatic hydrolysis may influence the DH of the final hydrolysates. Therefore, yak bone collagen may be more substantially hydrolyzed by Alcalase and Flavourzyme.Interestingly, measuring the bioactivity of samples that were hydrolyzed by different enzymes revealed that papain and Neutrase hydrolysates, with lower DH, promoted a higher level of osteoblast proliferation than that of other enzymes (Fig. S2†). Moreover, after cell culture for 96 h, Neutrase hydrolysates showed higher activity of promoting osteoblast proliferation than that of papain (P < 0.05). These findings should be emphasized, because a dearth of research on the multifunctional properties of Neutrase hydrolysates has been reported, despite protein hydrolyzed by other enzymes possessing multiple functions.28 Guo et al. also reported that a higher DH did not lead to greater bioactivity.26 The structure–activity relationship of OPPA peptides has not yet been fully elucidated. Enzyme type plays an important role in the bioactivity of the produced hydrolysate.29 Neutrase is an endopeptidase and has no specific selectivity at the restriction site, which might have contributed to obtaining more peptides with greater OPPA. Accordingly, the phenomenon that Neutrase hydrolysate has a higher OPPA was highlighted, and more extensive investigations should be performed.
3.2 Optimization of hydrolysis conditions of YBCP
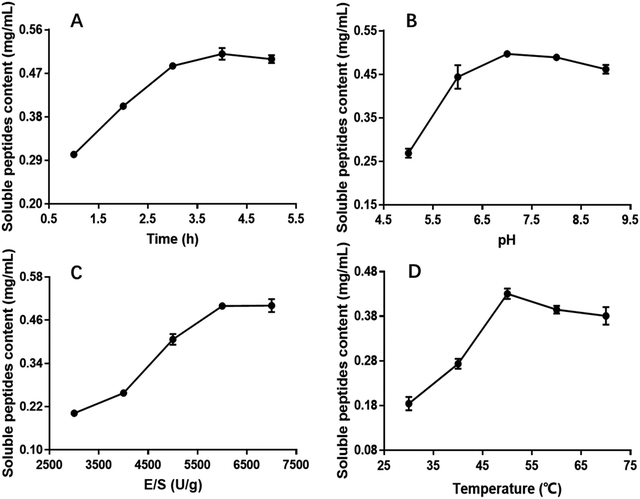
The influence of the initial pH on the hydrolysis efficiency might be due to the requirement of optimum pH for enzymatic reactions. Fig. 1B showed the effect of pH on soluble peptides content when other extraction parameters were set as follows: hydrolysis time of 4 h, E/S of 5000 U g−1 and temperature of 50 °C. The DH increased with increasing pH, reaching a maximum at an initial pH of 7.0 before decreasing, and this phenomenon was in good agreement with former research.32 At a hydrolysis pH of 5 to 7, the variance of the soluble peptides content was relatively rapid reaching a maximum of 0.4971 mg mL−1 and remaining at this level at approximately pH 7.0. Therefore, 7.0 was considered to be the optimal pH.
The effect of E/S (3000, 4000, 5000, 6000 and 7000 U g−1) on the CSP was investigated when the other three factors i.e. hydrolysis time, pH and temperature were fixed at 4 h, 7.0 and 50 °C, respectively. Fig. 1C showed that E/S had a significant effect on the CSP. The content values increased with increasing E/S between 3000 and 6000 U g−1. Then, the value was basically unchanged or slightly. Although the soluble peptides content was also high at 7000 U g−1, increasing the enzyme concentration results in more expensive industrial hydrolysis process. Therefore, 6000 U g−1 was sufficient and adopted in the present study.
Hydrolysis temperature is one of the important variables affecting the yield of soluble peptides, and it is necessary to select an optimum hydrolysis temperature to assure maximum soluble peptides content. Therefore, hydrolysis was carried out using the following temperatures: 30, 40, 50, 60 and 70 °C. The other three hydrolysis parameters were set at a hydrolysis time of 4 h, pH of 7.0 and E/S of 6000 U g−1 (Fig. 1D). The results indicated that the soluble peptides content significantly increased from 0.18 to 0.43 mg mL−1 when the temperature varied from 30 to 50 °C, and then slightly decreased when temperature exceeded 50 °C. Similar results were observed in a previous study.33 Thus, hydrolysis at 50 °C was deemed favorable for producing maximum soluble peptides content.
According to the single parameter studies, hydrolysis time of 4 h, E/S 6000 of U g−1, pH of 7.0 and temperature of 50 °C were adopted for RSM experiments.
The regression, credibility of the model and ANOVA for the response surface quadratic model were analyzed and are shown in Table 1. Based on the P-value of the model and the low probability value (P < 0.0001), the model was highly significant. As shown in Table 1, the linear coefficients (A, B, C and D), quadratic term coefficients (A2, B2, C2 and D2) and interaction coefficients (AB, AC, AD, BC, BD and CD) were all found to be significant (P < 0.05). The F-value represents the contribution rate of factors. The order of the impact strength of the factors was hydrolysis temperature > hydrolysis time > E/S > initial pH, which signified that the impact of hydrolysis temperature and hydrolysis time on the yield of soluble peptides content during hydrolysis were significantly higher than the other two factors (E/S and initial pH). The initial pH showed the least impact on the yield. A similar trend has been reported for peptides from cod bone.29
| Source | Sum of squares | Coefficient estimate | Degree of freedom | Standard error | Mean square | F-value | P-value |
|---|---|---|---|---|---|---|---|
| a Std. Dev., standard deviation; C.V., coefficient of variation. | |||||||
| Model | 0.073 | 0.49 | 14 | 2.765 × 10−3 | 5.204 × 10−3 | 136.14 | <0.0001 significant |
| A-time (h) | 0.010 | −0.029 | 1 | 1.785 × 10−3 | 0.010 | 270.12 | <0.0001 |
| B-temperature (°C) | 0.032 | 0.052 | 1 | 1.785 × 10−3 | 0.032 | 847.79 | <0.0001 |
| C-E/S (U g−1) | 1.070 × 10−3 | −9.442 × 10−3 | 1 | 1.785 × 10−3 | 1.070 × 10−3 | 27.99 | 0.0001 |
| D-pH | 5.031 × 10−4 | −6.475 × 10−3 | 1 | 1.785 × 10−3 | 5.031 × 10−4 | 13.16 | 0.0027 |
| AB | 5.176 × 10−4 | −0.011 | 1 | 3.091 × 10−3 | 5.176 × 10−4 | 13.54 | 0.0025 |
| AC | 5.176 × 10−4 | 0.011 | 1 | 3.091 × 10−3 | 5.176 × 10−4 | 13.54 | 0.0025 |
| AD | 8.151 × 10−4 | 0.014 | 1 | 3.091 × 10−3 | 8.151 × 10−4 | 21.32 | 0.0004 |
| BC | 6.300 × 10−4 | 0.013 | 1 | 3.091 × 10−3 | 6.300 × 10−4 | 16.48 | 0.0012 |
| BD | 1.257 × 10−3 | −0.018 | 1 | 3.091 × 10−3 | 1.257 × 10−3 | 32.88 | <0.0001 |
| CD | 4.906 × 10−4 | 0.011 | 1 | 3.091 × 10−3 | 4.906 × 10−4 | 12.84 | 0.0030 |
| A2 | 7.707 × 10−3 | −0.034 | 1 | 2.428 × 10−3 | 7.707 × 10−3 | 201.63 | <0.0001 |
| B2 | 0.011 | −0.041 | 1 | 2.428 × 10−3 | 0.011 | 278.44 | <0.0001 |
| C2 | 0.015 | −0.048 | 1 | 2.428 × 10−3 | 0.015 | 385.82 | <0.0001 |
| D2 | 2.267 × 10−3 | −0.019 | 1 | 2.428 × 10−3 | 2.267 × 10−3 | 59.31 | <0.0001 |
| Residual | 5.351 × 10−4 | 14 | 3.822 × 10−5 | ||||
| Lack of fit | 3.496 × 10−4 | 10 | 3.496 × 10−5 | 0.75 | 0.6746 not significant | ||
| Pure error | 1.855 × 10−4 | 4 | 4.638 × 10−5 | ||||
| Cor total | 0.073 | 28 | |||||
| Std. Dev. | 6.183 × 10−3 | R-squared | 0.9927 | ||||
| Mean | 0.44 | Adj R-squared | 0.9854 | ||||
| C.V. (%) | 1.42 | Rred R-squared | 0.9686 | ||||
| Press | 2.304 × 10−3 | Adeq precision | 44.755 | ||||
The precision of the model can be verified by the determination coefficient (R2) and the correlation coefficient (R). The R2 implied that 98.5% of the sample variation for yak bone collagen could be attributed to the independent variables, whereas only approximately 1.5% of the total variation cannot be explained by the model, which means the model fit the experimental data well.32,34 In general, a regression model with an R2 value greater than 0.9 is considered to have a very high correlation. An R value (correlation coefficient) close to 1 is associated with a better correlation between the experimental and predicted values. The value of R indicated a close agreement between the experimental results and the theoretical values predicted by the model equation. The ANOVA also demonstrated that there was a non-significant (P > 0.05) lack of fit, which further validated the model.
Response surface figures can directly reflect the impact of different variables on the response values. Based on the quadratic polynomial fitting equation, the Design Expert software was used to draw two-factor response surface maps where the effects of the two-factor interaction on the CSP could be observed (Fig. 2). The purpose of optimization was to determine the hydrolysis conditions that gave the maximum predicted soluble peptides content. The plots present the response as a function of every two factors, maintaining the other variables constant at their middle level (center value of the testing ranges).30 The maximum predicted value indicated by the surface was confined in the smallest ellipse in the contour diagram.31 The smallest ellipse in the contour plots illustrated that there was an optimal interaction between the independent variables. The variables that greatly influenced the hydrolysis efficiency were steep, and the response value changed substantially, while for variables that minimally influenced the hydrolysis efficiency, the response value changed minimally.
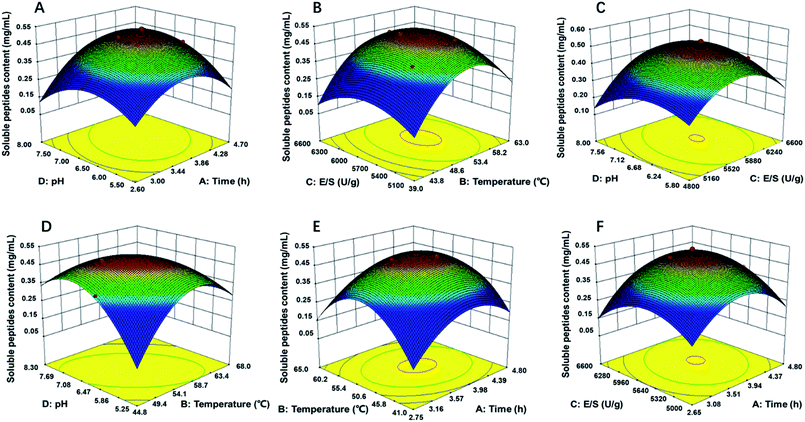 | ||
| Fig. 2 Response surface plots (3D) for the effects of variables on OPPA: (A) pH and time; (B) E/S and temperature; (C) pH and E/S; (D) pH and temperature; (E) temperature and time; (F) E/S and time. | ||
As showed in Fig. 2B, D and E, the yield of soluble peptides increased rapidly with increasing hydrolysis temperature, which indicated that temperature was one primary factor during hydrolysis procedure. Surface response, contour plots and the F-value of the two-factor interaction treatments showed that the interaction between any two factors that affected the soluble peptides content were in the following order: BD > AD > BC > AB > CD; the interaction of AB and AC was similar (Fig. 2E and F). By applying multiple regression analysis to the experimental data, the mathematical model demonstrated that the stationary point for a first order partial derivative presenting the maximum CSP had the following critical values: hydrolysis time of 3.6 h, E/S of 5637 U g−1, initial pH of 6.12 and temperature of 54 °C. Under optimal conditions, the predicted CSP was 0.5189 mg mL−1. The experimentally obtained mean content value of 0.5169 mg mL−1 (not significant at a 5% confidence level) validated that the RSM model was satisfactory and accurate.
3.3 Proliferation of osteoblasts
The OPPA of YBCP (<3 kDa) at different culture times was measured by the MTT method. MC3T3-E1 cells were cultured for 24, 48, 72 and 96 h with different concentrations (0.1 g mL−1, 0.2 g mL−1 and 0.5 g mL−1) (Fig. S3†). Significant increases in cell proliferation were observed in the group of 0.2 mg mL−1 and 0.5 mg mL−1 YBCP after 48 h culture time (P < 0.05). After 96 h of treatment, the cell proliferation rate increased to its highest values of 175.4% for the concentration of 0.5 mg mL−1. Herein, YBCP possessed excellent OPPA when compared with the control group. In addition, it demonstrated a good dose–response relationship.3.4 Identification of peptides sequences by mass spectrometry
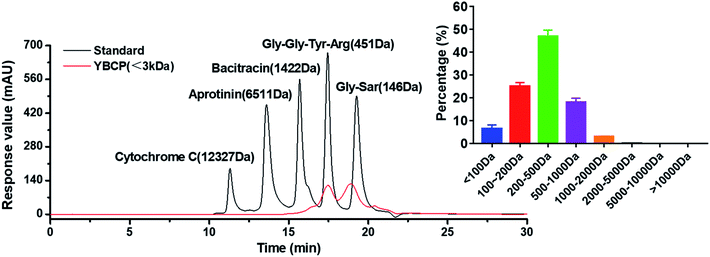
Food-derived bioactive peptides represent potential functional food ingredients. The most biologically potent peptides are commonly short in length, comprising 2 to 9 amino acids, and possess MW lower than 3 kDa.35,36 As shown in Fig. 3, the fraction from YBCP had many low MW peptides < 2000 Da, and 96.8% of the peptides' MW were lower than 1000 Da. A total of 59 peptides were identified based on the HPLC-MS/MS and Mascot analysis (Fig. 4). It is well known that the molecular structure of collagen type I consists of three polypeptide alpha-chains (two α1 chains and one α2 chain) twisted together to form a triple helix.37 Herein, 28 peptides were from the α1 chain, and 31 peptides were from the α2 chain, which accounted for 47 and 53% of all identified peptides, respectively. These peptides comprised 7–24 amino acids and molecular masses from 715.3575 Da to 2160.1036 Da (Tables 2 and 3). The amino acid composition and sequences affect the peptides bioactivity. However, very little research has focused on the OPPA of yak bones collagen peptides, thereby greatly limiting their exploitation and application.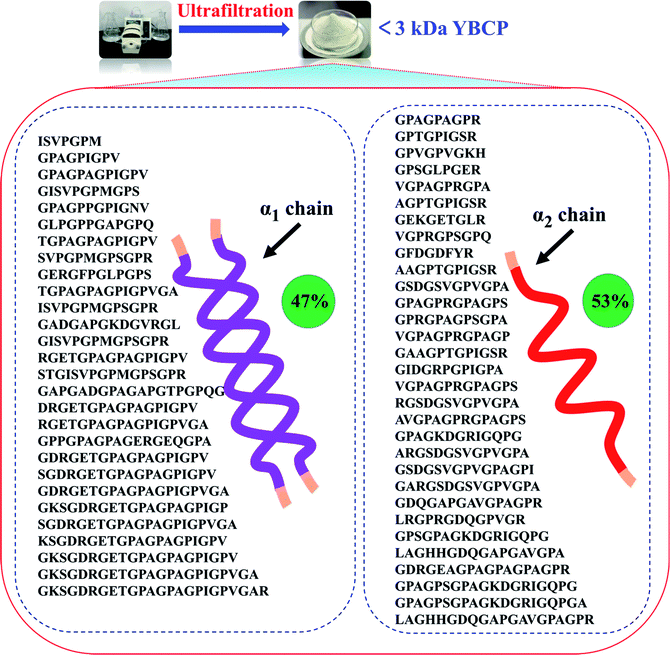 | ||
| Fig. 4 Peptides sequences identified from YBCP (<3 kDa) by HPLC-MS/MS and Swiss-Prot database search. | ||
| Rank | Amino acid sequence | Mr Calc. | Site range | Length | Score | -CE (kcal mol−1) |
|---|---|---|---|---|---|---|
| a Mr Calc., molecular calculation; CE, CDOCKER_energy. | ||||||
| 1 | GKSGDRGETGPAGPAGPIGPVGAR | 2160.1036 | 1060–1083 | 24 | 75.43 | 158.080 |
| 2 | GKSGDRGETGPAGPAGPIGPVGA | 2004.0025 | 1060–1082 | 23 | 63.23 | 132.928 |
| 3 | SGDRGETGPAGPAGPIGPVGA | 1818.8861 | 1062–1082 | 21 | 75.10 | 131.953 |
| 4 | GKSGDRGETGPAGPAGPIGP | 1776.8755 | 1060–1079 | 20 | 42.04 | 119.609 |
| 5 | GKSGDRGETGPAGPAGPIGPV | 1875.9439 | 1060–1080 | 21 | 69.04 | 117.383 |
| 6 | GADGAPGKDGVRGL | 1268.6473 | 751–764 | 14 | 42.97 | 117.184 |
| 7 | GDRGETGPAGPAGPIGPVGA | 1731.8540 | 1063–1082 | 20 | 69.81 | 108.143 |
| 8 | SGDRGETGPAGPAGPIGPV | 1690.8275 | 1062–1080 | 19 | 91.32 | 107.348 |
| 9 | KSGDRGETGPAGPAGPIGPV | 1818.9224 | 1061–1080 | 20 | 77.53 | 105.362 |
| 10 | GDRGETGPAGPAGPIGPV | 1603.7955 | 1063–1080 | 18 | 99.84 | 98.992 |
| 11 | GPPGPAGPAGERGEQGPA | 1600.7594 | 619–636 | 18 | 56.78 | 98.003 |
| 12 | GAPGADGPAGAPGTPGPQG | 1530.7063 | 934–952 | 19 | 62.80 | 89.869 |
| 13 | DRGETGPAGPAGPIGPV | 1546.7740 | 1064–1080 | 17 | 76.90 | 86.238 |
| 14 | STGISVPGPMGPSGPR | 1511.7403 | 171–186 | 16 | 58.51 | 85.072 |
| 15 | RGETGPAGPAGPIGPVGA | 1559.8056 | 1065–1082 | 18 | 45.22 | 83.832 |
| 16 | TGPAGPAGPIGPVGA | 1217.6405 | 1068–1082 | 15 | 51.32 | 75.836 |
| 17 | RGETGPAGPAGPIGPV | 1431.7470 | 1065–1080 | 16 | 56.63 | 65.216 |
| 18 | GERGFPGLPGPS | 1169.5829 | 967–978 | 12 | 45.82 | 62.140 |
| 19 | GISVPGPMGPSGPR | 1307.6656 | 173–186 | 14 | 73.13 | 59.186 |
| 20 | ISVPGPMGPSGPR | 1266.6391 | 174–186 | 13 | 76.32 | 58.233 |
| 21 | TGPAGPAGPIGPV | 1089.5819 | 1068–1080 | 13 | 50.86 | 54.586 |
| 22 | SVPGPMGPSGPR | 1153.5550 | 175–186 | 12 | 60.96 | 48.188 |
| 23 | GISVPGPMGPS | 997.4903 | 173–183 | 11 | 41.83 | 46.853 |
| 24 | GPAGPAGPIGPV | 988.5342 | 1069–1080 | 12 | 63.19 | 45.080 |
| 25 | GPAGPPGPIGNV | 1031.5400 | 844–855 | 12 | 93.84 | 42.997 |
| 26 | GPAGPIGPV | 763.4229 | 1072–1080 | 9 | 42.18 | 34.299 |
| 27 | ISVPGPM | 715.3575 | 174–180 | 7 | 51.80 | 29.412 |
| 28 | GLPGPPGAPGPQ | 1043.5400 | 187–198 | 12 | 47.00 | 29.395 |
| Rank | Amino acid sequence | Mr Calc. | Site range | Length | Score | -CE (kcal mol−1) |
|---|---|---|---|---|---|---|
| a Mr Calc., molecular calculation; CE, CDOCKER_energy. | ||||||
| 1 | LAGHHGDQGAPGAVGPAGPR | 1820.9030 | 1032–1051 | 20 | 47.16 | 156.365 |
| 2 | GPAGPSGPAGKDGRIGQPG | 1674.8438 | 1052–1070 | 19 | 94.54 | 124.349 |
| 3 | LAGHHGDQGAPGAVGPA | 1510.7277 | 1032–1048 | 17 | 44.03 | 123.166 |
| 4 | GPAGPSGPAGKDGRIGQPGA | 1745.8809 | 1052–1071 | 20 | 75.30 | 119.225 |
| 5 | GPSGPAGKDGRIGQPG | 1449.7325 | 1055–1070 | 16 | 63.63 | 112.205 |
| 6 | GDRGEAGPAGPAGPAGPR | 1588.7706 | 689–706 | 18 | 47.26 | 104.467 |
| 7 | GEKGETGLR | 945.4879 | 653–661 | 9 | 49.29 | 96.716 |
| 8 | GPAGKDGRIGQPG | 1208.6262 | 1058–1070 | 13 | 57.87 | 91.074 |
| 9 | LRGPRGDQGPVGR | 1363.7433 | 816–828 | 13 | 42.69 | 86.889 |
| 10 | GFDGDFYR | 975.4087 | 1109–1116 | 8 | 48.05 | 84.994 |
| 11 | ARGSDGSVGPVGPA | 1225.6051 | 231–244 | 14 | 55.10 | 82.368 |
| 12 | GDQGAPGAVGPAGPR | 1305.6426 | 1037–1051 | 15 | 45.42 | 78.154 |
| 13 | GARGSDGSVGPVGPA | 1282.6266 | 230–244 | 15 | 63.71 | 74.196 |
| 14 | RGSDGSVGPVGPA | 1154.5680 | 232–244 | 13 | 47.55 | 73.005 |
| 15 | AVGPAGPRGPAGPS | 1189.6204 | 1044–1057 | 14 | 51.75 | 69.936 |
| 16 | GSDGSVGPVGPA | 998.4669 | 233–244 | 12 | 40.73 | 69.381 |
| 17 | GAAGPTGPIGSR | 1039.5411 | 596–607 | 12 | 62.81 | 65.858 |
| 18 | GSDGSVGPVGPAGPI | 1265.6252 | 233–247 | 15 | 46.84 | 62.994 |
| 19 | AAGPTGPIGSR | 982.5196 | 597–607 | 11 | 49.21 | 60.065 |
| 20 | GIDGRPGPIGPA | 1105.5880 | 470–481 | 12 | 40.33 | 56.523 |
| 21 | GPVGPVGKH | 846.4712 | 965–973 | 9 | 43.32 | 53.582 |
| 22 | GPSGLPGER | 868.4403 | 635–643 | 9 | 47.00 | 52.185 |
| 23 | AGPTGPIGSR | 911.4825 | 598–607 | 10 | 45.83 | 52.097 |
| 24 | GPTGPIGSR | 840.4454 | 599–607 | 9 | 45.82 | 51.547 |
| 25 | VGPAGPRGPAGPS | 1118.5833 | 1045–1057 | 13 | 46.23 | 48.694 |
| 26 | VGPRGPSGPQ | 950.4934 | 991–1000 | 10 | 41.21 | 47.609 |
| 27 | GPRGPAGPSGPA | 1019.5148 | 1049–1060 | 12 | 53.78 | 43.976 |
| 28 | VGPAGPRGPAGP | 1031.5512 | 1045–1056 | 12 | 49.80 | 43.232 |
| 29 | GPAGPRGPAGPS | 1019.5148 | 1046–1057 | 12 | 46.12 | 43.228 |
| 30 | VGPAGPRGPA | 877.4770 | 1045–1054 | 10 | 49.71 | 42.472 |
| 31 | GPAGPAGPR | 778.4086 | 698–706 | 9 | 53.21 | 35.436 |
3.5 Molecular docking of YBCP and EGFR
The EGFR is a transmembrane glycoprotein, the signaling of which may affect bone formation through regulation of endochondral ossification.25 The inactivation of EGFR decreased expression levels of matrix metalloproteinases accompanied by increases in collagen fibrils and reduced degradation of extracellular matrix products in the growth plate.20 The EGFR can be activated by the EGF-related peptide growth factor family and multiple ligands.38 The activation of EGFR may play an essential role in the remodeling of the extracellular matrix.39 Therefore, bioactive peptides that activated the EGFR were considered to promote bone formation. Under normal physiological conditions, osteoblasts are responsible for new bone formation during bone development and remodeling. In the present study, yak bone collagen peptides can significantly promote osteoblasts proliferation, which may due to the fact that yak bone collagen peptides can regulate the body's physiological functions (promoting bone formation or reducing degradation of extracellular matrix products) by interacting with EGFR.In this study, we selected CDOCKER as the suitable docking algorithm for further investigation based on the number of poses and the RMSD value.12 CDOCKER is a scoring function to dock ligands into receptor binding site using a CHARMm force field based molecular dynamics scheme, which offers all the advantages of full ligand flexibility (including bonds, angles, dihedrals), the CHARMm family of force fields, the flexibility of the CHARMm engine, and reasonable computation times.12,20 Based on the traditional molecular mechanics of force field, the function ‘‘CDOCKER” was generally used due to more accurate than other docking algorithms. The higher affinity of ligand and receptor shows, the higher the score will be.20 The OPPA of the peptides would be indicated by the score of the affinity energy, i.e.‘‘-CDOCKER Energy”. The value of “-CDOCKER Energy” was calculated and listed in Tables 2 and 3. The “-CDOCKER Energy” ranged from 29.395 to 158.080 kcal mol−1. The peptide GKSGDRGETGPAGPAGPIGPVGAR showed the highest affinity among all the peptides with a “-CDOCKER Energy” of 158.080 kcal mol−1 compared with the other peptides. Of peptides with a “-CDOCKER Energy” >100 kcal mol−1, seven peptides from the α1 chain shared the same amino acid sequence (GDRGETGPAGPAGPIGPV) and three peptides from the α2 chain shared the same amino acid sequence (GPSGPAGKDGRIGQPG). Therefore, peptides GPSGPAGKDGRIGQPG (GP-16) and GDRGETGPAGPAGPIGPV (GD-18) were selected for docking to investigate the underlying mechanisms of interactions.
As shown in Fig. 5, the interacting amino acid residues of GP-16 with EGFR were Lys13, Thr15, Gln16, Gly18, Leu325 and Asp355, and the interacting amino acid residues of GD-18 with EGFR were Ser11, Asn12, Thr15, Gln16, Leu17, Leu325, Val350, Asp355, Phe357 and His409. The non-bonded interactions between the amino acid residues of the EGFR and the two peptides (GP-16 and GD-18) in their docking poses included H-bonds, interpolated charge interactions and hydrophobic interactions. GP-16 formed eleven H-bonds with EGFR residues Lys13, Thr15, Gln16, Gly18 and Asp355, and GD-18 formed nine H-bonds with EGFR residues Ser11, Asn12, Thr15, Gln16, Asp355 and His409. These interaction sites were similar with EGF and both of them were located at sites of Val350 and Phe357 in the EGFR.25 Moreover, Shi, et al. reported that 9 key amino acid residues, namely Lys13, Leu14, Thr15, Gln16, Tyr45, Leu98, Ser99, His409 and Ser418, were present in the interaction sites of the peptide ENLPEKADRDQYEL and EGFR, which included three same amino acid residues (Thr15, Gln16 and His409) demonstrated in the present study.12 They demonstrated that the peptides GP-16 and GD-18 may promote osteoblast proliferation with a molecular mechanism similar to the ENLPEKADRDQYEL sequence and EGF.
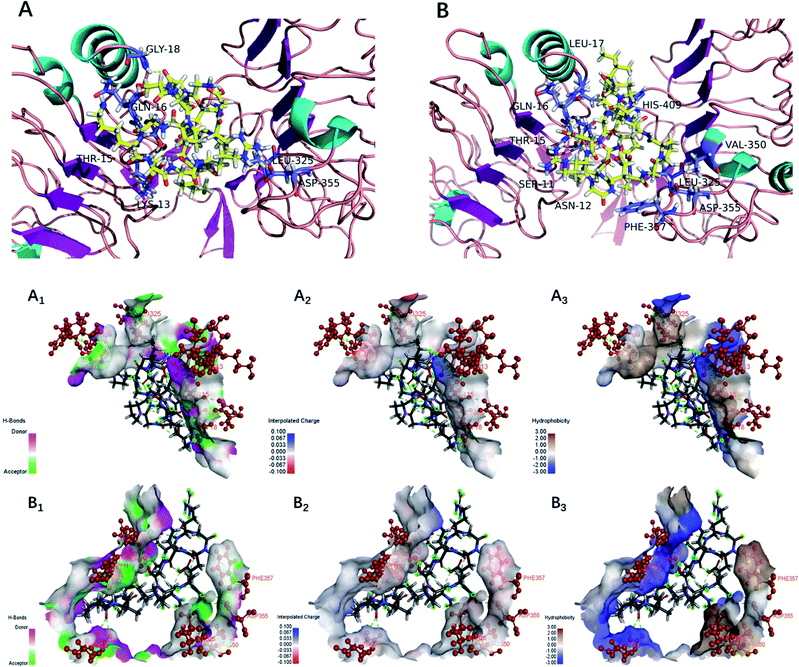 | ||
| Fig. 5 Docking for the interaction of YBCP with EFGR (PDB: 1IVO). (A) 3D structure of GP-16 and EFGR complex after docking; (B) 3D structure of GD-18 and EFGR complex after docking; (A1, B1), (A2, B2) and (A3, B3) represented H-bonds interaction, interpolated charge interaction and hydrophobic interaction, respectively. | ||
The higher number of H-bonds could explain the greater interaction between the EGFR and peptides GP-16 and GD-18. Additionally, GP-16 also has an alkyl hydrophobic interaction with Leu325 and a charge–charge interaction with Asp355; and GD-18 has four alkyl hydrophobic interactions with Leu17, Leu325, Val350 and His409, which also contributed to the stabilization of the EGFR and the peptides. These results suggest that the greater interactions of peptides GP-16 and GD-18 with the EGFR may contribute to the stronger stimulation potency on osteoblasts proliferation. Therefore, the fraction of YBCP promoting osteoblast proliferation may partly be attributed to the peptides GP-16 and GD-18 and/or alike peptides (with the same amino acid sequences) potently stimulating binding to the EGFR active site.
3.6 OPPA analysis of synthetic peptides (GP-16 and GD-18)
The OPPA of the synthetic peptides (GP-16 and GD-18) were determined at different culture time by the MTT method (Fig. S4†). MC3T3-E1 cells were cultured for 24, 48, 72 and 96 h in presence of the synthetic peptide with different concentrations (0.01 mg mL−1, 0.02 mg mL−1 and 0.05 mg mL−1). Significant increase of cell proliferation was observed in the group of 0.02 mg mL−1 and 0.05 mg mL−1 synthetic peptides (GP-16 and GD-18) at 48, 72 and 96 h culture time (P < 0.05). GP-16 and GD-18 increased the survival of osteoblasts had demonstrated. Herein, GP-16 and GD-18 possessed excellent OPPA when compared with the control group, which was consistent with the results of molecular docking. Moreover, after 96 h of treatment, the cell proliferation rate of GP-16 and GD-18 increased to their highest values of 135.0% and 149.2% for the concentration of 0.05 mg mL−1. It indicated that yak bones collagen peptides may take longer time to stimulate osteoblasts proliferation via receptor and signaling pathways.124 Conclusion
The findings of this work suggest that yak bones collagen may be one promising origin for the generation of osteogenic peptides by Neutrase hydrolysis. The OPPA of YBCP (<3 kDa) was evaluated by the MTT assay and a significant increase (175.4%) of osteoblasts proliferation could be observed. A total of 59 peptides in YBCP (<3 kDa) were identified by HPLC-MS/MS. The osteogenic activity of these peptides was then predicted by a molecular docking study performed on the EGFR protein (PDB: 1IVO). GP-16 and GD-18 were selected for an in-depth analysis of the interactions exhibited in their docked form with EGFR. It revealed that OPPA of GP-16 and GD-18 was mainly due to the formation of very strong hydrogen bonds with the EGFR. The YBCP (<3 kDa) promoting osteoblast proliferation may partly be attributed to the peptides GP-16 and GD-18 and/or alike peptides (with the same amino acid sequences) potently stimulating binding to the EGFR active site. This work established a theoretical foundation for the application of YBCP (<3 kDa) in functional foods. However, much work is obviously required on the dose–response relationship and structure–activity relationship of different peptides responsible for higher OPPA before such ingredients are confidently accepted by the functional and health promoting food markets.Conflicts of interest
All of the authors declare that there are no conflicts of interest of this work.Acknowledgements
This work was financially supported by the Science and Technology Special Project of Tibet Autonomous Region (Characteristic Agro-products Processing Technologies and Product Development Program) (XZ201801NA04), Ten Thousand Experts Plan, Yuandu Industry Leading Talent Program, Innovation Program and Inner Mongolia Science and Technology Major Project (The Activity Mechanism and Targeted Enzymatic Hydrolysis Technology of Animal-derived Peptides), and National Agricultural Science and Technology Innovation Program.References
- J. Aaseth, G. Boivin and O. Andersen, J. Trace Elem. Med. Biol., 2012, 26, 149–152 CrossRef CAS PubMed.
- P. D. Ottewell, J. Bone Oncol., 2016, 5, 124–127 CrossRef PubMed.
- Z. Amso, R. Kowalczyk, M. Watson, Y.-E. Park, K. E. Callon, D. S. Musson, J. Cornish and M. A. Brimble, Org. Biomol. Chem., 2016, 14, 9225–9238 RSC.
- S. Calabria, E. Cinconze, M. Rossini, E. Rossi, A. P. Maggioni, A. Pedrini and M. De Rosa, Patient Prefer. Adherence, 2016, 10, 523–530 CrossRef CAS PubMed.
- L. Forte, S. Sarda, C. Combes, F. Brouillet, M. Gazzano, O. Marsan, E. Boanini and A. Bigi, Colloids Surf., B, 2017, 160, 493–499 CrossRef CAS PubMed.
- H. Nakayama, H. Toho and T. Sone, Osteoporos Sarcopenia, 2018, 4, 29–32 CrossRef PubMed.
- Y. S. Park, J. Y. Lee, J. S. Suh, Y. M. Jin, Y. Yu, H. Y. Kim, Y. J. Park, C. P. Chung and I. Jo, Biomaterials, 2014, 35, 9747–9754 CrossRef CAS PubMed.
- E. Yamachika, Y. Matsui, M. Matsubara, T. Matsumura, N. Nakata, N. Moritani, A. Ikeda, H. Tsujigiwa, N. Ohara and S. Iida, J. Dent. Sci., 2017, 12, 333–339 CrossRef PubMed.
- B. Balkhi, E. Seoane-Vazquez and R. Rodriguez-Monguio, Saudi Pharm. J., 2018, 26, 238–243 CrossRef PubMed.
- A. Sila and A. Bougatef, J. Funct. Foods, 2016, 21, 10–26 CrossRef CAS.
- G. Chen, L. Cheng, H. Xu, H. Song, Y. Lv, C. Yang, T. Zhu and N. Sun, Int. J. Food Prop., 2011, 14, 1136–1141 CrossRef CAS.
- P. Shi, M. Liu, F. Fan, H. Chen, C. Yu, W. Lu and M. Du, Food Biosci., 2018, 22, 19–25 CrossRef CAS.
- S. C. Pigossi, M. C. Medeiros, S. Saska, J. A. Cirelli and R. M. Scarel-Caminaga, Int. J. Mol. Sci., 2016, 17, 1885–1899 CrossRef PubMed.
- A. B. Plonka, B. Khorsand, N. Yu, J. V. Sugai, A. K. Salem, W. V. Giannobile and S. Elangovan, Gene Ther., 2017, 24, 31–39 CrossRef CAS PubMed.
- M. Du, W. Xu, H. Yi, X. Han, C. Wang and L. Zhang, Mol. Nutr. Food Res., 2011, 55, 220–228 CrossRef CAS PubMed.
- S. G. Guo, Enzymatic hydrolysis of porcine hemoglobin and antioxidant activity of its hydrolysate, Doctoral Dissertation, South China University of Technology, 2007.
- L. L. Canabady-Rochelle, C. Harscoat-Schiavo, V. Kessler, A. Aymes, F. Fournier and J. M. Girardet, Food Chem., 2015, 183, 129–135 CrossRef CAS PubMed.
- M. L. Timón, V. Parra, J. Otte, J. M. Broncano and M. J. Petrón, LWT–Food Sci. Technol., 2014, 57, 359–365 CrossRef.
- F. Toldra, M. Reig, M. C. Aristoy and L. Mora, Food Chem., 2018, 267, 395–404 CrossRef CAS PubMed.
- G. Wu, L. D. Robertson, C. Brooks III and M. Vieth, J. Comput. Chem., 2003, 24, 1549–1562 CrossRef CAS PubMed.
- S. C. Ko, J. Jang, B. R. Ye, M. S. Kim, I. W. Choi, W. S. Park, S. J. Heo and W. K. Jung, Process Biochem., 2017, 54, 180–187 CrossRef CAS.
- P. Kittiphattanabawon, S. Benjakul, W. Visessanguan, T. Nagai and M. Tanaka, Food Chem., 2005, 89, 363–372 CrossRef CAS.
- C. Zhang, Y. Zhang, Z. Wang, S. Chen and Y. Luo, J. Funct. Foods, 2017, 35, 224–235 CrossRef CAS.
- W. Jia, W. Liu, S. Mi, C. Zhang, X. Li, T. Wu and Q. Yu, Food Anal. Method, 2017, 10, 3496–3507 CrossRef.
- H. Ogiso, R. Ishitani, O. Nureki, S. Fukai, M. Yamachika, J. Kim, K. Saito, A. Sakamoto, M. Inoue, M. Shirouzu and S. Yokoyama, Cell, 2002, 110, 775–787 CrossRef CAS PubMed.
- Y. Guo, D. Pan and M. Tanokura, Food Chem., 2009, 114, 328–333 CrossRef CAS.
- Y. Liu, X. Li, Z. Chen, J. Yu, F. Wang and J. Wang, Food Chem., 2014, 151, 459–465 CrossRef CAS PubMed.
- P. A. Harnedy, M. B. O'Keeffe and R. J. FitzGerald, Food Chem., 2015, 172, 400–406 CrossRef CAS PubMed.
- S. S. Wang, The preparation of collagen peptides and active calcium from cod bone and its preventive effects on osteoporosis, Doctoral Dissertation, Ocean University of China, 2013.
- R. Mohammadi, M. A. Mohammadifar, A. M. Mortazavian, M. Rouhi, J. B. Ghasemi and Z. Delshadian, Food Chem., 2016, 190, 186–193 CrossRef CAS PubMed.
- M. H. Ertop and Y. Coskun, J. Food Process. Preserv., 2018, 42, 13650–13659 CrossRef.
- L. Wang, B. Zhang, J. Han, Y. Zheng, J. Li and A. Shan, Int. Biodeterior. Biodegrad., 2015, 104, 112–117 CrossRef CAS.
- S. Tahmouzi, Carbohydr. Polym., 2014, 106, 238–246 CrossRef CAS PubMed.
- M. A. Bezerra, R. E. Santelli, E. P. Oliveira, L. S. Villar and L. A. Escaleira, Talanta, 2008, 76, 965–977 CrossRef CAS PubMed.
- Z. Khiari, M. Ndagijimana and M. Betti, Poultry Sci., 2014, 93, 2347–2362 CrossRef CAS PubMed.
- K. Lin, L. W. Zhang, X. Han and D. Y. Cheng, J. Funct. Foods, 2017, 32, 266–277 CrossRef CAS.
- Y. Zhang, X. Wu, C. Liang, P. Bao, X. Ding, M. Chu, C. Jia, X. Guo and P. Yan, Gene, 2018, 650, 41–48 CrossRef CAS PubMed.
- C. D. Angelmosqueda, Y. Gutierrezpuente, A. P. Lopezlozano, R. E. Romerozavaleta, A. Mendiolajimenez, C. E. M. D. La Garza, M. Marquezm and M. A. D. La Garzaramos, Head Face Med., 2015, 11(1), 29–35 CrossRef PubMed.
- S. Kawabata, M. C. Hollander, J. P. Munasinghe, L. R. Brinster, J. R. Mercadomatos, J. Li, L. Regales, W. Pao, P. A. Janne and K. Wong, Oncotarget, 2015, 6(13), 11357–11368 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra00945k |
| This journal is © The Royal Society of Chemistry 2019 |