Open Access Article
Open Access ArticleBiocatalytic oxidation of flavone analogues mediated by general biocatalysts: horseradish peroxidase and laccase†
Qingsong Yan‡
a,
Xiaoguang Tang‡c,
Baojing Zhanga,
Chunjie Wanga,
Sa Denga,
Xiaochi Maa,
Chao Wang a,
Dawei Li
a,
Dawei Li *b,
Shanshan Huanga and
Peipei Dong*a
*b,
Shanshan Huanga and
Peipei Dong*a
aCollege of Pharmacy, Research Institute of Integrated Traditional and Western Medicine, Dalian Medical University, Dalian 116044, P. R. China. E-mail: dongpeipei11@163.com
bThe First Affiliated Hospital of Dalian Medical University, No. 222 Zhongshan Road, Dalian 116011, P. R. China. E-mail: lidw87@163.com
cMedicial College of Chifeng University, Chifeng 02400, P. R. China
First published on 30th April 2019
Abstract
Horseradish peroxidase (HRP) and laccase are well known oxidases, which have been widely applied for the biosynthesis of organic compounds. In the present work, flavone analogues as an important type of bioactive natural product could be oxidized by HRP or laccase, which afforded dimeric and oxidative flavones. All of the flavone analogues usually possessing phenolic groups could be transformed using HRP. However, only flavonols, isoflavones and chalcones with phenolic groups and dihydroxylflavones were effective substrates of laccase. The radical reaction mechanism with the B-ring of flavone analogues as the radical reaction trigger was proposed for the oxidation of flavones. In silico molecular docking analyses for assaying the interaction between flavone analogues and oxidases indicated that the phenolic groups at the B rings of flavones docked into the HEME active pocket of HRP well. Kinetic behaviors of the oxidation for various flavone analogues mediated by HRP or laccase displayed Hill and substrate inhibition kinetic models. Therefore, in the present work, the oxidation of various flavone analogues mediated by HRP or laccase has been successfully characterized, which would be helpful for the preparation of flavone derivatives.
Introduction
Oxidases are well known functional enzymes, which play an important role for the life of animals, plants and microorganisms. Due to their highly oxidative capability and advances in green chemistry, peroxidases have been widely applied to address many of the challenges of modern synthetic organic chemistry.1–7 Oxidases are known as a series of hemoproteins, possessing copper or ferric ions in their active sites to catalyze various oxidative transformations of organic and inorganic compounds using H2O2 and other related oxides as oxidizing agents. In general, stable radicals formed in the presence of oxidases could undergo several coupling or reoxidation reactions, which further afforded the polyphenols.One of the most common peroxidases is horseradish peroxidase (HRP, EC 1.11.1.7) with a molecular weight of 40 kDa, which contains at least 30 kinds of HRP isoenzyme forms. The HRP isoenzyme C (HRP-C) is the most abundant one, which consists of 308 amino acid residues, 4 disulphide bridges between cysteine residues as well as a heme group [iron(III) protoporphyrin IX] and two calcium atoms.3,8 It is able to utilize hydrogen peroxide to catalyze the one-electron oxidation for a wide range of aromatic substrates with the formation of various polymers (Fig. 1).
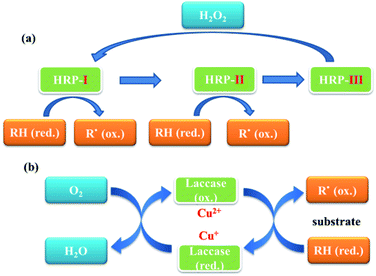 | ||
| Fig. 1 Proposed catalytic pathways of horseradish peroxidase (HRP), (a) and laccase (b). HRP: horseradish peroxidase; RH: substrate; R: free radical of substrate. | ||
Distinguished from HRP, laccase (Lac. EC 1.10.3.2), known as multi-copper oxidase, is widely distributed in natural sources, such as fungi, bacteria, plant, insects and lichens, with the molecular mass range of 50–130 kDa.9,10 Laccases have a functional unit that consists of four coppers, which could catalyze the oxidation of various organic compounds, such as diphenols, polyphenols, diamines, aromatic amines, benzeneethiols, and substituted phenols (Fig. 1).1
Flavones known as plant phenols are widely distributed in natural resources, which displayed multiple biological effects. In addition, polyphenols with high molecular weight showed better bioactivities, such as the rutin polymer and polycatechin with excellent superoxide scavenging activity.11 In the previous studies, some flavones have been transformed by laccase or HRP to afford the oxidation derivatives, which have drawn more and more attention of researchers for the application in biosynthesis.12–16 However, the enzymatic oxidations of flavones catalyzed by oxidases had been reported rarely with limited hypothetical mechanism.
The present study comprehensively compared the oxidation reactions of different flavone analogues mediated by HRP or laccase, respectively. The oxidative products have been prepared and identified by various spectral methods. Furthermore, the enzymatic oxidation characteristics have been studied on the basis of the flavone structure diversities and oxidase species. The oxidation mechanisms of various flavones mediated by HRP or laccase have been discussed on the basis of radical reaction, molecular docking and kinetics analyses.
Results and discussion
Oxidation of flavone analogues mediated by oxidases
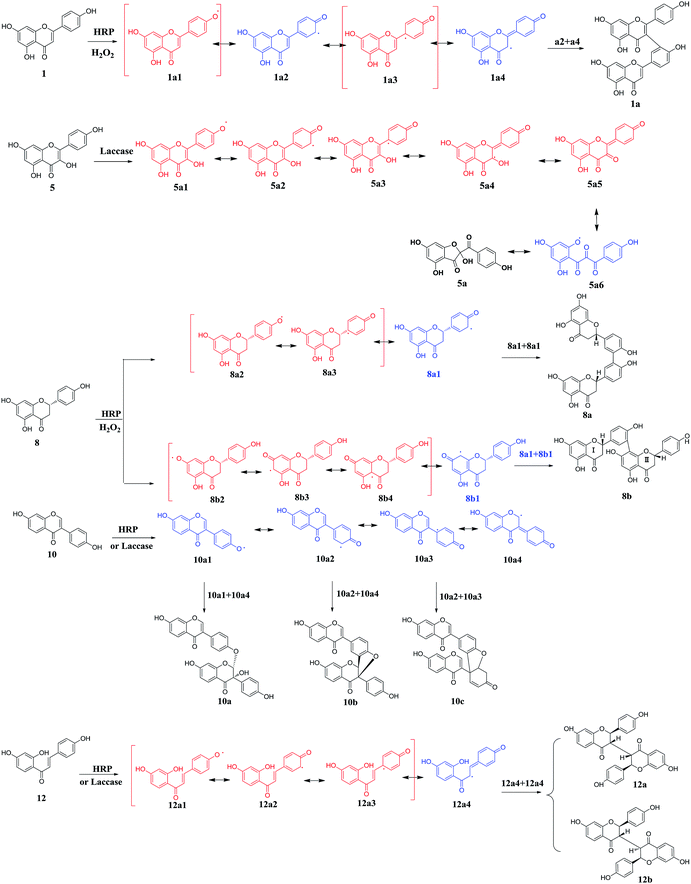
Flavones also known as 2-benzyl chromone, are obtained from plants as the bioactive substances.16 According to the difference of sub-structure, the flavone family mainly included flavones, flavonols, flavanones, isoflavones and chalcones, respectively (Fig. 2). Due to the phenolic groups, flavones are classified to be plant phenols. In the present study, various flavone analogues were used as the substrates to evaluate the oxidation capabilities of two oxidases HRP and laccase. The screening experiment was performed in primary reaction system with the products detected by HPLC-DAD. On the basis of the detected products of various flavone analogues, the capabilities of HRP and laccase to catalyze the oxidation reaction have been analyzed in detail. HRP could mediate the oxidation of most flavone analogues possessing one or more phenolic groups, except for the flavone glycoside (4) (Fig. 2). The saccharide group enlarges the molecular mass of the substrate, which leading to the steric effect between flavone substrate and HRP. For the laccase, more interesting features were observed according to the structure distinction. When flavone (3) possesses two adjacent phenolic groups, the oxidation of these substrates could be catalyzed by laccase. Flavonols (5, 6, 7), isoflavones (10, 11) and chalcone (12) with one or more phenolic groups were the effective substrates of laccase. On the other hand, flavanones (8, 9) could not be oxidated by laccase. On the basis of the screening experiment, HRP displayed the stronger capability to mediate the enzymatic oxidation of flavone analogues, than that of laccase.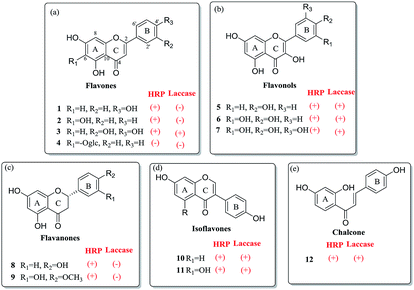 | ||
| Fig. 2 The screening oxidation of flavone analogues mediated by peroxidases: HRP and laccase. (a) Flavones; (b) flavonols; (c) flavanones; (d) isoflavones; (e) chalcones. | ||
Preparation and identification of the oxidated products
In order to investigate the enzymatic mechanism, the present study attempted to prepare the oxidated products of various flavone analogues mediated by HRP or laccase. When these flavonoid substrates with multiple or unstable products were got rid, the oxidation of these flavone analogues 1, 5, 8, 10 and 12 transformed by HRP or laccase yielded the oxidated products or polyflavones. On the basis of spectroscopic data (ESI-MS, HR-ESI-MS, 1H-NMR, 13C-NMR, 2D-NMR) and literatures,17–21 the products were determined as dimers 1a (yield 37%), 8a (yield 18%), 8b (yield 45%), 10a (yield 17%), 10b (yield 8%), 10c (yield 12%), 12a (yield 19%), 12b (yield 21%) mediated by HRP and oxidated flavonol 5a (yield 31%) mediated by laccase (Scheme 1). The 1H and 13C NMR data were assigned in Tables S1 and S2 (ESI)† respectively. In the screening experiment, substrates 1, 8, 10 and 12 could be catalyzed by HRP to afford the dimers, and 10, 12 gave the same products in the laccase enzymatic systems. For flavonol 5, multiple products were observed with HRP as the mediated enzyme, and the major product 5a was obtained using laccase as the catalyst. The structures of diflavones suggested that the polymerization positions located at the rings B and C generally. The oxidation of kaempferol (5) also occurred at the ring C of chromone. The oxidation of phenolic substrates catalyzed by oxidase (HRP or laccase) was known as phenolic radical reaction through electronic transfer. According to the chemical structures of various substrates (1, 5, 8, 10, and 12) and the corresponding products, the radical reaction courses were hypothesized as shown in Scheme 1.As shown in Scheme 1, the proposed radical reactions were initiated at the phenolic groups of ring B in the flavone structures, which indicated that the O–H bond in ring B was inclined to be dissociated in the presence of HRP or laccase.22 The bond energies of different phenolic groups of various flavones were calculated as shown in Table 1. The phenolic groups at C-5 position of flavones 1, 5 and 8 were not calculated for the intramolecular hydrogen bond between 5-OH and 4-ketone. Thus, it was obvious that the 4′-OH in various flavones had smaller bond energies than 7-OH, which indicated that the O–H (C-4′) bonds were inclined to be dissociated catalyzed by HRP or laccase. So, the radical reactions of flavones were predicted as shown in Scheme 1, and 4′-OH was the trigger of radical reaction.
| Flavones | Phenolic group | H–O energy (kcal mol−1) | Flavones | Phenolic group | H–O energy (kcal mol−1) |
|---|---|---|---|---|---|
| 1 | 4′-OH | 67.6 | 10 | 4′-OH | 68.6 |
| 7-OH | 72.5 | 7-OH | 72.6 | ||
| 5 | 4′-OH | 68.3 | 12 | 4′-OH | 66.8 |
| 7-OH | 72.2 | 7-OH | 69.7 | ||
| 8 | 4′-OH | 75.6 | |||
| 7-OH | 70.3 |
Comparison of the flavones transformation mediated by HRP and laccase
As the peroxidases, HRP and laccase could catalyze the various oxidation of phenolic substrates. However, the different capabilities of HRP and laccase were related to their oxidation–reduction potential (rH). When the substrates displayed the lower oxidation–reduction potential (rH) than HPR (<−0.2070 V) or laccase (0.5–0.8 V), the oxidation could be catalyzed by peroxidases. On the basis of the structures of various substrates and their corresponding phenolic radicals, the oxidation–reduction potentials (rH) of these flavone analogues were calculated as 1 (ΔE = −0.027 V), 5 (ΔE = −0.026 V), 8 (ΔE = −0.027 V), 10 (ΔE = −0.027 V), 12 (ΔE = −0.026 V) using Nernst equation. All of the flavones displayed similar and lower oxidation–reduction potentials than this of HRP, which confirmed the oxidation of these flavones mediated by HRP.It was known that the crystal structures of HRP and laccase were unambiguous, with the active centers HEME and copper ions respectively. The present work has performed the molecular docking to analysis the reaction mechanism between flavone analogues and HPR and laccase. Molecular docking revealed that flavone analogues 1, 5, 8, 10, and 12 could dock into the HEME active pocket of HRP well with the high probabilities as 98%, 93%, 90%, 100% and 90%, respectively.
As shown in Fig. 3, all of the flavone analogues bound to HPR with the optimal conformations, which displayed the ring B of these flavones docked into the HEME active pocket of HRP. So, the 4′-OH groups of these flavones were the nearest phenolic groups to the Fe atoms of HEME, further indicating the preferred formation of C-4′ phenolic radicals for the enzymatic oxidation of flavones. The binding energies calculation also suggested that all of the flavones bound to horseradish peroxidase with comparable binding affinity. Thus, HRP displayed a strong capability to catalyze the oxidation of flavones with the favorable C-4′ phenolic radicals.
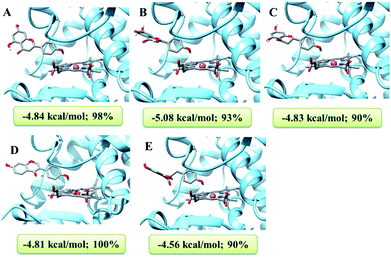 | ||
| Fig. 3 Docking analysis of flavones and HRP together with the binding energy and docking probability. (A) 1; (B) 5; (C) 8; (D) 10; (E) 12; brown ball is Fe atom and green ball is Ca ion of HEME. | ||
For the laccase, molecular docking revealed that most of the flavones bound to the surface of laccase (Fig. 4). And, few molecular could dock into to the active pocket of laccase possessing Cu ions. Among these flavones, more molecula of kaemferol (5) and isoliquiritigenin (12) bound to the sites close to the Cu ions active center of laccase, which suggested the preferred substrates 5 and 12 of laccase. Therefore, the molecular docking results also could confirm the low reaction efficiency of flavones catalyzed by laccase.
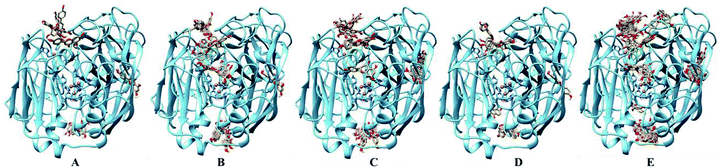 | ||
| Fig. 4 Docking analysis of flavones and laccase. (A) 1; (B) 5; (C) 8; (D) 10; (E) 12; brown balls are Cu ions. | ||
Kinetics analysis of the oxidation of flavones catalyzed by HRP and laccase
In the present work, kinetics analyses have been carried out to investigate the oxidation characteristics of HRP and laccase towards the various flavone analogues. For HRP, the kinetics behaviors of 1, 8, 10 and 12 to form corresponding products mediated by HRP were studied as shown in Fig. 5. Flavone apigenin (1), and isoflavone daidzein (10) with different products followed the Hill kinetic model (Fig. 5A, D–F), as well as the kinetic parameters were also analyzed systematically (Table 2). Especially, at the low concentration of 1, the obvious substrate active process was observed for Hill kinetic model (n = 2.944). The Km values also indicated that apigenin (Km 163.5 μM) had the better affinity towards HRP than daidzein. On the other hand, flavanone naringenin (8) and chalcone isoliquiritigenin (12) displayed substrate inhibition kinetics characteristics within the range of substrate concentrations tested in the kinetic exhibited a stronger substrate inhibitory effect for the oxidation catalyzed by HRP. The kinetic behaviors of the oxidation of the substrate 5, 10 and 12 transformed by laccase were also investigated as shown in Fig. 6. Flavonol 5 followed substrate inhibition kinetic model as well as 10 and 12 followed the Hill-kinetic model. The kinetic parameters exhibited that flavonol 5 had small affinity towards laccase (Km 8288 ± 401 μM) and weak inhibitory effect on laccase (Ki 1656 μM) (Table 3). The kinetic parameters analysis displayed big Km and small Vmax for these substrates, which suggested the inefficient oxidation of these flavone analogues mediated by laccase compared with the oxidation of flavone analogues mediated by HRP.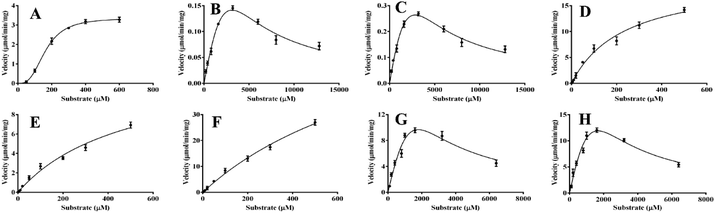 | ||
| Fig. 5 The enzymatic kinetics of the oxidation of flavones mediated by HRP. (A) 1a; (B) 8a, (C) 8b; (D) 10a; (E) 10b; (F) 10c; (G) 12a; (H) 12b. | ||
| Products | Vmax (μmol min−1 mg−1) | Km (μM) | n | Ki (μM) |
|---|---|---|---|---|
| 1a | 3.358 ± 0.6 | 163.5 ± 3.7 | 2.944 | |
| 8a | 1.790 ± 0.2 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 131 ± 164 131 ± 164 |
533.0 | |
| 8b | 2.489 ± 0.8 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 988 ± 143 988 ± 143 |
673.7 | |
| 10a | 25.64 ± 1.3 | 404.6 ± 5.8 | 0.8561 | |
| 10b | 15.08 ± 1.2 | 775.0 ± 21 | 0.8266 | |
| 10c | 56.45 ± 2.5 | 694.2 ± 36 | 0.9460 | |
| 12a | 143.5 ± 4.1 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 477 ± 1790 477 ± 1790 |
262.7 | |
| 12b | 165.0 ± 4.7 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 196 ± 45 196 ± 45 |
266.1 |
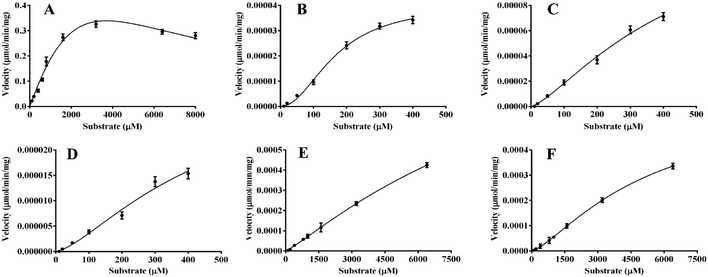 | ||
| Fig. 6 The enzymatic kinetics behaviors of the oxidation of flavones mediated by laccase. (A) 5a; (B) 10a; (C) 10b; (D) 10C; (E) 12a; (F) 12b. | ||
| Products | Vmax (μmol min−1 mg−1) | Km (μM) | n | Ki |
|---|---|---|---|---|
| 5a | 1.856 ± 0.3 | 8288 ± 401 | 1656 | |
| 10a | 4.069 ± 0.4 × 10−5 | 167.6 ± 10.6 | 2.035 | |
| 10b | 3.333 ± 0.3 × 10−5 | 427.9 ± 23 | 1.452 | |
| 10c | 1.523 ± 0.6 × 10−4 | 430.9 ± 32 | 1.363 | |
| 12a | 1.084 ± 0.2 × 10−3 | 9166 ± 615 | 1.224 | |
| 12b | 5.601 ± 0.7 × 10−4 | 4810 ± 373 | 1.418 |
Experimental
Apparatus
1H NMR and 13C NMR spectra were acquired on Bruker 501 spectrometer. ESIMS and LC-MSn were performed on an API 3200 Triple Quadrupole Mass spectrometer (MS/MS) equipped with an electrospray ionization (ESI) source (AB, Sciex). HPLC analysis were carried out on an Ultimate 3000 system with quaternary delivery system, degasser, autosampler, UV-detector, and YMC ODS (4.6 × 150 mm, 5 μm) analytical column. Preparative HPLC experiments were performed using an Agel instrument with a UV detector and a YMC-Pack ODS-A column (250 × 20 mm, 5 μm). Constant temperature oscillator (ZHWY-2102C) was produced by Shanghai Zhicheng Analysis Instrument Manufacturing Co., Ltd (China). The constant temperature mixing apparatus (MSC-100) was purchased from Hangzhou Allsheng Instruments Co., Ltd (China).Materials
All chemical solvents, including dichloromethane, ethyl acetate, petroleum ether (60–90 °C), acetone, methanol, and chemical reagents, such as hydrogen peroxide (H2O2), citric acid, disodium hydrogen phosphate (Na2HPO4), and dimethyl sulfoxide (DMSO), were A.R. grade and obtained from Sinopharm Chemical Reagent Beijing Co., Ltd. Dimethyl sulfoxide-d6 (DMSO-d6) was produced by Cambridge Isotope Laboratories, Inc. Methanol and acetonitrile for HPLC analysis were of chromatographic grade (SIGMA, USA). Horseradish peroxidase (HRP, >250 U mg−1) was purchased from J&K Scientific Ltd. Laccase from Trametes versicolor (120 U g−1) was obtained from Yuanye Biological Scientific Ltd (Shanghai, China).Biotransformation of flavones by HRP
Screen scale biotransformation was conducted in a 1 mL reaction system. 0.25 mg of HRP was dissolved in 94 μL of buffer (citric acid–hydrogen phosphate buffer solution, pH 3.0). One milligram of substrate (flavonoid) was dissolved in 376 μL of water, 376 μL of acetone, and 150 μL of buffer. The HRP solution and substrate solution was mixed up, which was incubated at 400 rpm at 25 °C. Five minutes later, 4 μL of a 10% H2O2 aqueous solution was added to the above mixture. The reaction was terminated by the addition of 1 mL ice-acetonitrile after 30 minutes followed by centrifugation at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min. The supernatant was analyzed by HPLC-DAD.
000 × g for 20 min. The supernatant was analyzed by HPLC-DAD.
Biotransformation of flavones by laccase
The incubation system for the laccase biotransformation contained 0.5 mg laccase (dissolved in 650 μL citric acid–hydrogen phosphate buffer solution, pH 4.0) and 0.5 mg flavonoid (dissolved in 350 μL acetone) in a final volume of 1 mL. After 6 h incubation at 30 °C, the reaction was terminated by the addition of 1 mL ice-acetonitrile, followed by centrifugation at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min. The supernatant was subjected to HPLC-DAD for analysis.
000 × g for 20 min. The supernatant was subjected to HPLC-DAD for analysis.
Preparation of oxidated flavones
The preparative biotransformation was conducted under the above-mentioned conditions, but 200 mg of the substrates were used. The reaction solution was extracted with CH2Cl2 and purified by using the pre-HPLC instrument with a C18 ODS column (detected at 210 nm, 8 mL min−1). Compound 1a was obtained at retention time 20.4 min with CH3CN–water (45%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 55%, 0.3% trifluoroacetic acid v/v). Compound 5a were isolated at retention time 28.8 min eluted by CH3CN–water (20%
55%, 0.3% trifluoroacetic acid v/v). Compound 5a were isolated at retention time 28.8 min eluted by CH3CN–water (20%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80%, 0.3% trifluoroacetic acid v/v). Compounds 8a and 8b were purified by CH3CN–water (45%
80%, 0.3% trifluoroacetic acid v/v). Compounds 8a and 8b were purified by CH3CN–water (45%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 55%, 0.3% trifluoroacetic acid v/v) at retention time 36.6 min and 67.2 min, respectively. Compounds 10a, 10b and 10c were isolated with CH3CN–water (35%
55%, 0.3% trifluoroacetic acid v/v) at retention time 36.6 min and 67.2 min, respectively. Compounds 10a, 10b and 10c were isolated with CH3CN–water (35%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65%, 0.3% trifluoroacetic acid v/v) at retention time 30.5 min, 42 min and 54 min, respectively. Compounds 12a and 12b were obtained at retention time 36.5 min and 53 min, respectively eluted by CH3CN–water (33%
65%, 0.3% trifluoroacetic acid v/v) at retention time 30.5 min, 42 min and 54 min, respectively. Compounds 12a and 12b were obtained at retention time 36.5 min and 53 min, respectively eluted by CH3CN–water (33%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 67%, 0.3% trifluoroacetic acid v/v).
67%, 0.3% trifluoroacetic acid v/v).
Quantitative analysis of biotransformation for kinetics investigation
An Agilent 1200 HPLC system possessed quaternary delivery system, degasser, autosampler, UV-detector, and Elite SinoChorm ODS-BP (2.1 × 150 mm, 5 μm) analytical column was used for separation. The mobile phase consisted of acetonitrile–0.1% formic acid aqueous solution at a flow rate of 450 μL min−1. An Applied Biosystems MDS Sciex API 3200 Triple Quadrupole Mass Spectrometer (MS/MS) equipped with electrospray ionization (ESI) source was applied for the target products analysis. The system was operated in negative mode with the following values: compounds 1a (ions 538.8 → 387.2); 8a (540.8 → 415.0); 8b (540.8 → 389.0); 10a (523.0 → 269.0); 10b (504.8 → 369.0); 10c (504.9 → 252.0); 12a (509.1 → 403.1); 12b (509.0 → 403.0). The optimized ion spray voltage and temperature were set at 4500 V and 500 °C, respectively. The curtain gas (CUR) flow was 20 L min−1; gas 1 and gas 2 (nitrogen) were set at 30 and 40 L min−1, respectively, and dwell time was 150 ms. Nitrogen was used as both curtain and collision gas, controlled at 13 and 6 psi, respectively. Quantification assay was performed using multiple reaction monitoring.Molecular docking
The protein structures of horseradish peroxidase (PDB ID: 1H5A)23 and laccase (PDB ID: 1KYA)24 were represented by CHARMM 22 force field parameters25 in order to properly model the HEME motif of horseradish peroxidase, Cu coordination of laccase, and disulfide bonds within each protein. The initial ligand structure was taken from the result of DFT calculations, and prepared using the PRODRG server (http://davapc1.bioch.dundee.ac.uk/cgi-bin/prodrg).26AutoDock 4.2 programs27 were used to perform all dockings. Autogrid program was used to pre-calculate grid maps of interaction energies for various atom types. The grid box was set sufficiently large to encompass the entire protein surface. Blind docking was performed using the Lamarckian generic algorithm as the searching method, and the default parameters for other docking settings were applied. A total of 100 conformations of each ligand were searched relative to each protein structure. The occurrence frequency of the ligand in different binding sites of the protein was calculated as indicative of the occupation propensity. The lowest binding energy in terms of docking scoring functions was also shown.
Conclusions
Horseradish peroxidase and laccase as the important biosynthesis enzymes, could catalyze the various oxidation of phenolic compounds. As the vital phenolic substances, flavone analogues with abundant natural resources and significant bioactivities, could be modified to form the series of potential bioactive molecules. Therefore, oxidases were applied to effectively catalyze the oxidation of various flavone analogues in this work. After the screening experiments, HRP had the preferred capability to oxidize most of the phenolic flavones, but only flavonols, isoflavones and chalcones with phenolic groups and dihydroxylflavones could become the effective substrates of laccase to yield the similar products with that mediated by HRP. For these phenolic groups at different positions of flavones, the O–H bond energy analysis revealed the lowest bound energy of 4′-OH in the B-ring of flavones, which implying that the 4′-radical was the trigger of these radical reactions. Furthermore, 4′-OH were nearest groups to the HEME active pocket of HRP by in silico molecular docking. However, for laccase, due to most flavones docking into its surface, the laccase did not efficiently catalyze the oxidation of flavones. Kinetic behaviors of the oxidation for these flavone analogues mediated by peroxidases exhibited the Hill and substrate inhibition kinetic models. In summary, our present work systemically characterized the oxidation of flavone analogues mediated by HRP and laccase, and some novel dimers were successfully prepared and identified.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This research program was supported financially by the National Natural Science Foundation of China (No. 81803683 and 81872970) and Dalian Youth Science and Technology Star Project (2017RQ046).Notes and references
- S. Witayakrana and A. J. Ragauskas, Adv. Synth. Catal., 2009, 351, 1187–1209 CrossRef.
- M. Mogharabia and M. A. Faramarzia, Adv. Synth. Catal., 2014, 356, 897–927 CrossRef.
- G. R. Lopes, D. C. G. A. Pinto and A. M. S. Silva, RSC Adv., 2014, 4, 37244–37265 RSC.
- M. Kidwai, A. Jain, A. Sharmab and R. C. Kuhad, Catal. Sci. Technol., 2013, 3, 230–234 RSC.
- X. X. Gao, S. S. Huang, P. P. Dong, C. Wang, J. Hou, X. K. Huo, B. J. Zhang, T. H. Ma and X. C. Ma, Catal. Sci. Technol., 2016, 6, 3585–3593 RSC.
- L. Heap, A. Green, D. Brown, B. V. Dongen and N. Turner, Catal. Sci. Technol., 2014, 4, 2251–2259 RSC.
- N. L. McCombs, T. Smirnova and R. A. Ghiladi, Catal. Sci. Technol., 2017, 7, 3104–3118 RSC.
- N. C. Veitch, Phytochemicals, 2004, 65, 249–259 CrossRef CAS.
- N. Hakulinen and J. Rouvinen, Cell. Mol. Life Sci., 2015, 72, 857–868 CrossRef CAS PubMed.
- Z. C. Liu, T. Xie, Q. P. Zhong and G. G. Wang, Acta Crystallogr., Sect. F: Struct. Biol. Commun., 2016, 72, 328–335 CrossRef CAS PubMed.
- N. E. Es-Safi, S. Ghidouche and P. H. Ducrot, Molecules, 2007, 12, 2228–2258 CrossRef CAS PubMed.
- S. Ghidouche, N. E. Es-Safi and P. H. Ducrot, Tetrahedron Lett., 2008, 49, 619–623 CrossRef CAS.
- H. M. Awad, M. G. Boersma, J. Vervoort and I. M. C. M. Rietjens, Arch. Biochem. Biophys., 2000, 378, 224–233 CrossRef CAS PubMed.
- L. G. Fenoll, P. A. García-Ruiz, R. Varón and F. García-Cánovas, J. Agric. Food Chem., 2003, 51, 7781–7787 CrossRef CAS PubMed.
- N. C. Veitch and R. J. Grayer, Nat. Prod. Rep., 2008, 25, 555–611 RSC.
- V. Krishnamachari, L. H. Levine and P. W. Paré, J. Agric. Food Chem., 2002, 50, 4357–4363 CrossRef CAS PubMed.
- K. Mohammad, I. Mohammad, R. Wasiur, H. Noriko, O. Masayoshi and K. Nobusuke, J. Chem. Soc., Perkin Trans. 1, 1981, 553–559 Search PubMed.
- G. Sagrera and G. Seoane, Synthesis, 2010, 2776–2786 CAS.
- I. Ahmed, K. Ishratullah, M. Ilyas, W. Rahman, O. Seligmann and H. Wagner, Phytochemistry, 1981, 20, 1169–1170 CrossRef.
- C. Ichino, H. Kiyohara, N. Soonthornchareonnon, W. Chuakul, A. Ishiyama, H. Sekiguchi, M. Namatame, K. Otoguro, S. Omura and H. Yamada, Planta Med., 2006, 72, 611–614 CrossRef CAS PubMed.
- L. V. Jqrgensen, C. Cornett, U. Justesen, L. H. Skibsted and L. O. Dragsted, Free Radical Res., 1998, 29, 339–350 CrossRef.
- M. H. Vakarelska-Popovska and Z. Velkov, Comput. Theor. Chem., 2016, 1077, 87–91 CrossRef CAS.
- G. I. Berglund, G. H. Carlsson, A. T. Smith, H. Szöke, A. Henriksen and J. Hajdu, Nature, 2002, 417, 463–468 CrossRef CAS PubMed.
- T. Bertrand, C. Jolivalt, P. Briozzo, E. Caminade, N. Joly, C. Madzak and C. Mougin, Biochemistry, 2002, 41, 7325–7333 CrossRef CAS PubMed.
- A. D. Mackerell, M. Feig and C. L. Brooks, J. Comput. Chem., 2004, 25, 1400–1415 CrossRef CAS PubMed.
- A. W. Schüttelkopf and D. M. F. van Aalten, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 1355–1363 CrossRef PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra00470j |
| ‡ Q. S. Yan and X. G. Tang contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |