Open Access Article
Open Access ArticleDevelopment of double strand RNA mPEI nanoparticles and application in treating invasive breast cancer†
Rui Liu,
Li-Min Mu,
Jing Bai,
Ya-Fei Du,
Ying Xie and
Wan-Liang Lu *
*
State Key Laboratory of Natural and Biomimetic Drugs, Beijing Key Laboratory of Molecular Pharmaceutics and New Drug System, School of Pharmaceutical Sciences, Peking University, Beijing 100191, China. E-mail: luwl@bjmu.edu.cn; Fax: +86 10 82802683; Tel: +86 10 82802683
First published on 30th April 2019
Abstract
Triple negative breast cancer (TNBC) has been characterized as a very heterogeneous subtype, and is more invasive and non-expressing of the genes for the estrogen receptor (ER), progesterone receptor (PR) and HER2/neu, with poor prognosis, and hence the efficacy of regular chemotherapy is very limited. Here, we report a kind of double strand RNA (dsRNA) mPEI nanoparticle for treatment of invasive TNBC. The studies were performed on TNBC cells in vitro and in TNBC cancer-bearing mice. The results showed that dsRNA mPEI nanoparticles were able to effectively transfect cells, and demonstrated a strong capability in knocking-down the Fra-1 gene and down-stream MMP-1 and MMP-9 genes in TNBC cells and TNBC cancer-bearing mice, thereby inhibiting the invasion and migration of cells. After intratumoral injection, dsRNA mPEI nanoparticles exhibited a robust anticancer efficacy in TNBC cancer-bearing mice, and the anticancer efficacy was superior to that of paclitaxel. In conclusion, dsRNA mPEI nanoparticles are able to effectively treat aggressive TNBC, and the mechanism studies reveal that they take effect by knocking-down Fra-1 relevant genes, hence interfering in transcription and translation of the genes, which are necessary for growth and metastasis of TNBC. Therefore, the present study offers a new and promising formulation and strategy for effective treatment of TNBC.
1. Introduction
Triple negative breast cancer (TNBC) has been characterized as the breast cancer that does not express the genes for the estrogen receptor (ER), progesterone receptor (PR) and HER2/neu.1 This makes TNBC more difficult to treat2 because most hormone therapies target one of the three receptors, therefore TNBC often requires a combinational treatment. Moreover, TNBC comprises a very heterogeneous subtype; some types of triple-negative breast cancer are more aggressive, with poor prognosis, although other types have very similar treatment outcomes to hormone receptor positive breast cancers.3 The standard therapeutic strategies for TNBC are surgery with adjuvant/neoadjuvant chemotherapy and radiotherapy. However, TNBC is generally very susceptible to chemotherapy. In some cases, early complete response does not correlate with overall survival. This makes it particularly complicated to treat TNBC.4Herein, we hypothesized that the dsRNA mPEI nanoparticles could treat the aggressive triple negative breast cancer by inhibiting invasive target genes. In this study, double strand RNA (dsRNA) is used as a stable structure to be encapsulated into mPEI nanoparticles for silencing the aggressive genes of TNBC while mPEI is a newly synthesized safe cationic polymer material for delivery of dsRNA into TNBC cells.
dsRNA is a double strand RNA designed based on endogenous microRNA. Its sense strand is identical to the strand of microRNA, and the anti-sense strand is complementary to the sense strand. As an evolutionarily conserved class of small non-coding RNAs (of 18–24 nucleotides),5 microRNAs (miRNAs) have emerged as pivotal regulators of gene expression6 and are involved in a variety of cellular processes, including differentiation, growth control, and cell fate determination.7 miRNAs often negatively regulate gene expression at the post-transcriptional level, with each miRNA able to target several mRNA species.8 miRNAs function through the argonaute (AGO) proteins,9 which usually exist in a RNA-induced silencing complex (RISC) that contains both RNA binding domains and RNase H domains.10 As a kind of miRNA, miR-34a is recognized as a master regulator of tumor suppression.11 Similarly to the biogenesis of all miRNAs, miR-34a is transcribed as a long hairpin molecule (pri-miRNA), which is subsequently cleaved by an RNase III Drosha to an approximately 70-nucleotide long stem-loop precursor (pre-miRNA).12 Following nuclear export, the pre-miRNA is further cleaved by an RNase III Dicer into 22-nucleotide long mature strands, which are incorporated into RNA-induced silencing complex (RISC).13 This RNA/protein complex mediates down-regulation of target transcripts by mRNA degradation or inhibition of translation.14,15
As one member of the activator protein-1 (AP-1) family,16 FOS-related antigen 1 (FRA-1),17 is located on chromosome 11q13 and encodes a mature mRNA of 1.7 kb in length. It is emerging as a key regulator of invasion in cancer cells and overexpressed in several types of human cancers.18 Fra-1 regulates several genes, implicates in invasion, angiogenesis and cell proliferation19,20 and directly induces the promoter activity of matrix metalloproteinase (MMP-1 (ref. 21) or MMP-9).22 MMP family is a family of metalloproteinase-dependent proteolytic enzymes that can effectively degrade extracellular matrices and play an important role in tumor growth and metastasis.23 Tumor cells must penetrate the basement membrane and traverse the extracellular matrix in order to invade surrounding structures and metastasize to distant sites.24,25
Polyethylenimine (PEI) has been widely used as a polycationic material for loading miRNAs.26 It is divided into two types: linear and branched.27 Branched PEI has a large specific surface area and a large positive charge on the surface. It can highly compress nucleic acids to form complexes, which can be integrated into cells by fusion with negatively charged cell membranes.28 Although PEI has high transfection efficiency due to positive electricity, the resulting strong cytotoxicity is not negligible. Therefore, a number of studies focused on the modification of PEI to reduce cytotoxicity and increase miRNA uptake into the cell at the same time.29 Sodium alginate is a by-product of the extraction of iodine and mannitol from kelp or sargasso of brown algae. The molecule is linked by β-D-mannuronic acid and α-L-guluronic acid by 1–4 bonds. It is a natural polysaccharide with the stability and safety required for pharmaceutical preparations.
The objectives of the present study were to develop a stable dsRNA nanoparticle, confirm its function for treating the aggressive triple negative breast cancer, and reveal the relevant mechanism. In the study, we synthesized multiple aldehyde sodium alginate–polyethyleneimine (mPEI) nanoparticles to carry double strand microRNA (dsRNA). The dsRNA mPEI nanoparticles could increase dsRNA uptake into the triple negative breast cancer (TNBC) cells, provide lysosomal escape ability and enhance the knockdown effect on the target genes of the cancer cells in vitro and in vivo.
2. Results and discussion
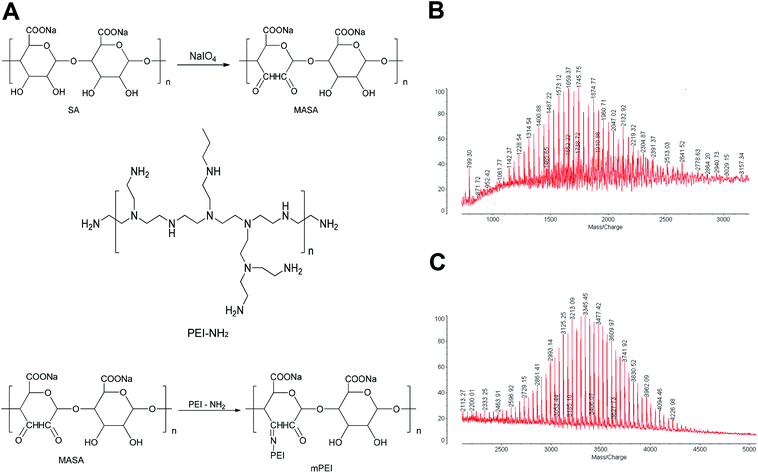
2.1. Synthesis and characterization of MASA and mPEI materials
To produce the reactive aldehyde group, multiple aldehyde sodium alginate (MASA) was firstly produced by oxidizing sodium alginate with sodium periodate under stirring at ambient temperature (Fig. 1A). The resultant powders were confirmed by MALDI-TOF-MS spectrum (Fig. 1B). It could be seen that the average mass of the product was m/z 1745.45.To synthesize mPEI, MASA was connected with PEI using aldehyde–amine condensation in a slightly alkaline environment (Fig. 1A). The obtained products were also confirmed by MALDI-TOF-MS spectrum (Fig. 1C). The results demonstrated that the average mass of the product was m/z 3345.45 and the mass distribution was concentrated.
By comparing the mass of the two products, the results showed that the difference of the average mass between the two was equal to that of PEI (m/z 1600), thus indicating a successful synthesis of mPEI by linking MASA with PEI. And mPEI was used as a gene carrier in subsequent experiments.
2.2. Preparation and characterization of dsRNA mPEI nanoparticles
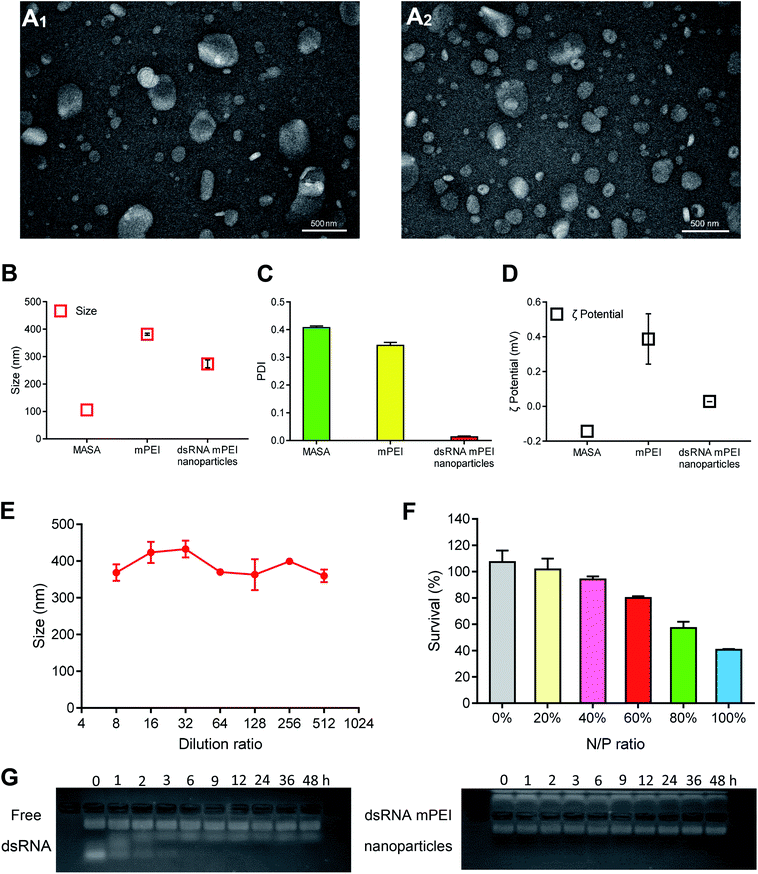
dsRNA and mPEI materials were able to self-assemble to nanoparticles by static attraction in the liquid state. When dsRNA was added into mPEI, dsRNA could be encapsulated into mPEI to form dsRNA mPEI nanoparticles.To characterize dsRNA mPEI nanoparticles, the morphologies of mPEI vector and dsRNA mPEI nanoparticles were observed by transmission electron microscopy (TEM). The average diameter of mPEI was about 350 nm (Fig. 2A1) and the average diameter of dsRNA mPEI nanoparticles was about 200 nm (Fig. 2A2). The results demonstrated that mPEI and dsRNA mPEI nanoparticles were both round in shape, and there were no obvious aggregations. Because the positive and negative charges interacted with each other to form nanoparticles, the size of dsRNA mPEI nanoparticle was smaller than mPEI.
Sizes and zeta potentials were further analyzed by dynamic light scattering (DLS). The particle sizes of MASA, mPEI and dsRNA mPEI nanoparticles were 105.5 ± 17.0, 381.6 ± 3.8, and 273.7 ± 14.4 nm, respectively (Fig. 2B). And the PDIs of MASA, mPEI and dsRNA mPEI nanoparticles were 0.407 ± 0.006, 0.343 ± 0.011 and 0.013 ± 0.003, respectively (Fig. 2C). When the positively charged mPEI bound to the negatively charged dsRNA, the particle size was slightly reduced due to electrostatic interaction. The sizes determined by TEM were smaller than that seen in DLS experiments, which might be due to dehydration effects that occurred during TEM sample preparation, leading to shrinking of the nanoparticles. The dilution stability results showed that the particle size was stable within a small range after dilution of different ratios (Fig. 2E). Since nanoparticles can be diluted up to 100 times by blood after entering the circulation of the body, a certain anti-dilution stability is necessary to ensure more accumulation of nanoparticles in the lesions and to exert effects.
After combined with negative dsRNA, dsRNA mPEI nanoparticles exhibited slightly positively charged (0.03 ± 0.00 mV) (Fig. 2D), showing a decrease in the electropositivity as compared to mPEI (0.39 ± 0.14 mV). The slight electropositivity may benefit for lowering the potential cytotoxicity of delivery material itself, and for cellular uptake by cancer cells due to a negatively charged cell membrane. It is characterized that PEI is often associated with significant cytotoxic effects.30 Therefore, it is necessary to evaluate the cell viability of nanoparticles when used as dsRNA delivery carriers. SRB staining assay was used to evaluate the safety of dsRNA mPEI nanoparticles at different N/P ratios (Fig. 2F). When the N/P ratios were set as a range of 0% (dsRNA only, dsRNA control)–100% (mPEI nanoparticles only, mPEI control), the cytotoxicity in MDA-MB-231 cells at 48 h incubation gradually increased. The results showed that the dsRNA mPEI nanoparticles at 60% N/P ratio had an acceptable cytotoxicity (>80% viability of cells) and were suitable for further experimental evaluations. Accordingly, the N/P ratio at 60% was selected for preparing dsRNA mPEI nanoparticles and used in the following evaluations due to the safety concern of nanoparticle materials.31
To evaluate the stability of dsRNA mPEI nanoparticles in blood, the protective effects of mPEI nanoparticles on dsRNA were analyzed by the gel retardation assay (Fig. 2G). It could be seen that free dsRNA would be degraded within 6 h. In contrast, intact dsRNA could be seen in the band of image for dsRNA mPEI nanoparticles at 48 h, demonstrated that the dsRNA mPEI nanoparticles could protect dsRNA from RNase degradation in serum and had a potential for future drug administration. There exist RNA degradation enzymes in physiological environment and cytoplasm of cancer cells, the protective effect of mPEI nanoparticles enables dsRNA to be less degraded during drug administration.
2.3. Invasiveness characterization of breast cancer cells
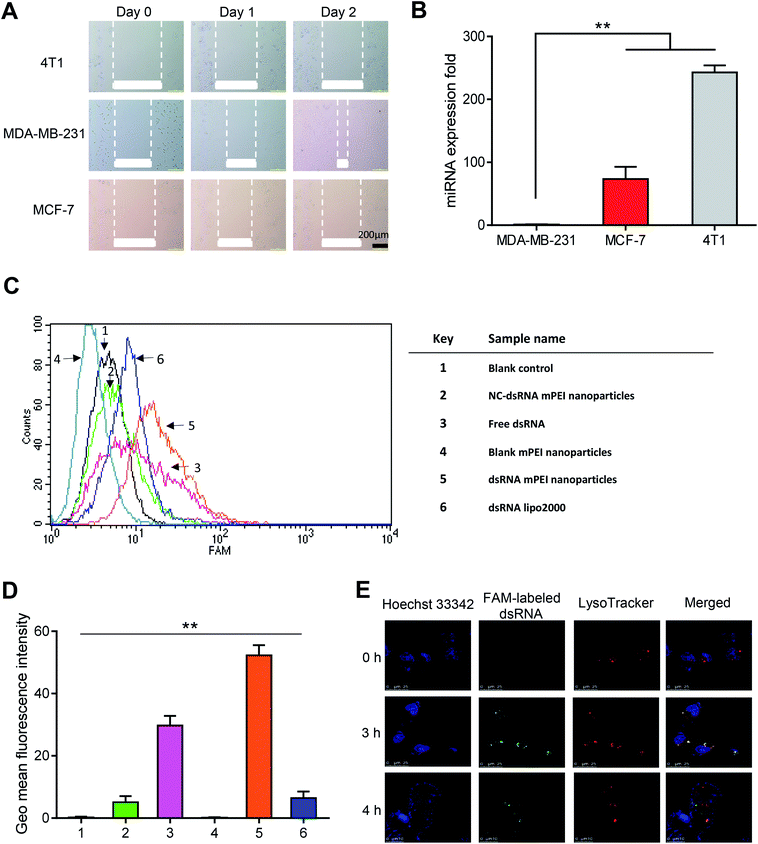
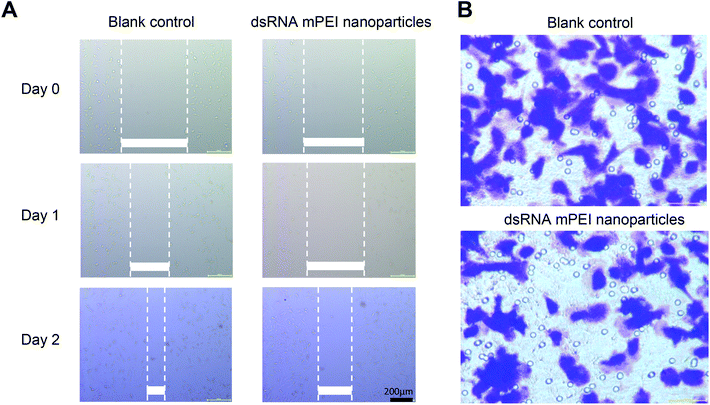
To characterize the invasiveness of different breast cancer cells, human TNBC MDA-MB-231 cells, human breast cancer MCF-7 cells and mouse breast cancer 4T1 cells were included for illustrating the invasiveness by using wound-healing assay and qRT-PCR assay.It could be seen that, after two days of incubation, the wounds in MDA-MB-231 cells were clearly healed, while the wounds in MCF-7 and 4T1 cells did not change obviously (Fig. 3A). The results from the wound-healing assay indicated that the MDA-MB-231 cells had the strongest invasiveness among the three breast cancer cells. Furthermore, the results from evaluation on miRNA expression by qRT-PCR showed that the miR-34a expression in MDA-MB-231 cells was the lowest among the three cells (Fig. 3B). As miR-34a, an intrinsic miRNA in the cells, has been associated with the inhibition of the invasiveness of the cancer,32,33 the result further indicated that the MDA-MB-231 cells were the strong invasive cancer cells, and accordingly, selected for further experimental evaluations in the present study.
2.4. Cellular uptake and lysosomal escape in TNBC cells
High cellular uptake of dsRNA is an initial step for gene transfection and for gene therapy effects.34 To evaluate the cellular uptake effect of dsRNA mPEI nanoparticles, FAM-labeled dsRNA was used as a fluorescence indicator to detect the intracellular fluorescence intensity by flow cytometry. The results showed that the dsRNA mPEI nanoparticles had the highest fluorescence intensity among varying control formulations at 4 h incubation with MDA-MB-231 cells, indicating that dsRNA mPEI nanoparticles were the most significantly internalized by the cells (Fig. 3C). As a quantitatively estimation by flow cytometry, the cellular uptake of dsRNA mPEI nanoparticles was 8-folds higher than that of dsRNA lipo2000 (Fig. 3D), demonstrating a very strong cellular uptake and transfection efficiency of dsRNA into breast cancer cells.The different cellular uptake effects of the varying formulations can be explained by the following aspects. In the image (Fig. 3D), the sample 1 is used as a blank control, and accordingly, there is no fluorescent signal detected for the cellular uptake; the sample 2 is used as a negative control, in which a meaningless dsRNA (negative effect) is labeled with FAM and encapsulated in mPEI nanoparticles. Since the meaningless dsRNA enters cells and can't be bound to the target mRNAs, it is cleared by the cells. Therefore, the fluorescent signal is less detected; the sample 3 is used as an isotype control, free dsRNA is degraded by nucleases after entering cells, hence weakening the fluorescent signal; the sample 4 is used as a compensation control, which doesn't contain FAM and therefore has no fluorescent signal, and is used to eliminate interference from impurities. In contrast, the sample 5 is used as a test sample, and the sample 6 is used as positive control. In comparison of sample 5 with sample 6, the results indicate that the dsRNA delivery efficiency of mPEI nanoparticles is significantly higher than that of lipo2000 in MDA-MB-231 cells.
To investigate the lysosomal escape after cellular uptake, laser confocal microscopy was used to directly observe the escape of dsRNA mPEI nanoparticles in MDA-MB-231 cells after incubation at different time points. The blue, red and green fluorescent colors were used to indicate Hoechst 33342 labeled nuclei, LysoTracker labeled lysosomes, and FAM-labeled dsRNA, respectively. The yellow color was used to indicate the co-localization of FAM-labeled dsRNA (green) and LysoTracker (red). The results showed that the dsRNA mPEI nanoparticles were captured by lysosomes of MDA-MB-231 cells at 3 h incubation (Fig. 3E). In viewing the separation of red and green color at 4 h incubation, dsRNA mPEI nanoparticles already escaped from the lysosomes at this time point, indicating a rapid lysosome escape.
The effective lysosomal escape of dsRNA mPEI nanoparticles can be explained by the proton sponge effect.35 In this process, the cationic nanoparticles are combined with the cell membrane, and enters the cells to form endosomes while the endosomes are fused with lysosomes, leading to the capture of dsRNA mPEI nanoparticles by lysosomes. In lysosomes, the unsaturated amino group on mPEI nanoparticles chelate with H+ provided by the proton pump, leading to continuously open of the proton pump on membrane of lysosomes. Each proton causes a Cl− and a water molecule to remain in lysosomes, causing swelling and rupturing of the lysosomes. And then dsRNA mPEI nanoparticles are released into cytoplasm. During the process of lysosomal escape, dsRNA could be also released from the intact or the raptured nanoparticles to take effect.
2.5. Knocking-down effect on the Fra-1 relevant genes in TNBC cells
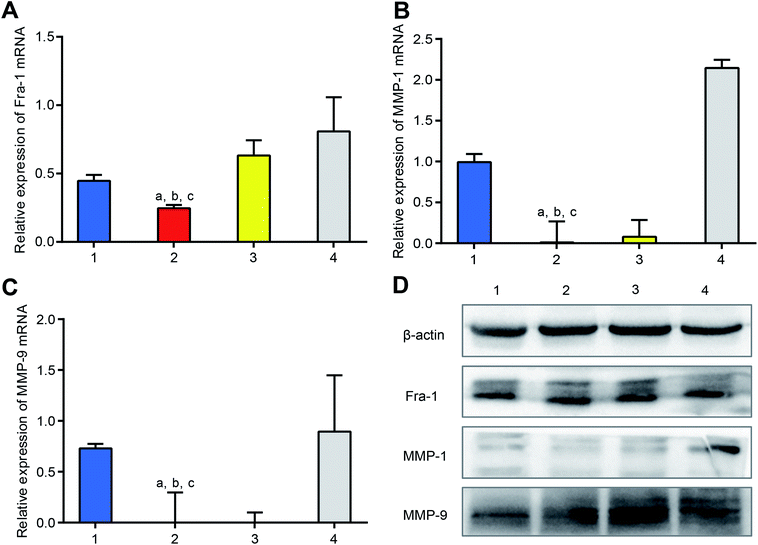
To study the knocking-down effect on mRNA in vitro, dsRNA mPEI nanoparticles were used to treat MDA-MB-231 cells, and the mRNAs of Fra-1, MMP-1 and MMP-9 in the cells were measured by qRT-PCR. The results showed that the dsRNA mPEI nanoparticles were significantly able to lower the mRNA levels of Fra-1 (Fig. 4A), MMP-1 (Fig. 4B) and MMP-9 (Fig. 4C). The knocking-down effect of dsRNA mPEI nanoparticles can be explained by the specific dsRNA, which is designed as a mimics sequence to supplement intrinsic miR-34a by which to inhibit the transcription of mRNA in cancer cells.36 Accordingly, extrinsic dsRNA bind with intrinsic mRNA, thereby interfering the transcription of mRNA.37As a positive control, dsRNA lipo2000 also lowered the mRNA levels of Fra-1, MMP-1 and MMP-9 but had a smaller effect that of dsRNA mPEI nanoparticles. The results indicate that the transfection effect of mPEI nanoparticles are stronger than that of the commercially available transfection agent lipo2000. On the contrary, ssRNA mPEI nanoparticles, which are used as the inhibitor control, obviously increase the mRNA levels of Fra-1, MMP-1 and MMP-9. The inhibitory effect of ssRNA mPEI nanoparticles is due to the fact that the ssRNA is also specifically designed according to the sequence of intrinsic miR-34a, which is capable of blocking the action of miR-34a. As for the negative control, NC-dsRNA mPEI nanoparticles did not influence the mRNA level of MMP-1 mRNA but slightly reduced mRNA levels of Fra-1 and MMP-9. The reason could be explained that the meaningless RNA (referring to as NC-dsRNA) may bind with minor part of mRNAs as well, hence slightly interfering the transcription of mRNA.
To evaluate the knocking-down effect on the target proteins in vitro, dsRNA mPEI nanoparticles were used to treat MDA-MB-231 cells, and the proteins of Fra-1, MMP-1 and MMP-9 in the cells were measured by western blotting (Fig. 4D). The results showed that both dsRNA mPEI nanoparticles and dsRNA lipo2000 were able to knock-down the protein expressions of Fra-1, MMP-1 and MMP-9. Among which, the protein expressions of MMP-1 and MMP-9 were more evident than that of Fra-1. This phenomenon could be explained by the regulation mechanism that the Fra-1 gene is an up-stream gene, which regulates the translation of down-stream genes (MMP-1 and MMP-9). Accordingly, the expressions of down-stream proteins (MMP-1, and MMP-9) are exhibited more significantly.
As an inhibitor control, ssRNA mPEI nanoparticles obviously increased the protein expressions of Fra-1, MMP-1 and MMP-9, demonstrating an evident inhibition to the knocking-down process. As a negative control, NC-dsRNA mPEI nanoparticles acted less effect these protein expressions.
The present design to knock-down Fra-1 and its relevant genes by dsRNA mPEI nanoparticles is based on the consideration that Fra-1 promotes the growth, invasion and metastasis of cancer cells.38 It is known that the intrinsic microRNA like miR-34 is able to inhibit the expression of Fra-1 gene in the normal cells. However, these intrinsic microRNAs are often insufficient to inhibit the higher expression of oncogenes (like Fra-1 gene) in the metastatic breast cancer and primary breast cancer with lymph node metastases.39 Accordingly, extrinsic supplement of active microRNA would assist to inhibit the growth and invasiveness of the cancer cells via knocking-down the oncogenes. The present study reveals that the dsRNA mPEI nanoparticles can reach such a purpose. In the design, dsRNA was synthesized according to the mimics of miR-34a, and mPEI nanoparticles could effectively deliver dsRNA into the TNBC cells to take effect.
As the downstream genes of Fra-1, MMP-1 and MMP-9 genes are closely associated with the migration and invasiveness of cancer, and exhibit over-expressions in the invasive breast cancer.40,41 The results from the present study as indicated in below section demonstrate that the dsRNA mPEI nanoparticles can effectively down-regulate the expression of MMP-1 and MMP-9 genes as well, hence contributing to the inhibition to growth, invasion and migration of TNBC cells.
2.6. Inhibition effect on the invasiveness of TNBC cells
To investigate inhibitory effect on the invasiveness of cancer cells, dsRNA mPEI nanoparticles were used to treat TNBC MDA-MB-231 cells, and the invasiveness was assessed by wound-healing assay and transwell invasion assay, respectively.In the wound-healing assay, dsRNA mPEI nanoparticles significantly inhibited the wound-healing rate within two days as compared to blank control (Fig. 5A). As wound healing assay has been used to indicate cell migration and cell–cell interaction,42 it could be a straightforward observation for rapid growth rate and distant invasiveness of the malignant cancer cells. Therefore, the inhibition of wound-healing of MDA-MB-231 cells in the study could suggest that the dsRNA mPEI nanoparticles inhibit not only the invasiveness but also the growth of the cells.
The invasiveness of TNBC cells were further evaluated by transwell invasion assay,43 and the results showed that the dsRNA mPEI nanoparticles significantly inhibited the migration of MDA-MB-231 cells from the upper chamber to lower one (Fig. 5B). As the migration is able to more clearly indicate the motion and invasion of the malignant cells, the results from this study could reveal the inhibitory effect on the invasiveness of MDA-MB-231 cells,44 and consequently, the inhibitory effect on the invasiveness of TNBC cells verify the function of dsRNA mPEI nanoparticles from in vitro aspect.
A unique nanoparticle formulation could achieve the specific purposes in the diagnosis and/or therapy for the refractory TNBC cancer. For examples, it is reported that the cRGD, a cyclic arginine–glycine–aspartic acid (cRGD) modified nanoparticles, UCNP@TTD-cRGD NPs,45 cRGD-MEH-PPV NPs46 and cRGD-siVEGF-TTD NPs47 have been used for targeted diagnosis and therapy of TNBC cells. These nanoparticle formulations demonstrate remarkable potentials in the imaging by a targeting molecule mediation and in the photodynamic therapy (PDT) by generating ROS under light irradiation.48,49 In the present study, dsRNA mPEI nanoparticles are developed for gene therapy by inhibiting the growth, invasion and migration of TNBC cells through knocking-down Fra-1 and its relevant genes. Albeit the differences in the action mechanism and the components of nanoparticles, the results from the present study reveal a promising new nanoparticle formulation to inhibit the growth and to overcome the invasiveness of the TNBC cancer.
2.7. Anticancer efficacy and inhibitory effect on Fra-1 relevant genes in TNBC cancer-bearing mice
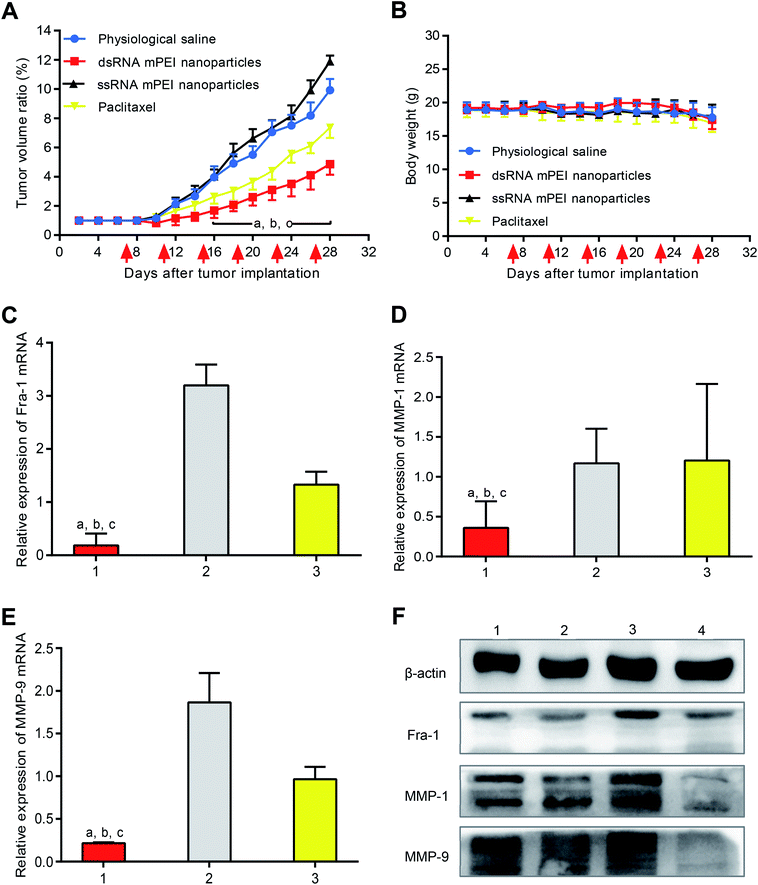
To evaluate the anticancer efficacy, MDA-MB-231 cells were inoculated into nude mice for establishing TNBC cancer-bearing mice model. The results showed that the tumor volume reached 180–200 mm3 at day 7 since the inoculation of cancer cells, indicating a successful establishment of the model. After administrations, both physiological saline group and ssRNA mPEI nanoparticles (inhibitor) were not able to inhibit growth of the tumor. In contrast, dsRNA mPEI nanoparticles exhibited the stronger anticancer efficacy than paclitaxel in the TNBC cancer-bearing mice (Fig. 6A), demonstrating a robust inhibitory effect to growth of the TNBC tumor.The result from in vivo anticancer efficacy indicate that the dsRNA mPEI nanoparticles have a strong cytotoxicity to TNBC cancer tissues. This result can successfully meet the expectation to inhibit the growth of TNBC cancer. However, it may also raise the safety concern that the dsRNA formulation may have a strong cytotoxicity to normal cells, tissues and organs. Actually, it may cause the cytotoxicity when the dsRNA formulation is given by a systemic administration, such as by intravenous injection. Nevertheless, the dsRNA formulation has little impact on the normal cells or tissues when it is given by topical administration via intra-tumoral injection. It is because the nanoparticles exhibit an enhanced permeability and retention (EPR) effect in tumor tissue over the range of 5–200 nm50 when administrated as an intravenous injection. In the case of intra-tumoral injection, bigger size (>200 nm) of the nanoparticles would be more beneficial for the “retention” effect, namely, nanoparticles could be accumulated into the tumor tissue.
After drug administration, the evaluation on body weight of mice could preliminarily indicate the safety of dsRNA mPEI nanoparticles. In this study, the changes of body weight were measured during the whole period of animal experiments (Fig. 6B). It could be seen that the body weights of tumor-bearing mice were stable, indicating that the dsRNA mPEI nanoparticles could be safe when used in vivo.
To confirm the knocking-down effect on mRNA in vivo, tumor tissues were removed from nude mice, and the mRNAs of Fra-1, MMP-1 and MMP-9 were measured by qRT-PCR. The results showed that dsRNA mPEI nanoparticles could significantly lower the mRNA levels of Fra-1 (Fig. 6C), MMP-1 (Fig. 6D) and MMP-9 (Fig. 6E). On the contrary, ssRNA mPEI nanoparticles increased the mRNA levels of target genes. For paclitaxel group, expression levels of mRNA remained unchanged.
To study the knocking-down effect on the target proteins in vivo, the proteins of Fra-1, MMP-1 and MMP-9 were measured by western blotting assay. The results showed that dsRNA mPEI nanoparticles were able to knock-down the protein expressions of Fra-1, MMP-1 and MMP-9. As an inhibitor control, ssRNA mPEI nanoparticles increased protein expressions. For paclitaxel group, the protein expressions were not affected.
These results exhibited that the results in vivo were more significant than those in vitro, it might because the concentration of mRNA and protein extracted from tumor tissue was much higher than that extracted from cells. Therefore, the results obtained were more significant. Combining vitro and vivo results, it could be concluded that the dsRNA mPEI nanoparticles had a significant inhibitory effect on expression of Fra-1, MMP-1 and MMP-9.
3. Experimental section
3.1. Materials
Sodium alginate, sodium periodate and ethylene glycol were purchased from J&K Scientific Ltd. (Beijing, China). Polyethyleneimine solution was obtained from Sigma-Aldrich Co. (St. Louis, MO, USA). Opti-MEM reduced-serum medium was supplied by Life Technologies (Carlsbad, CA, USA). Lipofectamine 2000 was provided by Invitrogen (New York, NY, USA). Sulforhodamine B was purchased from Macklin Ltd. (Shanghai, China). dsRNA (sense strand: 5′-UGGCAGUGUCUUAGCUGGUUGU-3′ and antisense strand: 5′-AACCAGCUAAGACACUGCCAUU-3′), ssRNA (inhibitor) (strand: 5′-ACAACCAGCUAAGACACUGCCA-3′), negative control (sense strand: 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense strand: 5′-ACGUGACACGUUCGGAGAATT-3′, named as NC), fluorescein-labeled dsRNA (5′ end of the sense strand, FAM-dsRNA), fluorescein-labeled ssRNA inhibitor (5′ end of the sense strand, FAM-ssRNA), and fluorescein-labeled negative control (5′ end of the sense strand, FAM-NC) were synthesized and purified with HPLC by Sangon Biotech Co., Ltd. (Shanghai, China). DMEM medium and PRMI 1640 medium were obtained from Macgene Technology Co., Ltd. (Beijing, China). The solvents were purchased from J&K Scientific Ltd. (Beijing, China).3.2. Cell lines and experimental animals
The human TNBC MDA-MB-231 cells, human breast cancer MCF-7 cells and mouse breast cancer 4T1 cells were obtained from Institute of Basic Medical Science, Chinese Academy of Medical Science (Beijing, China). MDA-MB-231 cells were cultured in Dulbecco's modified Eagle medium (DMEM) medium (Macgene, Beijing, China) supplemented with 10% fetal bovine serum (FBS), 100 U mL−1 penicillin, 100 μg mL−1 streptomycin (Macgene, Beijing, China). MCF-7 cells and 4T1 cells were cultured in Roswell Park Memorial Institute (RPMI 1640) (Macgene, Beijing, China) supplemented with 10% FBS, 100 U mL−1 penicillin, 100 μg mL−1 streptomycin. MDA-MB-231 cells and MCF-7 cells were cultured in humidified atmosphere of 5% CO2 at 37 °C, while 4T1 cells were cultured without 5% CO2. The cells for all experiments were in logarithmic phase of growth.Female BALB/c nude mice of 18–20 g (6–7 weeks old) were purchased from Vital River Laboratory Animal Center (Beijing, China) and maintained in a specific pathogen free (SPF) environment. All animal experiments were carried out in accordance with guidelines, and approved by the Committee for Experimental Animal Welfare of Biomedical Ethics of Peking University.
3.3. Synthesis of MASA
To obtain MASA, sodium alginate was oxidized by sodium periodate. Briefly, sodium alginate (10 g) was dispersed in 50 mL absolute ethanol to prepare a suspension (liquid A), and different amounts of sodium periodate was dissolved in 50 mL water to prepare a solution (liquid B). Adjusting the feed ratio of liquid B to liquid A so that the molar ratio of sodium periodate to sodium alginate monomer unit was 15%, 20%, 30%, 40%, respectively. Under stirring condition, liquid B was added into liquid A. After 30 min, ethylene glycol which had an equivalent molar ratio of sodium periodate was added to the system for terminating the reaction. The entire reaction process was protected from light. The reaction mixture was poured into a large amount of absolute ethanol (Vreaction mixture/Vabsolute ethanol = 1/5) under rapidly stirring to obtain sediment. Then the sediment was filtered and vacuum dried at 40 °C to obtain solid powders. The solid powders were transferred into a regenerated cellulose dialysis tubing (MWCO, 300 Da) and dialyzed against deionized water for 24 h to remove unreacted sodium periodate and ethylene glycol. Then the mixture was freeze dried to obtain MASA powders. The product was confirmed using MALDI-TOF-MS (Shimadzu, Japan).3.4. Synthesis of mPEI
To synthesize mPEI, MASA was connected with PEI using aldehyde–amine condensation. Briefly, MASA was dissolved in water to prepare solution I, and polyethyleneimine solution was dissolved in water to prepare solution II (pH 6–11, adjusting pH value with HCl). Solution I was dropped into solution II slowly under stirring condition in 0.5–2 h, and then the mixture was stirred for another 24 h. The mixture was transferred into a regenerated cellulose dialysis tubing and dialyzed against deionized water for 48 h to remove the solid insoluble matter. Then the mixture was freeze dried, and graft copolymer mPEI was obtained. The product was confirmed using MALDI-TOF-MS (Shimadzu, Japan).3.5. Preparation and characterization of dsRNA mPEI nanoparticles
dsRNA mPEI nanoparticles were prepared by a static incubation. Briefly, diethyl pyrocarbonate (DEPC) water was added into dsRNA freeze-dried powder to get dsRNA working solution. dsRNA working solution and mPEI were diluted with appropriate amount of Opti-MEM reduced-serum medium separately, then they were incubated for 5 min at room temperature, respectively. Afterwards, the diluted dsRNA solution was added to the diluted mPEI solution, vortexed, and then incubated for 20 min at room temperature to obtain the dsRNA mPEI nanoparticles.Morphology of the nanoparticles was observed by transmission electron microscopy (TEM; JEOL, Japan). Briefly, dsRNA mPEI nanoparticles samples at a concentration of 1 μM dsRNA were dropped on a copper grid (25 mm2). Then the sample was stained with 10 μL 2% uranyl acetate solution (Sigma-Aldrich) for 2 min and dried under vacuum overnight. Afterwards, a JOEL 100CX transmission electron microscope (100 kV) was used to observe the image of the sample.
Size and zeta potential of MASA, mPEI and dsRNA mPEI nanoparticles were determined using dynamic light scattering (DLS) instrument (Malvern Zetasizer Nano ZS, Malvern, UK) at 25 °C and at a scattering angle 90°. The final concentration of dsRNA was 100 nM. In order to verify the dilution stability of dsRNA mPEI nanoparticles, the nanoparticles were diluted with Opti-MEM reduced-serum mediums to different dilution ratios, and then the changes in particle sizes were measure by dynamic light scattering instrument.
3.6. Cytotoxicity of dsRNA mPEI nanoparticles at different N/P ratios
Sulforhodamine B (SRB) staining assay was used to evaluate the safety of dsRNA mPEI nanoparticles at different N/P ratios. Briefly, MDA-MB-231 cells were seeded in 96-well culture plates at a density of 8000 cells per well. After 24 h incubation, the cells were treated with 200 μL Opti-MEM reduced-serum medium containing dsRNA mPEI nanoparticles at different N/P ratios. After 4 h incubation, the culture medium was replaced by DMEM containing 10% FBS, and the cells were incubated for another 24 h. Afterwards, the culture medium was removed carefully, 100 μL 10% trichloroacetic acid (TCA) was added to each well of culture plates, respectively. The culture plates were incubated at 4 °C for 1 h, washed for five times with deionized water, and then dried in air. A volume of 200 μL SRB solution was added into each well and allowed to stand for 30 min in the dark. After removing the SRB solution, the culture plates were washed five times using 1% acetic acid. After air-dying, 100 μL 10 mM Tris base solution was added into each well to solubilize the protein-bound dye. The mixture was shaken on a shaker for 30 min. The absorbance of each well was measured using Infinite F50 Microplate Reader (Tecan Group Ltd., Beijing, China) at a wavelength of 540 nm. Survival rate was calculated with the formula: survival rate = (A540 nm of treated cells/A540 nm of blank medium control cells) × 100%, where A540 nm refers to absorbance at 540 nm.3.7. Gel retardation assay
Gel retardation assay was used to investigate the protective effect of the nanoparticles on dsRNA. Briefly, dsRNA mPEI nanoparticles at a concentration of 1 μM dsRNA concentration were mixed with FBS (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and then incubated at 37 °C for different time-points (0, 1, 2, 3, 6, 9, 12, 24, 36, and 48 h). After sampling, the samples were stored at −20 °C before the assay. The loading buffer of RNA (6×) was added to each sample, and then the sample was assayed by electrophoresis on 1% agarose gel containing 0.5 mg mL−1 Gelred (a special luminescent dye for staining RNA; Beyotime, Beijing, China). Electrophoresis was performed at 80 mV for 3 min, and subsequently 100 mV for 15 min. These resulting gels were photographed under the Imager Quant RTECL gel imaging system (Tanon, Shanghai, China). Free dsRNA was used as the control (dsRNA = 1 μM).
1, v/v) and then incubated at 37 °C for different time-points (0, 1, 2, 3, 6, 9, 12, 24, 36, and 48 h). After sampling, the samples were stored at −20 °C before the assay. The loading buffer of RNA (6×) was added to each sample, and then the sample was assayed by electrophoresis on 1% agarose gel containing 0.5 mg mL−1 Gelred (a special luminescent dye for staining RNA; Beyotime, Beijing, China). Electrophoresis was performed at 80 mV for 3 min, and subsequently 100 mV for 15 min. These resulting gels were photographed under the Imager Quant RTECL gel imaging system (Tanon, Shanghai, China). Free dsRNA was used as the control (dsRNA = 1 μM).
3.8. Invasiveness characterization of breast cancer cells
Wound-healing assay was used to compare invasiveness of the three cancer cells. Briefly, three breast cancer cells (MDA-MB-231 cells, MCF-7 cells and 4T1 cells) were counted, and seeded into 6-well culture plates at 2 × 105/well. When cell monolayer was formed, and a pipettor tip was used to create a scratch on each monolayer. Culture medium (1 mL) was used to wash each monolayer for removing the debris after the scratch, and then 2 mL culture medium was added into each well for further incubation. The monolayer was photographed at day 1 and day 2 since scratch, respectively.3.9. Expressions of target microRNA-34a in breast cancer cells
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to measure the expressions of microRNA in the three breast cancer cells. Briefly, MDA-MB-231, MCF-7 and 4T1 cells (2 × 105 per well) were seeded in 6-well culture plates. After 48 h incubation, total RNA was extracted using the Triol reagent (Macgene, Beijing, China), and then reversely transcribed into cDNA with a Bulge-Loop™ miRNA RT Kit (RiboBio Co. Ltd., Guangzhou, China) using thermal cycle PCR instrument (Techne, Princeton,NJ, USA).Afterwards, qPCR was performed using Bulge-Loop™ miRNA qPCR Kit (RiboBio Co. Ltd., Guangzhou, China) in triplicates, and data collection were performed on a real-time PCR amplifier (CFX connect, Bio-Rad, Singapore). U6 was selected as the housekeeping gene. The data were normalized according to the target microRNA-34a expression of MDA-MB-231 cells.
3.10. Cellular uptake and lysosomal escape of dsRNA mPEI nanoparticles
Flow cytometry was used to observe the cellular uptake of dsRNA mPEI nanoparticles by breast cancer cells. Briefly, MDA-MB-231 cells (2 × 105 per well) were seeded in 6-well culture plates. After 24 h incubation, the cells were treated with FAM-labeled dsRNA mPEI nanoparticles at a final concentration of 100 nM dsRNA and incubated at 37 °C for an additional 4 h. After incubation, the cells were harvested and washed three times with pre-cooled phosphate buffered saline (PBS pH 7.4; NaCl 137 mmol L−1, KCl 2.7 mmol L−1, Na2HPO4 10 mmol L−1 KH2PO4 2 mmol L−1), and intracellular fluorescence intensities were immediately detected by a FACS Calibur Flow Cytometer (Becton Dickinson, San Jose, CA, USA) with the excitation wavelength of 488 nm and the emission wavelength of 518 nm.Confocal microscopy was also used to observe organelle co-localization and lysosomal escape. Briefly, MDA-MB-231 cells were seeded into glass-bottom dishes at a density of 2 × 105 cells per dish. The cells were incubated for 24 h. Then the culture medium was replaced by 2 mL fresh Opti-MEM reduced-serum medium containing FAM-labeled dsRNA mPEI nanoparticles. The final concentration of FAM-labeled dsRNA was 100 nM. After transfected for 1 h, 3 h and 4 h respectively, the cells were washed three times with PBS (pH 7.4). Then, lysosome and nucleus were individually stained by lysosome tracker and Hoechst 33342 according to the manufacturer's instructions. The cells were visualized under a Leica TCS SP8 confocal fluorescence microscope (Leica Microsystems, Heidelberg, Germany).
3.11. In vitro gene knocking-down assays
qRT-PCR was used to measure the gene knocking-down effects of Fra-1, MMP-1 and MMP-9 at mRNA levels in the invasive MDA-MB-231 cells. Briefly, MDA-MB-231 cells (2 × 105 per well) were seeded in 6-well culture plates. After 24 h incubation, the cells were treated with dsRNA mPEI nanoparticles at a final concentration of 100 nM dsRNA for each, and incubated at 37 °C for an additional 4 h. After refreshing the cell culture medium, the cells were incubated for another 12 h for proliferation. Total RNA was extracted using the Triol reagent, and then reversely transcribed into cDNA with a Takara Reverse Transcription System (Takara Biomedical Technology Co., Ltd., Beijing, China) using thermal cycle PCR instrument (Techne, Princeton, NJ, USA).Afterwards, qPCR was performed using Takara qPCR Master Mix (Takara Biomedical Technology Co., Ltd., Beijing, China) in triplicates, and data collection were performed on a real-time PCR amplifier (CFX connect, Bio-Rad, Singapore). Glyceraldehyde-phosphate dehydrogenase (GAPDH) was selected as the housekeeping gene. The relative expression level for each target gene was calculated by 2−(Ct−Cc) (Ct and Cc were the mean threshold cycle differences after normalizing to GAPDH).
Western blotting was used to measure the gene knocking-down effects of Fra-1, MMP-1 and MMP-9 at protein levels in the invasive MDA-MB-231 cells.
Briefly, MDA-MB-231 cells (2 × 105 per well) were seeded in 6-well culture plates. After 24 h incubation, the cells were treated with dsRNA mPEI nanoparticles at a final concentration of 100 nM dsRNA for each, and incubated at 37 °C for an additional 4 h. After refreshing the cell culture medium, the cells were incubated for another 24 h for proliferation. The cells were washed with pre-cooled PBS (pH 7.4) for three times, digested with 0.25% trypsin and centrifuged in a 15 mL tube. After washing with PBS (pH 7.4), 500 μL RIPA (radio immunoprecipitation assay) lysis buffer supplemented with 2% protease inhibitor and 2% phosphatase inhibitor was added to resuspend the cells. Then, the cells were incubated on ice for 30 min and centrifuged in a 1.5 mL tube at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 30 min. The supernatant was collected into another centrifuge tube, and the concentration of protein was determined with a BCA protein assay kit (Beyotime, Beijing, China). Subsequently, loading buffer (5×) was added to protein samples and boiled them for 15 min to denature protein. Then 20 mg of total protein was loaded on a 10% sodium polyacrylamide gel and electrophoresed at 80 mV for 30 min and at 100 mV for 90 min. The proteins were transferred onto PVDF membranes at 100 mV for 80 min, and the membranes were blocked with 5% bovine serum albumin on a shaker for 2 h. The membranes were incubated with the Fra-1 (1
000g at 4 °C for 30 min. The supernatant was collected into another centrifuge tube, and the concentration of protein was determined with a BCA protein assay kit (Beyotime, Beijing, China). Subsequently, loading buffer (5×) was added to protein samples and boiled them for 15 min to denature protein. Then 20 mg of total protein was loaded on a 10% sodium polyacrylamide gel and electrophoresed at 80 mV for 30 min and at 100 mV for 90 min. The proteins were transferred onto PVDF membranes at 100 mV for 80 min, and the membranes were blocked with 5% bovine serum albumin on a shaker for 2 h. The membranes were incubated with the Fra-1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), MMP-1 (1
1000), MMP-1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), MMP-9 (1
1000), MMP-9 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) and β-actin (1
1000) and β-actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) (Biosynthesis Biotechnology Inc., Beijing, China) monoclonal antibody on a shaker overnight at 4 °C, respectively. The membranes were washed three times with TBST (tris-buffered saline and Tween 20), and then incubated with HRP-conjugated goat anti-rabbit antibodies (1
1000) (Biosynthesis Biotechnology Inc., Beijing, China) monoclonal antibody on a shaker overnight at 4 °C, respectively. The membranes were washed three times with TBST (tris-buffered saline and Tween 20), and then incubated with HRP-conjugated goat anti-rabbit antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) (Biosynthesis Biotechnology Inc., Beijing, China) at room temperature for 1 h. Finally, the membranes were washed three times with TBST and exposed using SageCapture™ MiniChemi610 System (Sagecreation, Beijing, China).
2000) (Biosynthesis Biotechnology Inc., Beijing, China) at room temperature for 1 h. Finally, the membranes were washed three times with TBST and exposed using SageCapture™ MiniChemi610 System (Sagecreation, Beijing, China).
3.12. Inhibition effect of dsRNA mPEI nanoparticles on invasiveness
Wound-healing assay was used to evaluate the inhibitory effect of dsRNA mPEI nanoparticles on the migration ability of cancer cells. Briefly, MDA-MB-231 cells (2 × 105 per well) were seeded in 6-well plates. For experimental group, the cells were transfected with dsRNA mPEI nanoparticles when at 50–60% confluence, and then incubated at 37 °C until cells reached 100% confluence to form a monolayer. For control group, the cells were incubated at 37 °C until cells reached 100% confluence to form a monolayer. Afterwards, pipettor tips were used to create a scratch on each cell monolayer. Culture medium (1 mL) was used to wash each monolayer for removing the debris after the scratch, and then 2 mL culture medium was added into each well for further incubation. The monolayer was photographed at day 1 and day 2 since scratch, respectively.Transwell invasion assay was used to observe the inhibitory effect of dsRNA mPEI nanoparticles on the invasiveness of cancer cells. Briefly, MDA-MB-231 cells were resuspended by DMEM containing 0.5% FBS to adjust cell density to 5 × 105/mL. The above cell suspension (100 μL) was added into the upper chamber of 8 μm transwells, and 700 μL DMEM containing 35% FBS was added into the lower chamber. For experimental group, dsRNA mPEI nanoparticles were added into the upper chamber. For blank control group, an equal amount medium was added into the upper chamber. After 10 h incubation, the filter side of the upper chamber was cleaned with a cotton swab and the cells were fixed with 4% paraformaldehyde at room temperature for 10 min. Then, the filter was stained with 0.1% crystal violet for 5 min and rinsed in PBS (pH 7.4). After washing the floating violet color, the samples were photographed under the microscope.
3.13. Anticancer efficacy and inhibitory effect on the Fra-1 relevant genes in mice
To study the anticancer efficacy of dsRNA mPEI nanoparticles in vivo, the cancer-bearing animals were xenografted with MDA-MB-231 cells in nude mice. Briefly, female BALB/c nude mice were kept in an SPF-grade sterile clean environment. The cancer-bearing mice were established by the subcutaneous injection of 2 × 106 cells in the right flank of mice. The cancer-bearing animals were randomly divided into four groups. The animals were administrated as the designed time-points (at day 7, 11, 15, 19, 23, and 27 since cancer cell inoculation) by intratumoral injection using the following: (1) 0.9% physiological saline; (2) dsRNA mPEI nanoparticles (1 mg kg−1); (3) ssRNA mPEI nanoparticles (inhibitor of dsRNA; 2 mg kg−1); (4) paclitaxel (5 mg kg−1). The tumor volume was measured using a Vernier caliper across its longest (L) and shortest (S) diameters, and calculated according to the formula, V = 0.5 × L (mm) × S2 (mm2). Administration was started when the tumor volume reached approximately 200 mm3.After the start of drug administration, the body weights of mice were measured every two days. After all administrations, the mice were sacrificed by cervical dislocation. The tumors were isolated, the volumes and weights of tumors were recorded.
To evaluate the gene knocking-down effects in the cancer-bearing mice, qRT-PCR assay and western blotting assay were performed on the isolated tumor tissues to determine mRNAs and proteins of the Fra-1, MMP-1 and MMP-9.
3.14. Statistical analyses
Data were expressed as the mean ± standard deviation (SD). Analysis of variance (ANOVA) was used to determine the significance among groups, after which, post hoc tests with the Bonferroni correction were used for multiple comparisons between individual groups. A value of P < 0.05 was considered as statistically significant.4. Conclusion
In sum, a novel type of dsRNA mPEI nanoparticles is successfully synthesized to treat aggressive TNBC cells. They are able to effectively transfect the cells, and demonstrate a strong capability in knocking-down up-stream Fra-1 gene, and down-stream MMP-1 and MMP-9 genes in TNBC cells and in TNBC cancer-bearing mice, thereby inhibiting the invasion and migration of the cells. After intratumoral injection, dsRNA mPEI nanoparticles demonstrate a robust anticancer efficacy in TNBC cancer-bearing mice, and the anticancer efficacy is superior to that of paclitaxel. In conclusion, dsRNA mPEI nanoparticles are able to effectively treat aggressive TNBC, and the mechanism studies reveal that they take effect by knocking-down of Fra-1 relevant genes, hence interfering transcription and translation of the genes, which are necessary for growth and metastasis of TNBC. Therefore, the present study offers a new and promising formulation and strategy for effective treatment of TNBC.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This study was funded by Beijing Natural Science Foundation (Key Grant No. 7181004) and National Natural Science Foundation of China (Grant 81673367, and 81874303).References
- R. Dent, M. Trudeau, K. I. Pritchard, W. M. Hanna, H. K. Kahn, C. A. Sawka, L. A. Lickley, E. Rawlinson, P. Sun and S. A. Narod, Clin. Cancer Res., 2007, 13, 4429–4434 CrossRef PubMed.
- S. E. Logue, E. P. McGrath, P. Cleary, S. Greene, K. Mnich, A. Almanza, E. Chevet, R. M. Dwyer, A. Oommen, P. Legembre, F. Godey, E. C. Madden, B. Leuzzi, J. Obacz, Q. Zeng, J. B. Patterson, R. Jager, A. M. Gorman and A. Samali, Nat. Commun., 2018, 9, 3267 CrossRef PubMed.
- M. C. Cheang, D. Voduc, C. Bajdik, S. Leung, S. McKinney, S. K. Chia, C. M. Perou and T. O. Nielsen, Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype, Clin. Cancer Res., 2008, 14, 1368–1376 CrossRef CAS PubMed.
- A. Albergaria, S. Ricardo, F. Milanezi, V. Carneiro, I. Amendoeira, D. Vieira, J. Cameselle-Teijeiro and F. Schmitt, Nottingham Prognostic Index in triple-negative breast cancer: a reliable prognostic tool?, BMC Cancer, 2011, 11, 299 CrossRef PubMed.
- R. Rupaimoole and F. J. Slack, MicroRNA therapeutics: towards a new era for the management of cancer and other diseases, Nat. Rev. Drug Discovery, 2017, 16, 203–222 CrossRef CAS PubMed.
- N. R. Christoffersen, R. Shalgi, L. B. Frankel, E. Leucci, M. Lees, M. Klausen, Y. Pilpel, F. C. Nielsen, M. Oren and A. H. Lund, p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC, Cell Death Differ., 2010, 17, 236–245 CrossRef CAS PubMed.
- V. K. Gangaraju and H. Lin, MicroRNAs: key regulators of stem cells, Nat. Rev. Mol. Cell Biol., 2009, 10, 116–125 CrossRef CAS PubMed.
- P. Bonetti, M. Climent, F. Panebianco, C. Tordonato, A. Santoro, M. J. Marzi, P. G. Pelicci, A. Ventura and F. Nicassio, Dual role for miR-34a in the control of early progenitor proliferation and commitment in the mammary gland and in breast cancer, Oncogene, 2019, 38, 360–374 CrossRef CAS PubMed.
- D. G. Zisoulis, Z. S. Kai, R. K. Chang and A. E. Pasquinelli, Autoregulation of microRNA biogenesis by let-7 and argonaute, Nature, 2012, 486, 541–544 CrossRef CAS PubMed.
- T. A. Farazi, J. I. Hoell, P. Morozov and T. Tuschl, microRNAs in human cancer, Adv. Exp. Med. Biol., 2013, 774, 1–20 CrossRef CAS PubMed.
- X. J. Li, Z. J. Ren and J. H. Tang, MicroRNA-34a: a potential therapeutic target in human cancer, Cell Death Dis., 2014, 5, e1327 CrossRef CAS PubMed.
- H. Hermeking, The miR-34 family in cancer and apoptosis, Cell Death Differ., 2010, 17, 193–199 CrossRef CAS PubMed.
- E. Slabáková, C. Zoran, J. Remšík and K. Souček, Alternative mechanisms of miR-34a regulation in cancer, Cell Death Dis., 2017, 8, e3100 CrossRef PubMed.
- L. Yu, J. Xiong, L. Guo, L. Miao, S. Liu and F. Guo, The effects of lanthanum chloride on proliferation and apoptosis of cervical cancer cells: involvement of let-7a and miR-34a microRNAs, BioMetals, 2015, 28, 879–890 CrossRef CAS PubMed.
- Y. S. Lee and A. Dutta, MicroRNAs in cancer, Annu. Rev. Pathol.: Mech. Dis., 2009, 4, 199–227 CrossRef CAS PubMed.
- M. Rokavec, H. H. Li, L. C. Jiang and H. Hermeking, The p53/miR-34 axis in development and disease, J. Mol. Cell Biol., 2014, 6, 214–230 CrossRef CAS PubMed.
- E. Henckels and R. Prywes, Fra-1 regulation of Matrix Metallopeptidase-1 (MMP-1) in metastatic variants of MDA-MB-231 breast cancer cells, F1000Research, 2013, 2, 229 Search PubMed.
- J. L. Pakay, J. Diesch, O. Gilan, Y. Y. Yip, E. Sayan, W. Kolch, J. M. Mariadason, R. D. Hannan, E. Tulchinsky and A. S. Dhillon, A 19S proteasomal subunit cooperates with an ERK MAPK-regulated degron to regulate accumulation of Fra-1 in tumour cells, Oncogene, 2012, 31, 1817–1824 CrossRef CAS PubMed.
- A. E. Sayan, R. Stanford, R. Vickery, E. Grigorenko, J. Diesch, K. Kulbicki, R. Edwards, R. Pal, P. Greaves, I. Jariel-Encontre, M. Piechaczyk, M. Kriajevska, J. K. Mellon, A. S. Dhillon and E. Tulchinsky, Fra-1 controls motility of bladder cancer cells via transcriptional upregulation of the receptor tyrosine kinase AXL, Oncogene, 2012, 31, 1493–1503 CrossRef CAS PubMed.
- A. S. Dhillon and E. Tulchinsky, FRA-1 as a driver of tumour heterogeneity: a nexus between oncogenes and embryonic signalling pathways in cancer, Oncogene, 2015, 34, 4421–4428 CrossRef CAS PubMed.
- A. Philips, C. Teyssier, F. Galtier, C. Rivier-Covas, J. M. Rey, H. Rochefort and D. Chalbos, FRA-1 expression level modulates regulation of activator protein-1 activity by estradiol in breast cancer cells, Mol. Endocrinol., 1998, 12, 973–985 CrossRef CAS PubMed.
- H. Yin, K. J. Kauffman and D. G. Anderson, Delivery technologies for genome editing, Nat. Rev. Drug Discovery, 2017, 16, 387–399 CrossRef CAS PubMed.
- K. Belguise, N. Kersual, F. Galtier and D. Chalbos, FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells, Oncogene, 2005, 24, 1434–1444 CrossRef CAS PubMed.
- O. Fundyler, M. Khanna and B. R. Smoller, Metalloproteinase-2 expression correlates with aggressiveness of cutaneous squamous cell carcinomas, Mod. Pathol., 2004, 17, 496–502 CrossRef CAS PubMed.
- D. Q. Matus, X. Y. Li, S. Durbin, D. Agarwal, Q. Chi, S. J. Weiss and D. R. Sherwood, In vivo identification of regulators of cell invasion across basement membranes, Sci. Signaling, 2010, 3, ra35 Search PubMed.
- O. Boussif, F. Lezoualc'h, M. A. Zanta, M. D. Mergny, D. Scherman, B. Demeneix and J. P. Behr, A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 7297–7301 CrossRef CAS.
- A. C. R. Grayson, A. M. Doody and D. Putnam, Biophysical and structural characterization of polyethylenimine-mediated siRNA deliveryin vitro, Pharm. Res., 2006, 23, 1868–1876 CrossRef PubMed.
- O. Boussif, M. A. Zanta and J. P. Behr, Optimized galenics improve in vitro gene transfer with cationic molecules up to 1000-fold, Gene Ther., 1996, 3, 1074–1080 CAS.
- D. W. Pack, A. S. Hoffman, S. Pun and P. S. Stayton, Design and development of polymers for gene delivery, Nat. Rev. Drug Discovery, 2005, 4, 581–593 CrossRef CAS PubMed.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Non-viral vectors for gene-based therapy, Nat. Rev. Genet., 2014, 15, 541–555 CrossRef CAS PubMed.
- C. V. Pecot, G. A. Calin, R. L. Coleman, G. Lopez-Berestein and A. K. Sood, RNA interference in the clinic: Challenges and future directions, Nat. Rev. Cancer, 2011, 11, 59–67 CrossRef CAS PubMed.
- X. An, C. Sarmiento, T. Tan and H. Zhu, Regulation of multidrug resistance by microRNAs in anti-cancer therapy, Acta Pharm. Sin. B, 2017, 7, 38–51 CrossRef PubMed.
- L. Kang, J. Mao, Y. Tao, B. Song, W. Ma, Y. Lu, L. Zhao, J. Li, B. Yang and L. Li, MicroRNA-34a suppresses the breast cancer stem cell-like characteristics by downregulating Notch1 pathway, Cancer Sci., 2015, 106, 700–708 CrossRef CAS PubMed.
- L. K. Medina-Kauwe, J. Xie and S. Hamm-Alvarez, Intracellular trafficking of nonviral vectors, Gene Ther., 2005, 12, 1734–1751 CrossRef CAS PubMed.
- D. A. Zajchowski, M. F. Bartholdi, Y. Gong, L. Webster, H. L. Liu, A. Munishkin, C. Beauheim, S. Harvey, S. P. Ethier and P. H. Johnson, Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells, Cancer Res., 2001, 61, 5168–5178 CAS.
- K. Gao and L. Huang, Achieving efficient RNAi therapy: progress and challenges, Acta Pharm. Sin. B, 2013, 3, 213–225 CrossRef.
- D. P. Bartel, MicroRNAs: genomics, biogenesis, mechanism, and function, Cell, 2004, 116, 281–297 CrossRef CAS PubMed.
- M. G. Annis, V. Ouellet, J. P. Rennhack, S. L'Esperance, C. Rancourt, A. M. Mes-Masson, E. R. Andrechek and M. Peter, Integrin-uPAR signaling leads to FRA-1 phosphorylation and enhanced breast cancer invasion, Breast Cancer Res., 2018, 20, 9 CrossRef PubMed.
- S. Yang, Y. Li, J. Gao, T. Zhang, S. Li, A. Luo, H. Chen, F. Ding, X. Wang and Z. Liu, MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1, Oncogene, 2013, 32, 4294–4303 CrossRef CAS PubMed.
- Q. M. Wang, L. Lv, Y. Tang, L. Zhang and L. F. Wang, MMP-1 is overexpressed in triple-negative breast cancer tissues and the knockdown of MMP-1 expression inhibits tumor cell malignant behaviors in vitro, Oncol. Lett., 2019 Feb, 17, 1732–1740 Search PubMed.
- M. Liu, Y. Hu, M. F. Zhang, K. J. Luo, X. Y. Xie, J. Wen, J. H. Fu and H. Yang, MMP1 promotes tumor growth and metastasis in esophageal squamous cell carcinoma, Cancer Lett., 2016, 377, 97–104 CrossRef CAS PubMed.
- C. C. Liang, A. Y. Park and J. L. Guan, In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro, Nat. Protoc., 2007, 2, 329–333 CrossRef CAS PubMed.
- P. Tibarewal, G. Zilidis, L. Spinelli, N. Schurch, H. Maccario, A. Gray, N. M. Perera, L. Davidson, G. J. Barton and N. R. Leslie, PTEN protein phosphatase activity correlates with control of gene expression and invasion, a tumor-suppressing phenotype, but not with AKT activity, Sci. Signaling, 2012, 5, ra18 CrossRef PubMed.
- P. Pandya, J. L. Orgaz and V. Sanz-Moreno, Modes of invasion during tumour dissemination, Mol. Oncol., 2017, 11, 5–27 CrossRef PubMed.
- G. R. Jin, R. Y. He, Q. Liu, M. Lin, Y. Q. Dong, K. Li, B. Z. Tang, B. Liu and F. Xu, Near-infrared light-regulated cancer theranostic nanoplatform based on aggregation-induced emission luminogen encapsulated upconversion nanoparticles, Theranostics, 2019, 9, 246–264 CrossRef PubMed.
- G. R. Jin, R. Y. He, Q. Liu, Y. Q. Dong, M. Lin, W. F. Li and F. Xu, Theranostics of triple-negative breast cancer based on conjugated polymer nanoparticles, ACS Appl. Mater. Interfaces, 2018, 10, 10634–10646 CrossRef CAS PubMed.
- G. R. Jin, G. X. Feng, W. Qin, B. Z. Tang, B. Liu and K. Li, Multifunctional organic nanoparticles with aggregation-induced emission (AIE) characteristics for targeted photodynamic therapy and RNA interference therapy, Chem. Commun., 2016, 52, 2752–2755 RSC.
- J. Zhao, G. R. Jin, G. J. Weng, J. J. Li, J. Zhu and J. W. Zhao, Recent advances in activatable fluorescence imaging probes for tumor imaging, Drug Discovery Today, 2017 Sep, 22, 1367–1374 Search PubMed.
- J. M. Williford, J. Wu, Y. Ren, M. M. Archang, K. W. Leong and H. Q. Mao, Recent Advances in Nanoparticle-Mediated siRNA Delivery, Annu. Rev. Biomed. Eng., 2014 Jul 11, 16, 347–370 Search PubMed.
- H. M. Chen, W. Z. Zhang, G. Z. Zhu, J. Xie and X. Y. Chen, Rethinking cancer nanotheranostics, Nat. Rev. Mater., 2017, 2, 17024 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra01889a |
| This journal is © The Royal Society of Chemistry 2019 |