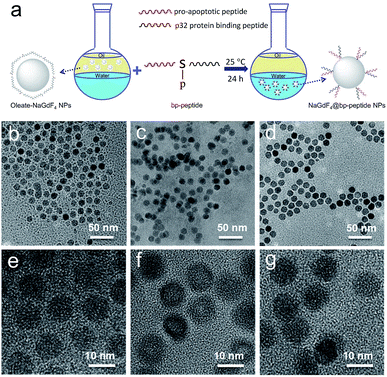
Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Peptide-functionalized NaGdF4 nanoparticles for tumor-targeted magnetic resonance imaging and effective therapy†
Yixin Chena,
Yu Fua,
Xiaodong Lia,
Hongda Chen *b,
Zhenxin Wang
*b,
Zhenxin Wang b and
Huimao Zhang*a
b and
Huimao Zhang*a
aDepartment of Radiology, The First Hospital of Jilin University, Changchun 130021, P. R. China. E-mail: huimaozhanglinda@163.com; Fax: +86-431-88783300
bState Key Laboratory of Electroanalytical Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, 130022, P. R. China. E-mail: chenhongda@ciac.ac.cn; Fax: +86-431-85262756
First published on 31st May 2019
Abstract
Metallic nanoparticles showed potent efficacy for diagnosis and therapy of cancer, but their clinical applications are limited by their poor tumor-targeting ability. Herein, peptide-functionalized 9 nm NaGdF4 nanoparticles (termed as, NaGdF4@bp-peptide NPs) have been synthesized through the Gd–phosphate coordination reaction of the spherical NaGdF4 nanoparticles with phosphopeptides (sequence: KLAKLAKKLAKLAKG(p-S)GAKRGARSTA, p-S means phosphorylated serine) including a p32 protein binding motif incorporating a cell-penetrating function, and a proapoptotic domain. The NaGdF4@bp-peptide NPs are ready to be efficiently internalized by cancer cells; they show a much higher cytotoxicity in MCF-7 breast cancer cells than the casein phosphopeptide (CPP) modified NaGdF4 nanoparticles (termed as, NaGdF4@CPP NPs). Using mouse-bearing MCF-7 breast cancer as a model system, the in vivo experimental results demonstrate that NaGdF4@bp-peptide NPs have integration of T1-weighted magnetic resonance imaging (MRI) contrast and tumor-targeting functionalities, and are able to suppress tumor growth without causing systemic toxicity.
Introduction
Nanotheranostics (also known as nanomedicines), integrating diagnostic and therapeutic functions into one nanosized material, have many advantages over the conventional molecular medicines, such as prolonged blood circulation time, controlled clearance pathways, and tunable physical properties for multimodal imaging.1–7 To date, numerous nanomaterials, including inorganic nanoparticles, organic nanoparticles, nanohydrogels, and organic–inorganic hybrid nanomaterials have been developed as promising nanotheranostics for precise treatment of cancer (as well as many other diseases). Although a few nanotheranostics have been approved by the U. S. Food and Drug Administration (FDA) for drug delivery (e.g., Doxil and Abraxane) or contrast agents (e.g., Feridex IV) for magnetic resonance imaging (MRI), clinical translation efforts of have been hampered by their low tumor delivery efficiency.8–10 For instance, Chan and co-authors found that only 0.59% injected dose of trastuzumab-coated nanoparticles reached the mouse-bearing SKOV-3 xenograft tumor through intravenous administration.10 The tumor delivery efficiency of nanomaterials is strongly determined by their own physicochemical properties including size, surface modification, and stimuli-responsiveness to tumor microenvironment.11–16 Therefore, there is a strong desire for the development of new modification strategies for generating nanomaterials with improved diagnostic and therapeutic capabilities through increasing their tumor accumulating amounts.Because of its low ionizing radiation, magnetic resonance imaging (MRI) enables noninvasive detecting and assessing disease progression at relatively frequent time intervals.17–19 However, it has been clinically found that the sensitivity of MRI is poor, resulting in difficulty in accurate diagnosis of tumor at early stage. In order to overcome detection limitation of MRI, contrast agents (CAs) are usually used for improvement of the signal-to-noise ratios (SNRs) in the imaging process through altering the relaxation time of local water molecules in lesion regions.20,21 Gadolinium (Gd) chelates (i.e., Gd-DTPA and Gd-DOTA) are currently clinical used CAs, which suffer low tumor accumulation efficiency and short circulation time. Benefiting from their unique physicochemical properties, the application of nanoparticles in bioimaging field has been demonstrated as a very promising strategy for circumventing the inherent defects of traditional small molecular CAs in terms of bioavailability and targeting ability.21–30 Versatile nanoparticle-based CAs have been constructed for MRI, computed tomography (CT), photoacoustic imaging (PAI) and fluorescence imaging (FI) to enhance the spatial resolution and detection sensitivity of these bioimaging modes. For example, nanoparticles containing Gd such as gadolinium oxide (Gd2O3) nanoparticles and sodium tetrafluoro gadolinium (NaGdF4) nanoparticles are appropriate CAs of MRI for visualization of tumor details and/or MRI-guided therapy of tumors.31,32
Peptides are one kind of important biological materials, which normally present structure-dependent functions. Peptides and peptide derivatives have been extensively employed for constructing multifunctional nanotheranostics since they exhibit chemical versatility and enable to specifically recognize other biomacromolecules.33–51 In particular, conjugation of nanoparticles with tumor-homing peptides and/or therapeutic peptides can generate novel nanotheranostics for highly sensitive tumor imaging and effective tumor-targeted therapy. For example, Ruoslahti and coauthors have been developed a theranostic nanosystem, which consists of iron oxide nanoworms conjugated with a composite peptide with proapoptotic domain (i.e., therapy motif) and cell surface p32 protein binding domain (i.e., tumor-homing motif).52,53 The in vivo experimental results indicate that the as-prepared theranostic nanosystem has excellent homing and penetrating activity in mouse-bearing breast tumor models, and enables to effectively retard tumor growth. Very recently, we also synthesized a kind of peptide mixture-modified NaGdF4 nanodot with active tumor targeting ability, which can be used as high efficient T1-weighted MRI CA for tracking small drug induced orthotopic colorectal tumor (c.a., 195 mm3 in volume) in mouse.54
Herein, a tumor-specific multifunctional nanotheranostic composed of NaGdF4 nanoparticle together with phosphopeptides has been developed for T1-weighted MRI guided cancer therapy. Strategically, the p32 protein binding peptide (sequence: AKRGARSTA) and the pro-apoptotic peptide (sequence: KLAKLAKKLAKLAK) were linearly linked together to form a new peptide (sequence: KLAKLAKKLAKLAKG(p-S)GAKRGARSTA, named as bp-peptide) through the phosphorylated tripeptide (–G(p-S)G–: –glycine–phosphorylated serine–glycine–). Benefiting from the strong coordination reaction between Gd(III) ion and phosphate, the hydrophilic bp-peptide conjugated NaGdF4 nanoparticles (NaGdF4@bp-peptide NPs) can be easily achieved through replacing initial hydrophobic oleate ligand by the bp-peptide molecules due to the phosphopeptides can be selectively enriched by the rare-earth materials through the coordination reactions of rare-earth ions with phosphate moieties in the peptides.54,55 Both of in vivo and in vitro experimental results demonstrated statistically significant improvement in tumor-targeted uptake and tumor suppression of NaGdF4@bp-peptide NPs over these of passive tumor-targeting the casein phosphopeptide (CPP) modified NaGdF4 nanoparticles (NaGdF4@CPP NPs).
Experimental section
Materials
Tryptone (casein phosphopeptide, CPP), 1-octadecene (ODE, 90%), oleic acid (OA, 90%) were obtained from Sigma-Aldrich Co. (St. Louis, USA). The Gd2O3 (99.99%) were reacted with excess HCl (6.0 mol L−1) to form GdCl3 solution. After dried completely at 30 °C, the resulting powder was redispersed in H2O to yield GdCl3 stocking solution (1.5 mol L−1). DMEM basic medium and fetal bovine serum (FBS) were purchased from Gibco Co. (New York, USA). Trypsin–EDTA digest and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide were purchased from Beijing Dingguo Biotechnology Ltd. (Beijing, China). The peptides were synthesized by Shanghai Synpeptide Ltd. (Shanghai, China). Other reagents (analytical grade) were purchased from Beijing Chemical Reagent Co. (Beijing, China). Deionized H2O (18.2 MΩ cm) were used in all experiments. All animal procedures were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals of the People's Republic of China and approved by the Animal Ethics Committee of Jilin University. The mice had free access to food and water and were raised on a 12 h light/12 h dark cycle at 20 °C.Synthesis of NaGdF4 nanoparticles
NaGdF4 nanoparticles were synthesized by literature reported procedure with slight modification.54 Briefly, 1 mL GdCl3 stocking solution was dried at 100 °C. Subsequently, 6 mL OA and 22.5 mL ODE were added and well mixed at 160 °C in Ar atmosphere. After cooled to 60 °C, 15 mL CH3OH solution containing 6 mmol NH4F and 3.75 mmol NaOH were added dropwise, and vigorously mixed for 12 h. After evaporating CH3OH completely at 90 °C, the temperature of mixture was slowly (10 °C per minute) increased to 250 °C and maintained for 10 minutes in Ar atmosphere. After cooling to room temperature, the as-prepared NaGdF4 nanoparticles were purified by repeated centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, three times) and redispersed in 10 mL cyclohexane.
000 rpm, three times) and redispersed in 10 mL cyclohexane.
Ligand-exchange of NaGdF4 nanoparticles
12 mL NaGdF4 nanoparticle cyclohexane solution (1 mg mL−1) were vigorously mixed with 32 mL peptide (bp-peptide or CPP) aqueous solution (2 mg mL−1) and stirred at room temperature for 12 h. The aqueous solution was separated, and the peptide modified NaGdF4 nanoparticles (NaGdF4@bp-peptide NPs or NaGdF4@CPP NPs) were purified by centrifugation repeated centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, three times) and redispersed in H2O.
000 rpm, three times) and redispersed in H2O.
Cytotoxicity evaluation
Human breast cancer cells (MCF-7) and normal kidney tissue cells (293) were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences. The MCF-7 cells and 293 cells were co-cultured with various concentrations of NaGdF4@bp-peptide NPs and NaGdF4@CPP NPs in DMEM (containing 10% (v/v) FBS and 100 U mL−1 penicillium–streptomycin) for 24 h, respectively. Then, the cells were washed and the MTT assays were used to detect cell viabilities. The normally cultured cells were used as control groups.In vitro study
The MCF-7 cells were co-cultured with various concentrations of NaGdF4@bp-peptide NPs and NaGdF4@CPP NPs in DMEM (containing 10% (v/v) FBS and 100 U mL−1 penicillium–streptomycin) for 4 h, respectively. Subsequently, the cells were washed with fresh culture medium and PBS (10 mM phosphate buffer (PB) with 137 mM NaCl, pH 7.4), respectively. The cells were digested by 1 mL trypsin (0.25 wt% in PBS) solution and collected by centrifugation (2000 rpm, 5 min), respectively. The NaGdF4@bp-peptide NPs and NaGdF4@CPP NPs stained cells were immobilized in 1 wt% agarose for in vitro MRI. The MRI was recorded with the Siemens 3.0 T MRI scanner (0.5 mm (slice thickness), 15 ms (TE), 358 ms (TR), 0.8 mm (in-plane resolution), and 50 mm × 50 mm (field of view)). After treated by 2 mL aqua regia, the amounts of Gd element in the NaGdF4@bp-peptide NPs and NaGdF4@CPP NPs stained cells were measured by inductively coupled plasma mass spectrometry (ICP-MS, ELAN 9000/DRC, PerkinElmer Co., USA), respectively.Biocompatibility analysis
Kunming mice with average body weight of 35 g were ordered from Liaoning Changsheng Biotechnology Ltd. (Liaoning, China). The mice were randomly divided into three groups: control group, NaGdF4@bp-peptide NPs treated group and NaGdF4@CPP NPs treated group. The mice in treated groups were injected intravenously with 100 μL NaGdF4@bp-peptide NPs or NaGdF4@CPP NPs (10 mg Gd per kg) in PBS through tail vein, respectively, while the mice in control group were only injected intravenously with 100 μL PBS. The bloods were taken for routine blood tests at 24 h post-injection. The body weights of mice were measured every 2 days until 31 days after injection. The mice were sacrificed, and main organs including heart, liver, spleen, lung and kidneys were collected for hematoxylin–eosin (H&E) staining analysis.In vivo study
Balb/c nude mice with average body weight of 20 g were purchased from Beijing Vital River Laboratory Animal Technology Ltd. (Beijing, China). The tumor model was constructed by subcutaneous injection of MCF-7 cells (1 × 106 cells in 100 μL PBS). The tumor size (V) was evaluated by the following formula: V = length × (width)2/2; the inhibition rate of tumor growth (IRT) was calculated as follows: (1 − VNPs/VCtrl) × 100% (VNPs: tumor volume of NaGdF4@bp-peptide NPs treated mouse; VCtrl: tumor volume of PBS treated mouse). The MCF-7 tumor-bearing Balb/c nude mice were anesthetized with 100 μL chloral hydrate (10 v/v%) while the size of tumor reached about 400 mm3 in volume. Subsequently, the mice were treated with 100 μL NaGdF4@bp-peptide NPs or NaGdF4@CPP NPs (10 mg Gd per kg) in PBS through intravenous injection via the tail vein, respectively. MRI of nude mice were performed at pre-injection (0 h), 1 h, 2 h, 4 h, 8 h and 24 h post-injection by Siemens 3.0 T MRI scanner with the following parameters: TR, 358 ms; TE, 15 ms; field of view, 120 mm × 72 mm and slice thickness, 2.0 mm. In addition, three mice were sacrificed at 24 h post-injection, and main organs and tumors were collected for ICP-MS measurement.Treatment efficiency evaluation
9 MCF-7 tumor-bearing nude mice were randomly divided into 3 groups which were treated by 100 μL PBS only (control group), 100 μL PBS containing NaGdF4@bp-peptide NPs (10 mg Gd per kg, NaGdF4@bp-peptide NPs treated group) and 100 μL PBS containing NaGdF4@CPP NPs (10 mg Gd per kg, NaGdF4@CPP NPs treated group) through tail vein, respectively. The tumor sites of mice were measured every 2 days until 31 days after injection.Result and discussion
Synthesis and characterization of nanoparticles
The hydrophobic OA-coated NaGdF4 nanoparticles (9.0 ± 1.0 nm in diameter) were synthesized by literature reported strategy with slight modification.54–56 In this case, the phosphopeptides (sequence: KLAKLAKKLAKLAKG(p-S)GAKRGARSTA, named as bp-peptide) were employed as a bifunctionalized ligand and phase transfer agent for preparing hydrophilic NaGdF4 nanoparticles (NaGdF4@bp-peptide NPs), and through formation of Gd3+–phosphate coordination bond in the ligand exchange reaction under mild experimental conditions because phosphopeptides have the ability to react with multivalent cations and form robust metal–phosphate coordination bonds (as shown in Fig. 1a).54,55,57 Strategically, the p32 protein binding peptide (AKRGARSTA) is linearly linked to the pro-apoptotic peptide (KLAKLAKKLAKLAK) through the phosphorylated tripeptide (glycine–phosphorylated serine–glycine). The p32 protein (HAPB1, gC1qR or C1qbp) is a mitochondrial protein which highly expresses on the cell surfaces of activated endothelial cells and various tumor cells.58,59 Therefore, the p32 protein binding peptide can improve the tumor accumulation ability of nanoparticles. For comparison, the NaGdF4@CPP NPs were also prepared by same strategy. After ligand exchange, there are negligible changes on morphology, size and crystalline nature of NaGdF4 nanoparticles (as shown in Fig. 1b–g). The successful exchange of OA with phosphopeptides (bp-peptide and/or CPP) was demonstrated FTIR and EDS. After conjugation with phosphopeptides, the nitrogen and phosphorus peaks are clearly observed at 0.39 keV and 1.99 keV in the EDS spectra of NaGdF4@bp-peptide NPs and NaGdF4@CPP NPs (as shown in Fig. S1 in ESI†). The antisymmetric bending mode of PO43− (1083 cm−1) and synthetic spectrum band of stretching vibration and bending vibration of PO43− (1657 cm−1) are clearly observed in FTIR spectra of NaGdF4@bp-peptide NPs and NaGdF4@CPP NPs (as shown in Fig. S2†). The results indicate that the phosphopeptides are successfully immobilized on the NaGdF4 nanoparticle surface. The NaGdF4@bp-peptide NPs exhibit slight positive surface charges (0.63 mV) in H2O because the bp-peptide has relatively high isoelectric points (PI). The slightly positive charged surface may enhance the cellular uptake of NaGdF4@bp-peptide NPs. The hydrodynamic diameter (HD) and zeta potential of NaGdF4@bp-peptide NPs are 269.5 nm and −0.31 mV in culture medium, while the HD and zeta potential of NaGdF4@CPP NPs are 110.1 nm and −5.23 mV in culture medium. The phenomenon suggests that the interactions of NaGdF4@bp-peptide NPs with components of culture medium are stronger than those of NaGdF4@CPP NPs with components of culture medium. However, the longitudinal relaxivity (r1) value (5.8 mM−1 s−1) of NaGdF4@bp-peptide NPs is lower than that (7.9 mM−1 s−1) of NaGdF4@CPP NPs (as shown in Fig. 2). The relative low r1 value of NaGdF4@bp-peptide NPs may due to the branch structure and rigidity of bp-peptide which prolongs the exchange rate of the intra-spherical water molecules around the Gd3+.60,61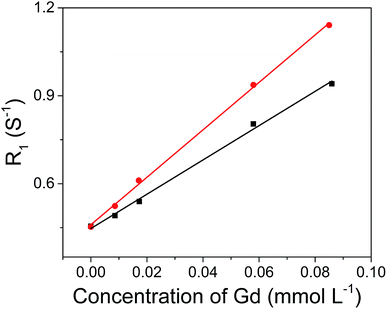 | ||
| Fig. 2 r1 relaxivities of NaGdF4@bp-peptide NPs (black line, r1 = 5.8 mM−1 s−1) and NaGdF4@CPP NPs (red line, r1 = 7.9 mM−1 s−1) as a function of the molar concentration of Gd3+ in the solution. | ||
Interactions of NaGdF4@bp-peptide NPs with cells
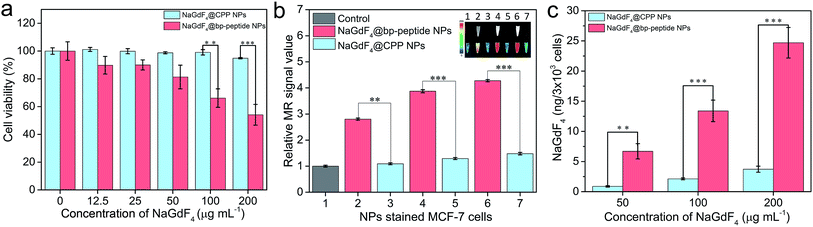
In order to test the cytotoxicity of as-prepared NaGdF4@bp-peptide NPs, the breast cancer cells (MCF-7) and human normal kidney tissue cells (293) were co-cultured the as-prepared NaGdF4@bp-peptide NPs. After incubated with up to 200 μg mL−1 NaGdF4@bp-peptide NPs for 24 h, the viability of MCF-7 cells is less than 60%, while viability of 293 cells is higher than 80% (as shown in Fig. 3a and S3†). The result suggests that NaGdF4@bp-peptide NPs have relatively low cytotoxicity to normal cells and could be used as antitumor agents. In addition, the NaGdF4@CPP NPs have low cytotoxicities to both of 293 cells and MCF-7 cells (as shown in Fig. 3a and S3†). The result indicates that the high cytotoxicity of NaGdF4@bp-peptide NPs towards MCF-7 cells originate from their ligands. The T1-weighted MR signal intensity of nanoparticle stained MCF-7 cell pellet is increased by increasing the concentration of nanoparticles in culture medium (as shown in Fig. 3b). Under same experimental condition, the MR signal intensity of NaGdF4@bp-peptide NPs stained MCF-7 cells is much stronger (≥2.9 times) that of NaGdF4@CPP NPs stained MCF-7 cells. In addition, the cellular internalization amount of NaGdF4@bp-peptide NPs is higher (≥6.7 times) than that of NaGdF4@CPP NPs (as shown in Fig. 3c). These results demonstrate that the binding affinity of NaGdF4@bp-peptide NPs with MCF-7 cells is much higher than that of NaGdF4@CPP NPs with MCF-7 cells.In vivo toxicity investigation
The healthy Kunming mice were intravenously injected a single dose (10 mg kg−1 of Gd) of NaGdF4@bp-peptide NPs or NaGdF4@CPP NPs, respectively. The blood routine analysis was used to test acute toxicities of nanoparticles. At 1 day post-injection, blood platelet count of NaGdF4@bp-peptide NPs treated mice is higher (2.44 times) than those of NaGdF4@CPP NPs treated mice and untreated mice, while other blood routine indicators of NaGdF4@bp-peptide NPs treated mice are comparable to those of NaGdF4@CPP NPs treated mice and untreated mice (as shown in Table S1†). The blood platelet count of NaGdF4@bp-peptide NPs treated mice is decreased with increasing the post-injection time. There is little difference in blood components among treated groups and control group at 7 day of post-injection. The long-term in vivo toxicities of nanoparticles were evaluated by monitoring the body weight changes of mice, histology analysis of major organs, and blood biochemical assays at 31 day of post-injection. As shown in Fig. S4,† the body weights of mice in all tested groups were increased steadily as the time prolonged. Comparing with the untreated mice, the main organs (e.g., heart, liver, spleen, lung, kidneys) of NaGdF4@bp-peptide NPs or NaGdF4@CPP NPs treated mice exhibit little abnormalities or lesions (as shown in Fig. 4). For blood biochemical assays, there is negligible difference among untreated mice and NaGdF4@bp-peptide NPs (10 mg Gd per kg body weight) treated mice at 31 day of post-injection (as shown in Table S1†). The results further confirm the good biocompatibility of NaGdF4@bp-peptide NPs. The results of in vivo acute and chronic toxicity analysis suggest that the NaGdF4@bp-peptide NPs have reasonable biocompatibility.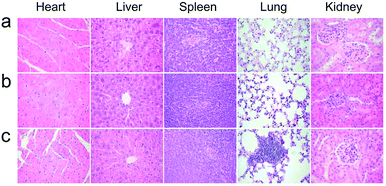 | ||
| Fig. 4 Histology analysis (H&E staining) of mice at 31 day post-injection. (a) Control, (b) NaGdF4@CPP NPs and (c) NaGdF4@bp-peptide NPs (10 mg Gd per kg body weight), respectively. | ||
In vivo tumor-targeting capacity of NaGdF4@bp-peptide NPs
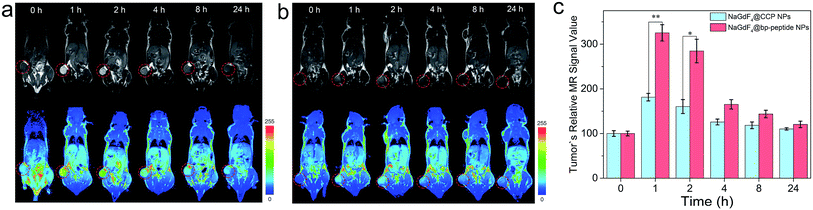
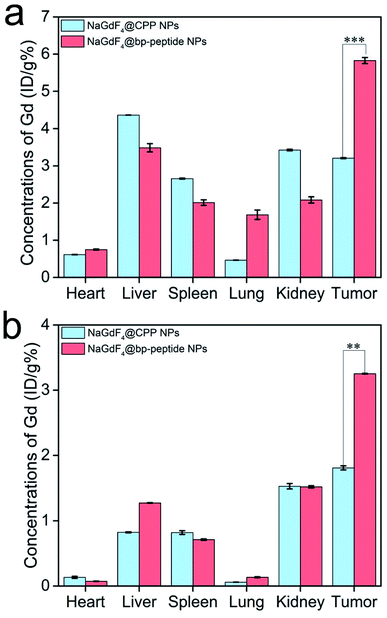
The MCF-7 tumor-bearing Balb/c nude mice were used to investigate the tumor-targeting capacities of NaGdF4@bp-peptide NPs and NaGdF4@CPP NPs. The nanoparticles were intravenously injected into mice through the tail vein, respectively. The T1-weighted MR images of mouse were recorded at different time points (pre-injection (0), 1, 2, 4, 8 and 24 h) of post-injection. As expected, strongly positive MR contrast enhancement in the tumor sites are observed within 0 to 24 h post-injection of NaGdF4@bp-peptide NPs, and the maximum MR signal enhancement (3.25 times) was achieved at 1 h post injection (as shown in Fig. 5a and c). The NaGdF4@CPP NPs show relatively poor MR contrast enhancement in the tumor site (as shown in Fig. 5b and c). The result of in vivo MRI indicates that NaGdF4@bp-peptide NPs have good tumor-targeting capacity. The relative strong MCF-7 tumor-targeting capacity of NaGdF4@bp-peptide NPs may due to high binding affinity of bp-peptide with MCF-7 cells.For further confirming tumor-targeting capacity of NaGdF4@bp-peptide NPs, the MCF-7 tumor-bearing Balb/c nude mice were sacrificed at 1 h or 24 h post-injection, respectively. The main organs and tumors of mice were collected, and the total amounts of Gd element in these tissues were measured by ICP-MS. As shown in Fig. 6, large amounts of Gd element were found in liver, spleen, kidneys and tumor. The Gd amounts in these organs at 1 h post-injection are higher than the Gd amounts in these organs at 24 h post-injection. The result indicates that both of NaGdF4@bp-peptide NPs and NaGdF4@CPP NPs are mainly accumulated in the liver, spleen, kidneys and tumor, and gradually excreted from body by hepatic and renal clearance pathways. Notably, the Gd amount in tumor of NaGdF4@bp-peptide NPs treated mouse is higher (1.82 times at 1 h post-injection and 1.78 times at 24 post-injection) than the Gd amount in tumor of NaGdF4@CPP NPs treated mouse. The result demonstrates that NaGdF4@bp-peptide NPs have excellent tumor-targeting capacity.
In vivo antitumor efficacy of NaGdF4@bp-peptide NPs
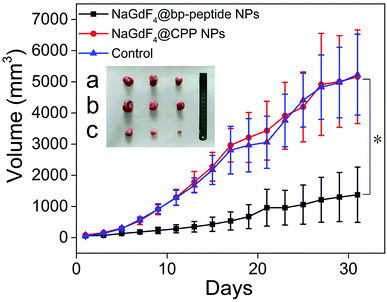
The MCF-7 tumor-bearing Balb/c nude mice were divided randomly into three groups (n = 3), control group, NaGdF4@bp-peptide NPs group, and NaGdF4@CPP NPs, which were treated by 100 μL PBS, and single dose (10 mg Gd per kg body weight) of NaGdF4@bp-peptide NPs and NaGdF4@CPP NPs through tail vein. After the different treatments were administered, the tumor sizes were measured with calipers every 2 days. Comparison with the PBS treated and NaGdF4@CPP NPs treated mice, the NaGdF4@bp-peptide NPs treated mice exhibit a delay in the tumor growth (as shown in Fig. 7 and S5†). The average tumor volume (1375 ± 885 mm3) of NaGdF4@bp-peptide NPs treated mice is much smaller than those of PBS treated (5229 ± 1296 mm3) and NaGdF4@CPP NPs treated (5160 ± 1501 mm3) mice. This result suggested that the NaGdF4@bp-peptide NPs have good antitumor bioactivity with the IRT as high as 73.7%.Conclusions
In summary, taking the advantage of strong interaction of phosphonate group with Gd3+, a new theranostic nanosystem (NaGdF4@bp-peptide NPs) has been constructed consisting of a NaGdF4 NPs core as the T1-weighted MR contrast agent and a peptide as the active-tumor targeting ligand as well as antitumor agent. In vitro and in vivo experiments demonstrate that the as-prepared NaGdF4@bp-peptide NPs have reasonable biocompatibility and excellent tumor targeting capacity. Furthermore, NaGdF4@bp-peptide NPs exhibit good anticancer efficacy with the IRT as high as 73.7% in subcutaneous MCF-7 breast tumor Balb/c nude mouse models. Owing to the diverse biological functionalities of peptides, this work provides us a facile preparation method to fabricate theranostic nanoparticles which have potential as anticancer agents for molecular imaging guided therapy.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank the National Natural Science Foundation of China (Grant No. 81571737 and 81871406) for financial support.References
- D.-E. Lee, H. Koo, I.-C. Sun, J. H. Ryu, K. Kim and I. C. Kwon, Chem. Soc. Rev., 2012, 41, 2656–2672 RSC.
- Y. Tao, M. Li, J. Ren and X. Qu, Chem. Soc. Rev., 2015, 44, 8636–8663 RSC.
- M. Yu and J. Zheng, ACS Nano, 2015, 9, 6655–6674 CrossRef CAS.
- M. W. Tibbitt, J. E. Dahlman and R. Langer, J. Am. Chem. Soc., 2016, 138, 704–717 CrossRef CAS PubMed.
- D. Kim, J. Kim, Y. I. Park, N. Lee and T. Hyeon, ACS Cent. Sci., 2018, 4, 324–336 CrossRef CAS PubMed.
- E. B. Ehlerding, P. Grodzinski, W. Cai and C. H. Liu, ACS Nano, 2018, 12, 2106–2121 CrossRef CAS PubMed.
- K. Sztandera, M. Gorzkiewicz and B. Klajnert-Maculewicz, Mol. Pharmaceutics, 2019, 16, 1–23 CrossRef CAS PubMed.
- S. Wilhelm, A. J. Tavares, Q. Dai, S. Ohta, J. Audet, H. F. Dvorak and W. C. W. Chan, Nat. Rev. Mater., 2016, 1, 16014 CrossRef CAS.
- Y. Liu, Z. Yang, X. Huang, G. Yu, S. Wang, Z. Zhou, Z. Shen, W. Fan, Y. Liu, M. Davisson, H. Kalish, G. Niu, Z. Nie and X. Chen, ACS Nano, 2018, 12, 8129–8137 CrossRef CAS.
- Q. Dai, S. Wilhelm, D. Ding, A. M. Syed, S. Sindhwani, Y. Zhang, Y. Y. Chen, P. MacMillan and W. C. W. Chan, ACS Nano, 2018, 12, 8423–8435 CrossRef CAS.
- V. P. Torchilin, Nat. Rev. Drug Discovery, 2014, 13, 813 CrossRef CAS PubMed.
- W. Dai, X. Wang, G. Song, T. Liu, B. He, H. Zhang, X. Wang and Q. Zhang, Adv. Drug Delivery Rev., 2017, 115, 23–45 CrossRef CAS PubMed.
- K. Han, Z. Ma and H. Han, J. Mater. Chem. B, 2018, 6, 25–38 RSC.
- A. M. E. Abdalla, L. Xiao, M. W. Ullah, M. Yu, C. Ouyang and G. Yang, Theranostics, 2018, 8, 533–548 CrossRef CAS PubMed.
- J. Lu, J. Wang and D. Ling, Small, 2018, 14, 1702037 CrossRef PubMed.
- H. V. T. Nguyen, A. Detappe, N. M. Gallagher, H. Zhang, P. Harvey, C. Yan, C. Mathieu, M. R. Golder, Y. Jiang, M. F. Ottaviani, A. Jasanoff, A. Rajca, I. Ghobrial, P. P. Ghoroghchian and J. A. Johnson, ACS Nano, 2018, 12, 11343–11354 CrossRef CAS.
- J. L. Major and T. J. Meade, Acc. Chem. Res., 2009, 42, 893–903 CrossRef CAS.
- C. Chen, N. Kang, T. Xu, D. Wang, L. Ren and X. Guo, Nanoscale, 2015, 7, 5249–5261 RSC.
- P. Brynolfsson, J. Yu, R. Wirestam, M. Karlsson and A. Garpebring, Magn. Reson. Med., 2015, 74, 1156–1164 CrossRef CAS.
- P. Caravan, J. J. Ellison, T. J. McMurry and R. B. Lauffer, Chem. Rev., 1999, 99, 2293–2352 CrossRef CAS.
- B. R. Smith and S. S. Gambhir, Chem. Rev., 2017, 117, 901–986 CrossRef CAS PubMed.
- V. Biju, Chem. Soc. Rev., 2014, 43, 744–764 RSC.
- S. Gai, C. Li, P. Yang and J. Lin, Chem. Rev., 2014, 114, 2343–2389 CrossRef CAS.
- B. Zhou, B. Shi, D. Jin and X. Liu, Nat. Nanotechnol., 2015, 10, 924 CrossRef CAS PubMed.
- L. Cole, R. Ross, J. Mr Tilley, T. Vargo-Gogola and R. Roeder, Nanomedicine, 2015, 10, 321–341 CrossRef CAS PubMed.
- N. Lee, D. Yoo, D. Ling, M. H. Cho, T. Hyeon and J. Cheon, Chem. Rev., 2015, 115, 10637–10689 CrossRef CAS PubMed.
- J. Weber, P. C. Beard and S. E. Bohndiek, Nat. Methods, 2016, 13, 639 CrossRef CAS.
- V. Gujrati, A. Mishra and V. Ntziachristos, Chem. Commun., 2017, 53, 4653–4672 RSC.
- K. J. McHugh, L. Jing, A. M. Behrens, S. Jayawardena, W. Tang, M. Gao, R. Langer and A. Jaklenec, Adv. Mater., 2018, 30, 1706356 CrossRef PubMed.
- M. Jiao, P. Zhang, J. Meng, Y. Li, C. Liu, X. Luo and M. Gao, Biomater. Sci., 2018, 6, 726–745 RSC.
- J. Fang, P. Chandrasekharan, X.-L. Liu, Y. Yang, Y.-B. Lv, C.-T. Yang and J. Ding, Biomaterials, 2014, 35, 1636–1642 CrossRef CAS PubMed.
- F. Liu, X. He, Z. Lei, L. Liu, J. Zhang, H. You, H. Zhang and Z. Wang, Adv. Healthcare Mater., 2015, 4, 559–568 CrossRef CAS PubMed.
- T. Zako, H. Nagata, N. Terada, A. Utsumi, M. Sakono, M. Yohda, H. Ueda, K. Soga and M. Maeda, Biochem. Biophys. Res. Commun., 2009, 381, 54–58 CrossRef CAS PubMed.
- S. Lee, J. Xie and X. Chen, Biochemistry, 2010, 49, 1364–1376 CrossRef CAS PubMed.
- J. M. Kinsella, R. E. Jimenez, P. P. Karmali, A. M. Rush, V. R. Kotamraju, N. C. Gianneschi, E. Ruoslahti, D. Stupack and M. J. Sailor, Angew. Chem., Int. Ed., 2011, 50, 12308–12311 CrossRef CAS.
- Y.-H. Kim, J. Jeon, S. H. Hong, W.-K. Rhim, Y.-S. Lee, H. Youn, J.-K. Chung, M. C. Lee, D. S. Lee, K. W. Kang and J.-M. Nam, Small, 2011, 7, 2052–2060 CrossRef CAS.
- D. Ghosh, Y. Lee, S. Thomas, A. G. Kohli, D. S. Yun, A. M. Belcher and K. A. Kelly, Nat. Nanotechnol., 2012, 7, 677 CrossRef CAS PubMed.
- L. Yang, H. K. Sajja, Z. Cao, W. Qian, L. Bender, A. I. Marcus, M. Lipowska, W. C. Wood and Y. A. Wang, Theranostics, 2014, 4, 106–118 CrossRef CAS.
- W. Poon, X. Zhang, D. Bekah, J. G. Teodoro and J. L. Nadeau, Nanotechnology, 2015, 26, 285101 CrossRef PubMed.
- N. Lu, Y. Tian, W. Tian, P. Huang, Y. Liu, Y. Tang, C. Wang, S. Wang, Y. Su, Y. Zhang, J. Pan, Z. Teng and G. Lu, ACS Appl. Mater. Interfaces, 2016, 8, 2985–2993 CrossRef CAS PubMed.
- H. Qin, Y. Ding, A. Mujeeb, Y. Zhao and G. Nie, Mol. Pharmacol., 2017, 92, 219–231 CrossRef CAS PubMed.
- A. A. P. Mansur, S. M. Carvalho, Z. I. P. Lobato, M. d. F. Leite, A. d. S. Cunha Jr and H. S. Mansur, Bioconjugate Chem., 2018, 29, 1973–2000 CrossRef CAS PubMed.
- P. Zhang, Y. Cui, C. F. Anderson, C. Zhang, Y. Li, R. Wang and H. Cui, Chem. Soc. Rev., 2018, 47, 3490–3529 RSC.
- C. D. Spicer, C. Jumeaux, B. Gupta and M. M. Stevens, Chem. Soc. Rev., 2018, 47, 3574–3620 RSC.
- G.-B. Qi, Y.-J. Gao, L. Wang and H. Wang, Adv. Mater., 2018, 30, 1703444 CrossRef PubMed.
- H. L. Chee, C. R. R. Gan, M. Ng, L. Low, D. G. Fernig, K. K. Bhakoo and D. Paramelle, ACS Nano, 2018, 12, 6480–6491 CrossRef CAS PubMed.
- Y. Shen, L. Liang, S. Zhang, D. Huang, R. Deng, J. Zhang, H. Qu, S. Xu, C. Liang and W. Xu, ACS Appl. Mater. Interfaces, 2018, 10, 7910–7918 CrossRef CAS PubMed.
- L. Zhu, H. Zhao, Z. Zhou, Y. Xia, Z. Wang, H. Ran, P. Li and J. Ren, Nano Lett., 2018, 18, 1831–1841 CrossRef CAS PubMed.
- F. Chen, X. Zhang, K. Ma, B. Madajewski, M. Benezra, L. Zhang, E. Phillips, M. Z. Turkey, F. Gallazzi, O. Penate-Medina, M. Overholtzer, M. Pauliah, M. Gonen, P. Zanzonico, U. Wiesner, M. S. Bradbury and T. P. Quinn, ACS Appl. Mater. Interfaces, 2018, 10, 4379–4393 CrossRef CAS PubMed.
- H. Zhang, Z. Guo, B. He, W. Dai, H. Zhang, X. Wang and Q. Zhang, Adv. Healthcare Mater., 2018, 7, 1800269 CrossRef PubMed.
- Y. Jiang, X. Pang, R. Liu, Q. Xiao, P. Wang, A. W. Leung, Y. Luan and C. Xu, ACS Appl. Mater. Interfaces, 2018, 10, 31674–31685 CrossRef CAS PubMed.
- A. King, C. Ndifon, S. Lui, K. Widdows, V. R. Kotamraju, L. Agemy, T. Teesalu, J. D. Glazier, F. Cellesi, N. Tirelli, J. D. Aplin, E. Ruoslahti and L. K. Harris, Sci. Adv., 2016, 2, e1600349 CrossRef PubMed.
- S. Sharma, V. R. Kotamraju, T. Mölder, A. Tobi, T. Teesalu and E. Ruoslahti, Nano Lett., 2017, 17, 1356–1364 CrossRef CAS PubMed.
- H. Chen, X. Li, F. Liu, H. Zhang and Z. Wang, Mol. Pharmaceutics, 2017, 14, 3134–3141 CrossRef CAS PubMed.
- F. Liu, X. He, J. Zhang, H. Zhang and Z. Wang, Small, 2015, 11, 3676–3685 CrossRef CAS PubMed.
- N. J. J. Johnson, W. Oakden, G. J. Stanisz, R. Scott Prosser and F. C. J. M. van Veggel, Chem. Mater., 2011, 23, 3714–3722 CrossRef CAS.
- F. Liu, X. He, L. Liu, H. You, H. Zhang and Z. Wang, Biomaterials, 2013, 34, 5218–5225 CrossRef CAS PubMed.
- V. Fogal, L. Zhang, S. Krajewski and E. Ruoslahti, Cancer Res., 2008, 68, 7210–7218 CrossRef CAS PubMed.
- L. Xu, N. Xiao, F. Liu, H. Ren and J. Gu, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 1530 CrossRef CAS PubMed.
- A. M. Ponsiglione, M. Russo, P. A. Netti and E. Torino, Interface Focus, 2016, 6, 20160061 CrossRef PubMed.
- M. Port, I. Raynal, L. V. Elst, R. N. Muller, F. Dioury, C. Ferroud and A. Guy, Contrast Media Mol. Imaging, 2006, 1, 121–127 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra02135c |
| This journal is © The Royal Society of Chemistry 2019 |