Open Access Article
Open Access ArticleKnockdown of circPVT1 inhibits progression of papillary thyroid carcinoma by sponging miR-126
Ling Taoa,
Li Yanga,
Ping Tiana,
Xiangyang Guob and
Yanping Chen *c
*c
aInstitute of Inspection Technology, Xinyang Vocational and Technical College, Key Laboratory of Geriatric Diseases of Xinyang, Xinyang, 464000, China
bDepartment of Endocrinology, Xinyang Central Hospital, Xinyang, 464000, China
cDepartment of Laboratory, Qingdao Women and Children's Hospital, No. 6 Tongfu Road, Shibei District, Qingdao, 266000, China. E-mail: chenyanping12019@163.com; Tel: +86-0532-68661155
First published on 30th April 2019
Abstract
Background: Papillary thyroid carcinoma (PTC) is the most common thyroid cancer. Recent studies have reported that circular RNAs (circRNAs) play essential roles in human cancers, including PTC. However, the roles of circRNA plasmacytoma variant translocation 1 (PVT1) in PTC progression and its potential mechanism remain largely unknown. Methods: The expressions of circPVT1 and microRNA-126 (miR-126) were measured in PTC tissues and cells by quantitative real-time polymerase chain reaction (qRT-PCR). Cell viability, apoptosis, migration and invasion were detected in PTC cells by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT), flow cytometry, Western blot or trans-well assays, respectively. The interaction between circPVT1 and miR-126 was explored by bioinformatics analysis, luciferase activity assay and RNA immunoprecipitation. A mouse xenograft model was established to investigate the role of circPVT1 in PTC progression in vivo. Results: High expression of circPVT1 was shown in PTC tissues and cells and was associated with poor outcomes of patients. Knockdown of circPVT1 suppressed viability, migration and invasion but induced apoptosis in PTC cells. miR-126 was bound to circPVT1 and reduced in PTC tissues and cells. Moreover, inhibition of miR-126 reversed the regulatory effect of the circPVT1 interference on viability, apoptosis, migration and invasion in PTC cells. Besides, circPVT1 knockdown attenuated tumor growth via up-regulating miR-126 in vivo. Conclusion: CircPVT1 knockdown inhibited PTC progression by sponging miR-126. This may indicate circPVT1 as a novel target for treatment of PTC.
Introduction
Thyroid cancer is one of the most common cancers with rising incidence worldwide.1 Papillary thyroid carcinoma (PTC) accounts for ∼80–85% of thyroid malignancy and exhibits better prognosis.2 With the development of understanding the pathogenesis, diagnosis and treatment have gained great advances in thyroid cancer.3 However, its incidence is still growing. Hence, it is of great significance to study the pathogenesis and progression of PTC.Circular RNAs (circRNAs) are a novel class of noncoding RNAs, which display important biological functions in mammalian cells and play essential roles in diagnosis and treatment of cancers.4,5 Previous study has revealed that many circRNAs are dysregulated and may exhibit important roles in PTC pathogenesis.6 CircRNAs, with better stability than the linear RNA, could regulate cell proliferation, apoptosis, migration and invasion in PTC progression.7,8 CircRNA plasmacytoma variant translocation gene 1 (circPVT1) has been indicated as a potential prognostic marker in gastric cancer, which is derived from the regions in long noncoding RNA PVT1 locus, locating on chromosome 8q24.9 Moreover, Li et al. reported that circPVT1 contributes to cell proliferation and invasion by sponging miR-125b and regulating E2F transcription factor 2 in non-small cell lung cancer.10 Verduci et al. showed that circPVT1 promotes cell proliferation in head and neck squamous cell carcinoma.11 Previous studies have reported that PVT1 could promote PTC progression by multiple pathways.12–14 However, the role of circPVT1 and potential mechanism in PTC remain largely unknown.
MicroRNAs (miRNAs) are also a class of noncoding RNAs with approximately 22 nucleotides in length, which play essential roles in prognosis and treatment of PTC.15 Former work has reported that miR-126 is widely expressed and exhibits potential roles in diagnosis of human cancers.16 Importantly, miR-126 is indicated as a tumor suppressor in some types of cancers, including lung cancer, gastric cancer and pancreatic cancer.17–19 More particularly, miR-126 has been suggested to be lowly expressed in malignant thyroid cancer.20 However, little is known about the association of circPVT1 and miR-126 in PTC. Bioinformatics analysis predicts the binding sites of miR-126 and circPVT1. Hence, we hypothesized that circPVT1 might regulate PTC progression by miR-126. In this study, we detected the effect of circPVT1 on viability, apoptosis, migration and invasion in PTC cells and explored the interaction between circPVT1 and miR-126.
Materials and methods
Tissue samples
A total of 39 PTC patients without chemotherapy, radiotherapy or other treatments before surgical resection were recruited from Xinyang Central Hospital. Paired tumor tissues and adjacent normal samples were snap-frozen in liquid nitrogen and stored at −80 °C in key laboratory of geriatric diseases of Xinyang until used. The clinicopathologic features of patients were shown in Table 1. Moreover, the 5 year survival was analyzed in PTC patients. All procedures performed in this study were in accordance with the ethical standards of Xinyang Central Hospital and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The written informed consents were obtained from all participants involved in this research.| Clinicopathologic features | N (%) | CircPVT1 level | P value | |
|---|---|---|---|---|
| High (%) | Low (%) | |||
| Age (years) | P > 0.05 | |||
| ≥50 | 23 (59.0) | 12 (52.2) | 11 (47.8) | |
| <50 | 16 (41.0) | 9 (56.3) | 7 (43.7) | |
| Gender | P > 0.05 | |||
| Female | 28 (71.8) | 15 (53.6) | 13 (46.4) | |
| Male | 11 (28.2) | 6 (54.5) | 5 (45.5) | |
| T stage | P < 0.05 | |||
| T1–T2 | 17 (43.6) | 6 (35.3) | 11 (64.7) | |
| T3–T4 | 22 (56.4) | 15 (68.2) | 7 (31.8) | |
| Lymph node metastasis | P < 0.05 | |||
| No | 18 (46.2) | 5 (27.8) | 13 (72.2) | |
| Yes | 21 (53.8) | 16 (76.2) | 5 (23.8) | |
| ATA risk | P > 0.05 | |||
| Low | 14 (35.9) | 7 (50.0) | 7 (50.0) | |
| Intermediate | 22 (56.4) | 12 (54.5) | 10 (45.5) | |
| High | 3 (7.7) | 2 (66.7) | 1 (33.3) | |
| Status | P < 0.05 | |||
| Alive | 27 (69.2) | 11 (40.7) | 16 (59.3) | |
| Death | 12 (30.8) | 10 (83.3) | 2 (16.7) | |
Cell culture and transfection
Normal human thyroid follicular epithelial cell Nthy-ori-3-1 and PTC cell lines (TPC-1, BCPAP, BHP5-16 and BHP2-7 cells) were purchased from American Tissue Culture Collection (Manassas, VA, USA) and cultured in Dulbecco's Modified Eagle Medium (Gibco, Carlsbad, CA, USA) with 10% fetal bovine serum (Gibco), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (Invitrogen, Carlsbad, CA, USA) at 37 °C and 5% CO2.Small interfering RNA (siRNA) against circPVT1 (si-circPVT1), siRNA negative control (si-NC), pcDNA vector, circPVT1 overexpression vector (circPVT1), miR-126 mimic (miR-126), miRNA negative control (miR-NC), miR-126 inhibitor (anti-miR-126) and miRNA inhibitor negative control (anti-miR-NC) were obtained from Genepharma (Shanghai, China). TPC-1 and BCPAP cells were transfected with 40 nM of these oligonucleotides or 100 ng vectors via using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. After the culture of 24 h, cells were harvested for further use.
Quantitative real-time polymerase chain reaction (qRT-PCR)
TRIzol reagent (Invitrogen) was used to isolate total RNA. After the assay of quality, the RNA was reverse transcribed using cDNA Synthesis Kit (TransGen Biotech, Beijing, China) according to the manufacturer's instructions. qRT-PCR reaction was conducted by using SYBR green (Vazyme Biotech, Nanjing, China) on ABI7500 system (Applied Biosystems, Carlsbad, CA, USA) and every sample was prepared in triplicate. The amplified procedure was set as: 95 °C for 5 min and 40 cycles of 95 °C for 30 s and 60 °C for 1 min. The relative levels of miR-126 and circPVT1 were measured with U6 small RNA or GAPDH as internal control using 2−ΔΔCt method.21 The primers were listed as follows: miR-126 (forward, 5′-TCGUACCGTGAGTAATAATGCG-3′; reverse, 5′-CATTATTACTTTTGGTAC GCG-3′), U6 (forward, 5′-CTCGCTTCGGCAGCACATATACT-3′; reverse, 5′-ACGCTTCACGAATTTGCGTGTC-3′), circPVT1 (forward, 5′-GGTTCCACCA GCGTTATTC-3′; reverse, 5′-CAACTTCCTTTGGGTCTCC-3′), GAPDH (forward, 5′-AATGGGCAGCCGTTAGGAAA-3′; reverse, 5′-TGAAGGGGTCATTG ATGGCA-3′).Western blot
Total proteins were extracted from tissues or cells by using RIPA lysis buffer (Beyotime Biotech, Shanghai, China) and quantified by BCA protein assay kit (Beyotime Biotech). After the centrifugation and denaturation, equal amounts of proteins were separated on SDS-PAGE gel and then transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat milk for 1 h at room temperature, and then incubated with primary antibodies overnight at 4 °C against Bax (ab199677, Abcam, Cambridge, UK), Bcl-2 (ab196495, Abcam), PCNA (ab18197, Abcam) or β-actin (ab8227, Abcam) and horseradish peroxidase-conjugated secondary antibody (ab6721, Abcam) for 2 h at room temperature. β-Actin was used as loading control and the protein blots were visualized using enhanced chemiluminescence chromogenic substrate (Beyotime Biotech).Cell viability
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay was conducted to measure cell viability. TPC-1 and BCPAP cells were seeded into 96-well plates at a density of 3000 cells per well and every group was prepared in triplicate. After the culture for 0, 24, 48 or 72 h, cell medium was replaced with medium containing 0.5 mg ml−1 MTT solution (Thermo Fisher, Wilmington, DE, USA) and cultured for another 4 h. Subsequently, each well was added into 100 μl of dimethylsulfoxide (Thermo Fisher) for 10 min and the absorbance at 490 nm was measured using a microplate reader (Bio-Rad, Hercules, CA, USA).Cell apoptosis
The Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Yeasen, Shanghai, China) was used for cell apoptosis analysis via flow cytometry. After the culture for 72 h, TPC-1 and BCPAP cells were collected, washed with PBS and then resuspended in binding buffer. Subsequently, cells were stained with 5 μl Annexin V-FITC and 10 μl PI for 10 min in the dark. The stained cells were analyzed by using a flow cytometer (Becton Dickinson, San Jose, CA, USA).Trans-well assay
Cell migration and invasion were measured by trans-well assay. TPC-1 and BCPAP cells (1 × 105 cells) were cultured in the upper chambers (Corning, Corning, NY, USA) with serum-free medium at 37 °C in 5% CO2. The bottom chambers were added with normal medium containing 10% fetal bovine serum. After the culture for 12 h, cells on the upper side of the well were carefully removed using a cotton swab. Migrated cells on the basal side of the membrane were fixed with 100% methanol for 10 min and then stained with 1% crystal violet (Sigma, St. Louis, MO, USA). For invasion assay, the trans-well chambers were coated with Matrigel (Becton Dickinson) and experiment was conducted following the similar approach. Five visual fields were randomly selected and migrated or invasive cells were counted under a microscope (Olympus, Tokyo, Japan).Bioinformatics analysis and luciferase activity assay
The putative binding sites of circPVT1 and miR-126 were predicted by bioinformatics analysis using StarBase online. The pGL3 vectors (Promega, Madison, WI, USA) were used for synthesis of wild-type (WT) luciferase reporter vectors (circPVT1-WT). The mutant (MUT) luciferase reporter vectors (circPVT1-MUT) were generated by mutating the binding sites (UAAUAAU) to (GCGGUGC) using the Fast Site-Directed Mutagenesis Kit (Tiangen, Beijing, China). TPC-1 and BCPAP cells were co-transfected with 20 ng luciferase reporter vectors and 40 nM of miR-126 or miR-NC using Lipofectamine 2000 according to the manufacturer's protocols. After the transfection for 24 h, luciferase activity assay was analyzed by using a luciferase assay kit (Promega).RNA immunoprecipitation (RIP)
RIP assay was performed to validate the association between circPVT1 and miR-126 by using RNA-binding protein immunoprecipitation kit (Millipore) according to the manufacturer's protocols. In brief, TPC-1 and BCPAP cells were lysed in RIP buffer containing magnetic beads bound with Ago2 or IgG antibody. The enrichments of circPVT1 and miR-126 immunoprecipitated on beads were measured by qRT-PCR.Murine xenograft tumorigenesis assay
A total of 14 SPF BALB/c nude mice (male, five-week-old) were obtained from Vital River Laboratory Animal Technology (Beijing, China) and acclimatized to housing in specific cages with a 12 h light/dark cycle for one week. Every effort was made to minimize animals (n = 7/group) during the study. TPC-1 cells were transfected with the lentiviral vectors with short hairpin RNA against circPVT1 (sh-circPVT1) or corresponding negative control (sh-NC) constructed by GeneCopoeia (Rockville, MD, USA). Then stably transfected cells were subcutaneously injected into mice after acclimatization. Tumor volume was examined every week for a total of four weeks with the formula as (length × width2)/2. Reaching end points, mice were killed and tumor specimens were weighted and collected for further molecular analyses. Every animal experiment was performed in strict accordance with the Guidelines for Care and Use of Laboratory Animal and was approved by the Ethics Committee of Animal Research of Xinyang Central Hospital during the study.Statistical analysis
Data were presented as the mean ± standard deviation (SD) from three independent experiments. Student's t test or one-way analysis of variance (ANOVA) was conducted for statistical differences analyses by using GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA). The association between circPVT1 expression and clinicopathologic features od PTC patients was analyzed using χ2 test. The overall survival curve of patients was generated by Kaplan–Meier method and analyzed by log-rank test. Statistically significant was indicated when P < 0.05.Results
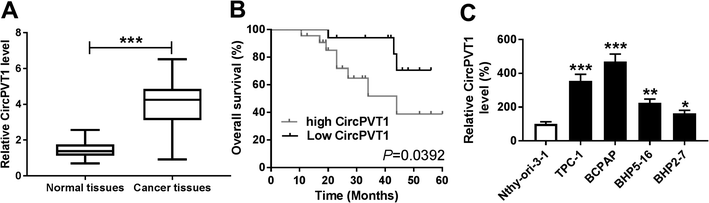
CircPVT1 expression is enhanced in PTC
To explore the potential role of circPVT1 in PTC progression, its expression was first measured in PTC samples. Compared with that in normal tissues, the abundance of circPVT1 was obviously elevated in PTC tissues (n = 39) (Fig. 1A). Then the clinical significance of circPVT1 was analyzed in PTC patients. The patients were classified as high (n = 21) or low circPVT1 expression (n = 18) group according the mean level of circPVT1 abundance. The clinical information analyses revealed that high expression of circPVT1 was associated with T stage, lymph node metastasis and survival status (P < 0.05), but not correlated with age, gender and ATA risk of patients (P > 0.05) (Table 1). Moreover, patients in high expression of circPVT1 group displayed relative lower survival than low expression group (P = 0.0392) (Fig. 1B). Besides, the level of circPVT1 was detected in PTC cells. qRT-PCR assay indicated higher expression of circPVT1 in PTC cells (TPC-1, BCPAP, BHP5-16 and BHP2-7) than that in Nthy-ori-3-1 cells (Fig. 1C). The TPC-1 and BCPAP cells with relative higher abundance of circPVT1 were used for subsequent experiments.Interference of circPVT1 promotes apoptosis and inhibits viability, migration and invasion in PTC cells
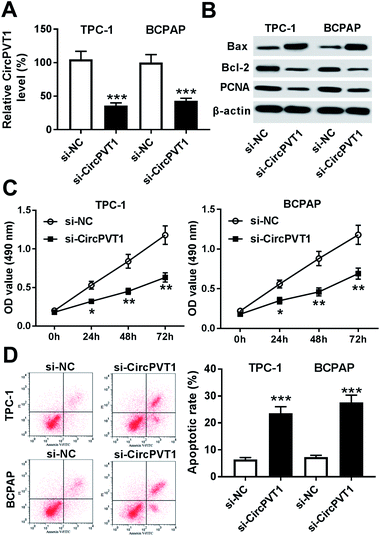
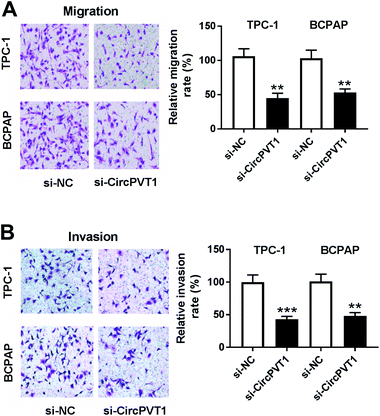
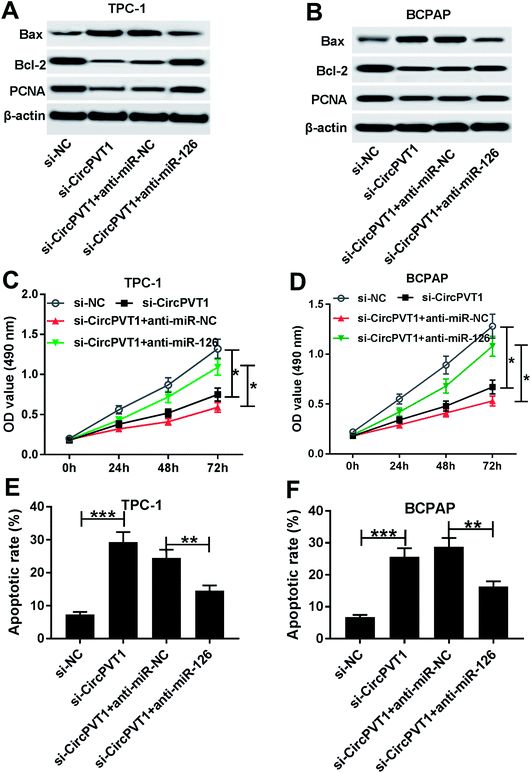
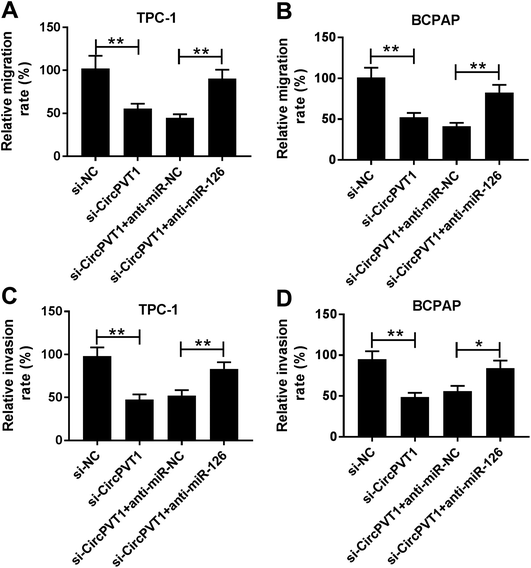
To investigate the effect of circPVT1 on PTC progression, TPC-1 and BCPAP cells were transfected with si-circPVT1 or si-NC for 24 h. After the transfection, the abundance of circPVT1 was effectively reduced 64% and 57% in TPC-1 and BCPAP cells transfected with si-circPVT1 compared with that in si-NC group, respectively (Fig. 2A). Moreover, Western blot assay showed that knockdown of circPVT1 led to obvious increase of Bax and reduction of Bcl-2 and PCNA at protein levels in TPC-1 and BCPAP cells (Fig. 2B). Furthermore, silencing circPVT1 significantly impaired viability of TPC-1 and BCPAP cells at 24, 48 and 72 h (Fig. 2C). Meanwhile, interference of circPVT1 resulted in a strong increase of apoptosis in TPC-1 and BCPAP cells at 72 h (Fig. 2D). Besides, knockdown of circPVT1 led to a 55% and 47% reduction of the migrated ability of TPC-1 cells and BCPAP cells compared with the respective NC group (Fig. 3A). Similarly, the ability of cell invasion was notably reduced 56% and 51% in TPC-1 and BCPAP cells transfected with si-circPVT1 compared with that in cells treated with si-NC (Fig. 3B).CircPVT1 is a decoy of mR-126
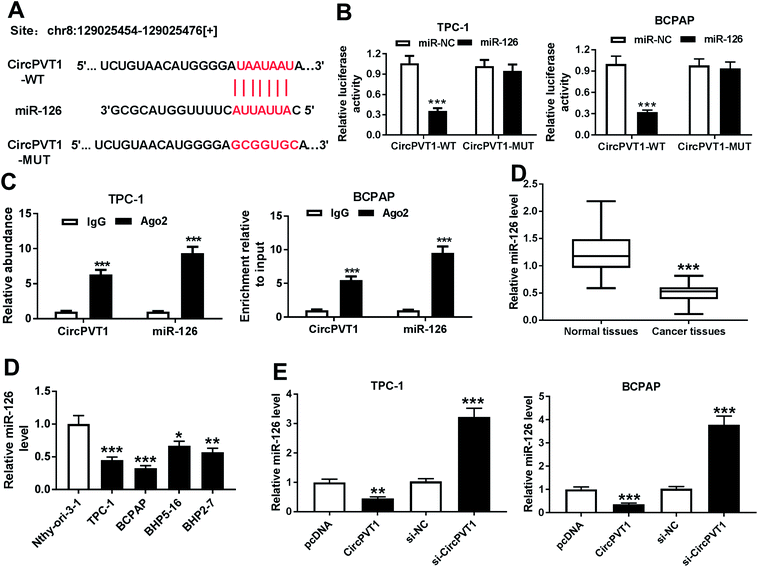
In order to elucidate the underlying mechanism allows circPVT1 participating in PTC progression, potential miRNAs bound to circPVT1 were explored by bioinformatics analysis using StarBase. The putative binding sites of circPVT1 and miR-126 were shown in Fig. 4A, suggesting that miR-126 might be bound to circPVT1. To validate this prediction, the WT or MUT luciferase reporter vectors were generated and transfected into PTC cells. At 24 h after the transfection, the luciferase activity was significantly reduced in TPC-1 and BCPAP cells transfected with miR-126 compared with that in cells treated with miR-NC in circPVT1-WT group, while it was not affected in circPVT1-MUT group (Fig. 4B). Furthermore, RIP assay revealed that circPVT1 and miR-126 in TPC-1 and BCPAP cells were markedly enriched by Ago2 RIP compared with those in IgG RIP group (Fig. 4C). Subsequently, the expression of miR-126 was measured in PTC tissues and cells. Results showed low expression of miR-126 in PTC tissues and cells compared with that in their corresponding control (Fig. 4D). In addition, the effect of circPVT1 on miR-126 expression was investigated in the two cells. The analysis of qRT-PCR revealed that the abundance of miR-126 was notably reduced by overexpression of circPVT1 and increased by circPVT1 knockdown in TPC-1 and BCPAP cells (Fig. 4E).CircPVT1 regulates apoptosis, viability, migration and invasion by sponging miR-126 in PTC cells
To identify whether miR-126 is required for circPVT1-mediated progression of PTC, TPC-1 and BCPAP cells were transfected with si-NC, si-circPVT1, si-circPVT1 and anti-miR-NC or anti-miR-126. As shown in Fig. 5A and B, knockdown of miR-126 weakened interference of circPVT1-induced increase of Bax and decrease of Bcl-2 and PCNA protein levels in TPC-1 and BCPAP cells. Furthermore, exhaustion of miR-126 reversed silencing circPVT1-induced viability suppression in TPC-1 and BCPAP cells (Fig. 5C and D). In addition, miR-126 knockdown attenuated silence of circPVT1-induced apoptosis induction in TPC-1 and BCPAP cells (Fig. 5E and F). Besides, trans-well analysis displayed that knockdown of miR-126 counteracted the suppressive effect of circPVT1 interference on migration and invasion in TPC-1 and BCPAP cells (Fig. 6A–D).Knockdown of circPVT1 attenuates tumor growth by regulating miR-126 in xenograft model
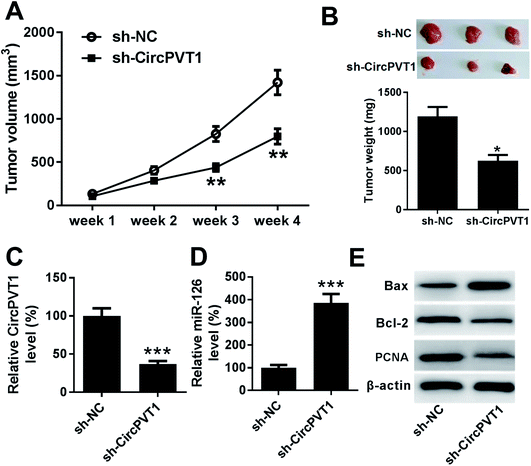
To further investigate the anti-cancer role of circPVT1 knockdown in PTC, TPC-1 cells stably transfected with sh-circPVT1 or sh-NC were injected into nude mice to establish murine xenograft model. After the cell injection, tumor volume was measured every week and results showed that the tumor volume was significantly reduced in sh-circPVT1 group compared with that in sh-NC group (Fig. 7A). Moreover, tumor weight was obviously smaller in sh-circPVT1 group than that in sh-NC group (Fig. 7B). Besides, the expressions of circPVT1, miR-126, Bax, Bcl-2 and PCNA were measured in tumor tissues of each group. As displayed in Fig. 7C and D, the expression of circPVT1 was markedly decreased and miR-126 was notably enhanced in tissues from mice in sh-circPVT1 group compared with that in sh-NC group. In addition, great increase of Bax and reduction of Bcl-2 and PCNA protein levels were described in samples of sh-circPVT1 group compared with those of sh-NC group (Fig. 7E).Discussion
Many genes could appear capable of producing circRNAs, which function as main biomarkers and master regulators of gene expression in multiple cancers.22 More particularly, previous studies have indicated circRNAs might play essential roles in tumorigenesis, progression, prognosis and treatment of PTC, such as circRNA-102171, circ-ITCH, circZFR and circ-0137287.7,8,23–25 However, the role of circPVT1 in PTC and its mechanism remain largely unknown. In this study, we first elucidated the oncogenic role of circPVT1 in PTC and explored the interaction between circPVT1 and miR-126 in vitro and in vivo.Previous work displayed PVT1 as an oncogenic lncRNA in thyroid cancer.14 Moreover, lncRNA PVT1 has been suggested to promote viability and invasion of PTC cells by acting as a competing endogenous RNA (ceRNA) of miR-30a to regulate insulin like growth factor 1 receptor.12 Zhou et al. reported that PVT1 promoted cell proliferation by regulating cell cycle process in PTC.13 CircPVT1 is known to be generated by circularization of an exon of the PVT1 gene.26 Accruing evidences have indicated circPVT1 as an oncogene to promote cancer progression and poor prognosis in non-small cell lung cancer, head and neck squamous cell carcinoma and gastric cancer.9–11 According to these reporters, we hypothesized that circPVT1 might also play as an oncogene in PTC. In this research, we first measured the expression of circPVT1 in PTC and found that circPVT1 was highly expressed in PTC tissues and cells compared with their corresponding control, respectively. Moreover, we found that high expression of circPVT1 was associated with T stage, lymph node metastasis and low survival rate, suggesting that circPVT1 might predict poor prognosis of PTC. Then the function of circPVT1 was investigated in PTC cells. In the present study, down-regulation of pro-proliferation protein PCNA and OD490 uncovered the inhibition of viability in PTC cells transfected with si-circPVT1. Furthermore, the elevated apoptosis in PTC cells induced by knockdown of circPVT1 was revealed by increase of pro-apoptotic protein Bax and apoptotic rate and reduction of anti-apoptotic protein Bcl-2. These findings indicated that circPVT1 contributed to cell growth by regulating apoptosis. Meanwhile, analysis of trans-well demonstrated that silencing circPVT1 suppressed PTC cells migration and invasion. These data of function assays reflected circPVT1 as an oncogene to contribute to PTC progression.
The emerging effort demonstrated that circRNAs were implicated in progression of multiple cancers via serving as miRNA sponges.27 In this study, qRT-PCR assay revealed that the expression of miR-126 was significantly decreased in PTC tissues and cells, which is also in agreement with former works.28–30 Moreover, Xiong et al. reported that miR-126 suppressed cell growth and metastasis by regulating solute carrier family 7 member 5 and a disintegrin and metalloproteinase 9 in thyroid cancer.28 Wen et al. suggested that miR-126 inhibited cell proliferation, colony formation, migration and invasion but induced apoptosis and cell cycle G1 stage arrest in PTC by mediating lipoprotein receptor-related protein 6.29 Additionally, Qian et al. showed that miR-126 repressed cell proliferation, migration and invasion by targeting C–X–C chemokine receptor type 4 in thyroid cancer.30 These efforts all disclosed that miR-126 could act as a tumor suppressor in PTC through modulating varying targets. To figure out whether miR-126 was associated with circPVT1-mediated progression of PTC, the interaction between them was explored in PTC cells, which was confirmed by luciferase activity analysis and Ago2 RIP. Furthermore, miR-126 expression was negatively regulated by circPVT1 and its abrogation alleviated the regulatory effect of circPVT1 silencing on viability, apoptosis, migration and invasion in TPC-1 and BCPAP cells, indicating that circPVT1 addressed progression of PTC by sponging miR-126 in vitro. Besides, pre-clinical studies are responsible for better understanding the potential mechanism of PTC pathogenesis. Therefore, we established the TPC-1 xenograft model to investigate the role of circPVT1 and mechanism in vivo. Knockdown of circPVT1 group exhibited reduction of tumor volume, weight, Bcl-2 and PCNA levels and increase of miR-126 and Bax levels compared with sh-NC group, which revealed that circPVT1 interference attenuated PTC tumor growth by up-regulating miR-126 in vivo. Functional miRNAs are known to regulate potential targets by binding their 3′ UTR. Although a large number of targets of miR-126 have been indicated, the promising targeted gene was absence in this research. Hence, more targets of miR-126 and potential signaling pathway should be explored in further study.
In conclusion, circPVT1 was up-regulated in PTC tissues and cells and indicated poor prognosis. Knockdown of circPVT1 inhibited PTC progression in vitro and in vivo. Moreover, this effect might be achieved by sponging miR-126. This indicated circPVT1/miR-126 network as promising avenue for treatment of PTC.
Conflicts of interest
The authors have no conflict of interest to declare.Abbreviations
| PTC | Papillary thyroid carcinoma |
| circRNA | Circular RNA |
| PVT1 | Plasmacytoma variant translocation 1 |
| ceRNA | Competing endogenous RNA |
| miRNA | MicroRNA |
| siRNA | Small interfering RNA |
| NC | Negative control |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| FITC | Fluorescein isothiocyanate |
| PI | Propidium iodide |
| WT | Wild-type |
| MUT | Mutant |
| RIP | RNA immunoprecipitation |
| SD | Standard deviation |
| ANOVA | Analysis of variance |
References
- M. E. Cabanillas, D. G. McFadden and C. Durante, Lancet, 2016, 388, 2783–2795 CrossRef CAS
.
- V. A. LiVolsi, Mod. Pathol., 2011, 24, S1–S9 CrossRef CAS PubMed
.
- D. F. Schneider and H. Chen, Ca-Cancer J. Clin., 2013, 63, 374–394 Search PubMed
.
- M. Zhang and Y. Xin, J. Hematol. Oncol., 2018, 11, 21 CrossRef PubMed
.
- Y. Wang, T. Lu, Q. Wang, J. Liu and W. Jiao, Oncol. Rep., 2018, 40, 3119–3135 CAS
.
- N. Peng, L. Shi, Q. Zhang, Y. Hu, N. Wang and H. Ye, PLoS One, 2017, 12, e0170287 CrossRef PubMed
.
- W. Bi, J. Huang, C. Nie, B. Liu, G. He, J. Han, R. Pang, Z. Ding, J. Xu and J. Zhang, J. Exp. Clin. Cancer Res., 2018, 37, 275 CrossRef PubMed
.
- H. Wei, L. Pan, D. Tao and R. Li, Biochem. Biophys. Res. Commun., 2018, 503, 56–61 CrossRef CAS PubMed
.
- J. Chen, Y. Li, Q. Zheng, C. Bao, J. He, B. Chen, D. Lyu, B. Zheng, Y. Xu, Z. Long, Y. Zhou, H. Zhu, Y. Wang, X. He, Y. Shi and S. Huang, Cancer Lett., 2017, 388, 208–219 CrossRef CAS PubMed
.
- X. Li, Z. Zhang, H. Jiang, Q. Li, R. Wang, H. Pan, Y. Niu, F. Liu, H. Gu, X. Fan and J. Gao, Cell. Physiol. Biochem., 2018, 51, 2324–2340 CrossRef CAS PubMed
.
- L. Verduci, M. Ferraiuolo, A. Sacconi, F. Ganci, J. Vitale, T. Colombo, P. Paci, S. Strano, G. Macino, N. Rajewsky and G. Blandino, Genome Biol., 2017, 18, 237 CrossRef PubMed
.
- K. Feng, Y. Liu, L. J. Xu, L. F. Zhao, C. W. Jia and M. Y. Xu, Biomed. Pharmacother., 2018, 104, 686–698 CrossRef CAS PubMed
.
- Q. Zhou, J. Chen, J. Feng and J. Wang, Tumor Biol., 2016, 37, 3105–3113 CrossRef CAS PubMed
.
- A. K. Murugan, A. K. Munirajan and A. S. Alzahrani, Endocr.-Relat. Cancer, 2018, 25, R59–R82 CAS
.
- J. C. Lee, J. S. Gundara, A. Glover, J. Serpell and S. B. Sidhu, Oncologist, 2014, 19, 1141–1147 CrossRef CAS PubMed
.
- J. Yan, S. Ma, Y. Zhang, C. Yin, X. Zhou and G. Zhang, Medicine, 2016, 95, e4644 CrossRef CAS PubMed
.
- B. Liu, X. C. Peng, X. L. Zheng, J. Wang and Y. W. Qin, Lung Cancer, 2009, 66, 169–175 CrossRef PubMed
.
- R. Feng, M. K. Beeharry, S. Lu, B. K. Sah, F. Yuan, M. Yan, B. Liu, C. Li and Z. Zhu, Cancer Biomarkers, 2018, 22, 119–126 CAS
.
- S. Hamada, K. Satoh, W. Fujibuchi, M. Hirota, A. Kanno, J. Unno, A. Masamune, K. Kikuta, K. Kume and T. Shimosegawa, Mol. Cancer Res., 2012, 10, 3–10 CrossRef CAS PubMed
.
- M. Kitano, R. Rahbari, E. E. Patterson, Y. Xiong, N. B. Prasad, Y. C. Wang, M. A. Zeiger and E. Kebebew, Ann. Surg. Oncol., 2011, 18, 3443–3452 CrossRef PubMed
.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed
.
- E. Arnaiz, C. Sole, L. Manterola, L. Lparraguirre, D. Otaegui and C. H. Lawrie, Semin. Cancer Biol., 2018, 12, 1–16 Search PubMed
.
- X. Lan, J. Xu, C. Chen, C. Zheng, J. Wang, J. Cao, X. Zhu and M. Ge, Cell. Physiol. Biochem., 2018, 47, 1122–1132 CrossRef CAS PubMed
.
- M. Wang, B. Chen, Z. Ru and L. Cong, Biochem. Biophys. Res. Commun., 2018, 504, 283–288 CrossRef CAS PubMed
.
- X. Lan, J. Cao, J. Xu, C. Chen, C. Zheng, J. Wang, X. Zhu, X. Zhu and M. Ge, J. Clin. Lab. Anal., 2018, 32, e22573 CrossRef PubMed
.
- A. C. Panda, I. Grammatikakis, K. M. Kim, S. De, J. L. Martindale, R. Munk, X. L. Yang, K. Abdelmohsen and M. Gorospe, Nucleic Acids Res., 2017, 45, 4021–4035 CrossRef CAS PubMed
.
- L. Qian, S. L. Yu, Z. Chen, Z. Q. Meng, S. L. Huang and P. Wang, Biochim. Biophys. Acta, Rev. Cancer, 2018, 1870, 247–260 CrossRef CAS PubMed
.
- Y. Xiong, S. Kotian, M. A. Zeiger, L. Zhang and E. Kebebew, PLoS One, 2015, 10, e0130496 CrossRef PubMed
.
- Q. Wen, J. Zhao, L. Bai, T. Wang, H. Zhang and Q. Ma, Oncol. Rep., 2015, 34, 2202–2210 CrossRef CAS PubMed
.
- Y. Qian, X. Wang, Z. Lv, C. Guo, Y. Yang, J. Zhang and X. Wang, Mol. Med. Rep., 2016, 14, 453–459 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2019 |