Open Access Article
Open Access ArticlePatient-derived bladder cancer xenograft models reveal VEGF and CDK4 enhancing tumor metastasis behavior†
Yong Zhao‡
a,
Mingjie An‡b,
He Zhanga,
Dengxu Tana,
Xue Chena,
Pengpeng Wua,
Weijun Qinc,
Caiqin Zhang*a and
Changhong Shi *a
*a
aDivision of Cancer Biology, Laboratory Animal Center, The Fourth Military Medical University, Xi'an, Shaanxi 710032, China. E-mail: changhong@fmmu.edu.cn; zhangcaiqin-beibei@163.com
bMBBS Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, 510080, China
cDepartment of Urology, Xijing Hospital, The Fourth Military Medical University, Xi'an, Shaanxi 710032, China
First published on 6th June 2019
Abstract
New strategies to treat advanced bladder cancer are urgently required. A patient-derived xenograft (PDX) model has become a useful tool to evaluate chemotherapeutics and investigate personalized cancer treatment options. In this study, fresh human bladder cancer specimens (C09303 and C21391) were transplanted subcutaneously into nude mice to establish PDX models. These models well retained pathological characteristics and molecular markers of the original tumor. Primary cells derived from C09303 were cultured and perfused into the bladders of nude mice to establish orthotopic xenograft models. Evident signals of lung and kidney metastases were detected by whole-body optical imaging. Gemcitabine displayed a significant therapeutic effect on the subcutaneous or orthotopic xenograft tumors of C09303. In vitro, matrigel invasion assays showed that C09303 primary culture cells are remarkably invasive. To study the metastatic properties of C09303, we sequenced the genomes of C09303 and C21391, and found that the expression of VEGF and CDK4 was significantly upregulated, and VEGF-knockdown or CDK4-knockdown C09303 cells showed attenuated invasiveness and migration. This PDX model revealed that VEGF and CDK4 enhanced tumor metastasis behavior, suggesting them as novel molecular therapeutic targets. This framework provides a tool for research on metastasis mechanism and individualized treatment for bladder cancer.
Introduction
Bladder cancer has the highest incidence among urinary malignancies and is one of the top ten most common tumors in humans.1 The standard treatment for advanced bladder cancer remains radical surgery followed by cisplatin (CDDP)-based chemotherapy.2 Unfortunately, a high proportion of the cases (50–70%) become metastatic, leading to extremely low survival rates.3 Multiple trials were developed to explore new therapeutic options for bladder cancer patients. However, few improvements have occurred in the last decades. Consequently, there is an urgent need for new strategies to treat advanced bladder cancer.Establishment of xenograft bladder cancer model mimicking clinical features would provide an effective tool for drug evaluation, biomarker identification, and personalized medicine.4 Conventional cell-derived xenograft (CDX) models are lack of tumor heterogeneity and unable to accurately reflect the patient's treatment status for assessments of metastatic tumors and drug sensitivity.5 Recently, patient-derived xenograft (PDX) models established from clinical fresh tumor specimens have been reported to well maintain the interactions between components of the tumor microenvironment, but also more accurately simulates the main features of bladder cancer patients.6,7 It provides in-depth information on the tumor biology and heterogeneity of individual human cancers,8 better represent the complexity of human tumors, and preserve the primary tumor architecture and gene expression patterns. Moreover, treatment of a PDX has been shown to reproduce the clinical outcomes observed in individual patients.9 Because most PDX models are established by subcutaneous transplantation, rare metastases can also occur that differ from those detected in clinical bladder cancer cases. Thus, we hypothesized that orthotopic transplantation might more faithfully simulate the structure and environment of tumor growth, and allow the tumor cells to metastasis.
Some immune checkpoint inhibitors display promising results in some bladder cancer cases, part of patients who shows objective responses to these therapies.10 The molecular portrait of bladder cancer has identified multiple alterations that could be actionable through molecularly targeted therapies.11 Thus, it is especially essential to explore new therapeutic targets for advanced bladder cancer. Accordingly, in this study, two PDX models of bladder cancer were successfully established. By bladder instillation of the primary culture of tumor cells from PDX model, we established an orthotopic transplantation model with higher metastasis characteristics. Therapeutic efficacy of intravesical instillation of gemcitabine was further validated, and the metastasis mechanism was analyzed at the molecular level by RNA sequencing in order to screen for novel molecularly therapeutic targets.
Experimental
Clinical specimens
Clinical bladder cancer specimens, C09303 and C21391, were obtained from the Department of Urology Surgery at Xijing Hospital. The histological aspect of the specimens were as follows: C09303, epithelial tumors with low differentiation and myometrial invasive, and C21391, epithelial tumors with medium differentiation and non-muscle invasive. The use of human tissue specimens was approved by the institutional review board (IRB, KY20173027-1) of the Fourth Military Medical University (FMMU). Tumor specimens were obtained during operations, aseptically removed, and xenografts were performed within 4 h. To establish the PDX models, fresh human bladder cancer specimens were transplanted subcutaneously into nude mice according to a published protocol.12Animals
Male BALB/c nude mice, 6–7 weeks old, were purchased from Vital River (Beijing, China), and bred in a barrier environment at the Laboratory Animal Center of FMMU. The mice were anesthetized with a combination of intravenous ketamine (10 mg kg−1) and xylazine (3 mg kg−1), and then maintained under isoflurane during surgery and imaging (RC2 Rodent gas anesthesia machine). All animal experiments were approved by the Laboratory Animal Ethics and Welfare Committee of FMMU (no. 20170109).Cell culture and lentiviral infection
Fresh tumor tissues derived from the PDX model were cut into 1 mm3 pieces with sterile scissors after washing with Hanks' solution. The tumor tissues were then digested with 0.25% trypsin/EDTA for 30 min, followed by culture in RPMI 1640 medium (Hyclone) supplemented with 20% fetal bovine serum (Gibco) in a humidified (37 °C, 5% CO2) incubator. The obtained C09303 cells were plated in 24-well plates (2 × 104 cells per well) and labeled with luciferase by lentiviral transfection (Luc-C09303) according to a published protocol.13Matrigel invasion assays and scratch test
Transwell chambers with polycarbonate membrane filters (6.5 mm diameter, 8 μm pore size) were coated with Matrigel. The number of cells invading through the Matrigel was counted by randomly selecting four visual fields, and the extent of invasion was expressed as the average number of cells per microscopic field detected at a magnification of 200×.14 Two blinded independent investigators independently interpreted the results of the Matrigel invasion assay. After dilution of Matrigel five times with RPMI 1640 medium, 70 μl of the solution was placed on the upper chamber of a transwell chamber, and incubated at 37 °C for 1 h. After the Matrigel coagulated, 600 μl of serum-free medium was added to the 24-well plate in the lower part of the chamber. After adding 100 μl of culture medium containing 10% fetal bovine serum and 5000 cells, the cells were incubated at 37 °C for 20 h. The transwell chamber was removed, fixed with methanol, washed with PBS, stained with 0.1% crystal violet, and the stained cells were counted under the microscope in triplicate.15For scratch test, C09303 and T24 were cultured in 6-well plates respectively until 100% confluence, the cells were then starved for the next 12 hours in RPMI-1640 without 10% FCS and scratched once vertically with a 200 μl pipette tip. Twenty-four hours after washing with PBS twice, migration was analyzed by microscopy, and expressed as the percentage of the wound width in comparison to the control sample.
Bladder instillation
Anesthetized mice were maintained under isoflurane during surgery and imaging. The microinjector was set to an injection rate of 30 μl/5 s, and connected to a polytetrafluoroethylene-coated intravenous catheter.16 The catheter instillation site was lubricated with sterile glycerol and inserted through the urethral opening into the bladder (with an insertion depth of approximately 3 cm). The bladder was pretreated with 100 μl of 0.1 mg ml−1 poly-L-lysine and allowed to remain in the bladder for 5 min.17 For in situ instillation, 30 μl of primary cultured cells of C09303 was used at 5 × 106 cells per ml. After the instillation was completed, the catheter was retained in the urinary tract for 5 min and then removed.BLI assay
Bioluminescent imaging (BLI) of the tumor xenografts, either in vivo or ex vivo, was performed after the mice received D-luciferin intraperitoneally (3 mg per mouse).18 Whole-body or organ-specific optical imaging was performed at 10 min after injection of D-luciferin using the Caliper Lumina II Small animal optical imaging system (PerkinElmer, MA, USA),19 and the fluorescence intensity was calculated per square centimeter of the tumor.Gemcitabine therapy
Ten female BALB/c nude mice bearing subcutaneous C09303 tumors were randomly assigned to two groups (n = 5): the control group and therapy group. When the tumor reached 100–150 mm3, the mice were intraperitoneally injected with 2 mg kg−1 of gemcitabine twice a week for 3 weeks.20 The tumor volume was measured every three days and calculated according to the formula: V = 1/2 a × b2, where a represents the tumor length, and b represents the tumor width. The control was treated with sterile saline. At the experimental end point, tumors were collected and fixed with 4% paraformaldehyde. After 21 days of the establishment of the in situ models of bladder cancer, the model was imaged in vivo to detect the intensity of luciferase at the tumor site.Similarly, female BALB/c nude mice bearing C09303 orthotopic tumors (n = 10) were randomly assigned to two groups: the control group and therapy group. Intravesical gemcitabine was administered transurethrally at 2%.21 Each administration was repeated 3 times at one-week interval for 3 weeks. The fluorescence intensity of luciferase at the tumor site was detected by in vivo imaging, and the effect of gemcitabine treatment was determined by the change in fluorescence intensity.
RNA sequencing and RT-PCR analysis
The sequencing library of each RNA sample was prepared using Ion Total RNA-Seq Kit v2 according to the manufacturer protocol (Life Technologies). After transcriptome sequencing, pathway analysis was used to analyze the signal pathways associated with tumor metastasis, and the upregulated genes in this pathway were selected (p < 0.05). qPCR analysis was conducted to evaluate the expression levels of VEGF and CDK4 with the primers indicated in Table 1. The data were analyzed by the ΔΔCt method: first, the ΔCt value of each tumor tissue was calculated (ΔCt = average Ct − average of housekeeping gene Ct), and the ΔΔCt value was calculated (ΔΔCt = ΔCt C09303 − ΔCt C21391) for each gene of the two tumors. Finally, the difference in expression of the corresponding genes of C09303 and C21391 was determined by 2−ΔΔCt.| Primer | Sequence |
|---|---|
| VEGF-F | GGAGTACCCTGATGAGATCGAG |
| VEGF-R | CATTTGTTGTGCTGTAGGAAGC |
| CDK4-F | GCTTTCTGTGGTCGGGTACA |
| CDK4-F | ACTTGCAGAGACAGTGGTCG |
| β-Actin-F | CATGTACGTTGCTATCCAGGC |
| β-Actin-F | CTCCTTAATGTCACGCACGAT |
Western blotting analysis
VEGF and CDK4 protein expression in the C09303 and C21391 samples was assessed by western blotting. Lysates of each tumor tissue were run on 10% sodium dodecyl sulfate-polyacrylamide gels and transferred onto nitrocellulose membranes. The membranes were blocked with TBST buffer containing 5% skimmed milk and incubated with rabbit anti-human VEGF (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, Proteintech) and anti-CDK4 (1
500, Proteintech) and anti-CDK4 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, CST) overnight, followed by the addition of HRP-linked anti-rabbit IgG and developed using an enhanced chemiluminescence substrate. The internal control was β-actin, and the gray value was measured relative to the control.
1000, CST) overnight, followed by the addition of HRP-linked anti-rabbit IgG and developed using an enhanced chemiluminescence substrate. The internal control was β-actin, and the gray value was measured relative to the control.
RNA interference
VEGF and CDK4 knockdown were performed using a targeting RNAi (GIEP1021083427 and GIEP1021053814) compared with control non-targeting RNAi purchased from Genechem (Shanghai, China). C09303 cells were seeded at 5.0 × 105 per well in six-well plates and incubated for 24 h.Statistical analysis
Statistical analysis was performed using SPSS v. 19 (SPSS Inc., Chicago, USA). All values in figures are presented as mean ± standard error of measurement for at least three independent experiments. Comparison of two groups was performed using Student's-test, and differences with p < 0.05 were considered to be statistically significant.Results
Establishment of PDX models
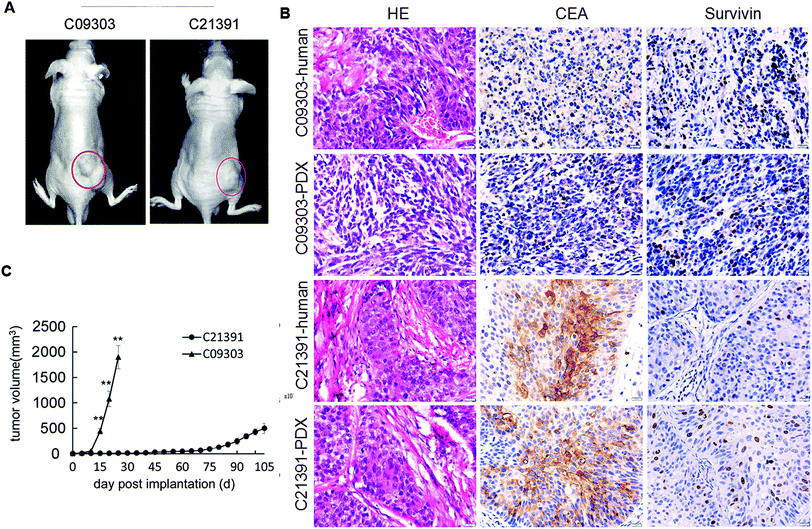
Computed tomography revealed the lesion in the bladder space, with nodule foci protruding in the bladder wall of two patients (C09303 and C21391). The histological subtype was transitional cell carcinoma based on H&E staining. Both fresh bladder cancer specimens were subcutaneously transplanted into the ventral region of nude mice to establish the PDX models (Fig. 1A). The primary cells derived from the tumor of C09303 PDX model were cultured, and further labeled with luciferase (Luc-C09303); however, the primary cells of C21391 could not be primary-cultured. H&E staining confirmed the identical histology in PDX tumor tissues with the original tumor, and IHC assay showed robust expression of CEA and survivin protein in both samples (Fig. 1B). A significant difference in the growth rate of the two PDX tumors was observed, C09303 reached the maximum volume at 4 weeks after implantation (Fig. 1C).Orthotopic transplantation of C09303 tumors display strong metastasis potential
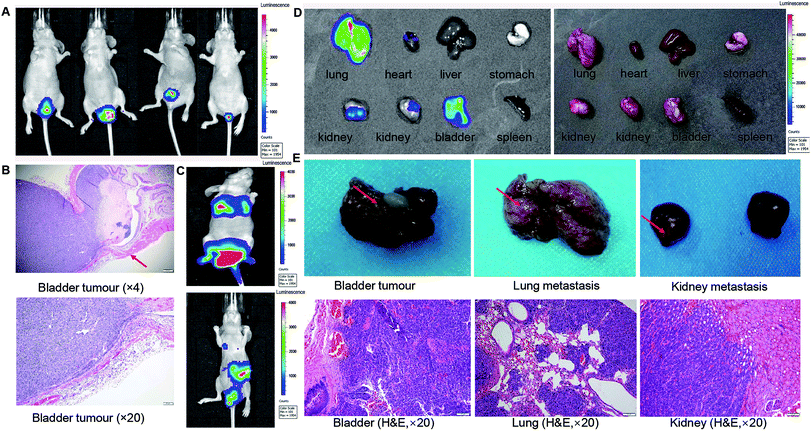
A total of 30 μl Luc-tagged C09303 cells (5 × 106 cells per ml) were perfused into the bladders of five nude mice to establish an orthotopic xenograft model. Two weeks later, the mice were subjected to whole-body optical imaging, and a strong BLI signal was detected in the bladder (Fig. 2A). Surgical isolation of the whole bladders of the mice with subsequent H&E staining showed that the intraepithelial tumor had non-invasive growth (Fig. 2B), which was consistent with the clinical features of patient C09303. One month later, two of the mice showed a strong BLI signal in the thoracic cavity (Fig. 2C). After sacrifice, we found strong BLI signals in the lung, kidney, and bladder of these mice as well (Fig. 2D), suggesting metastasis to these organs. Furthermore, the bladder tissue and metastasis foci were fixed with 4% polyoxymethylene, and H&E staining confirmed the pathological structure of the tumor (Fig. 2E).Gemcitabine inhibited tumor growth
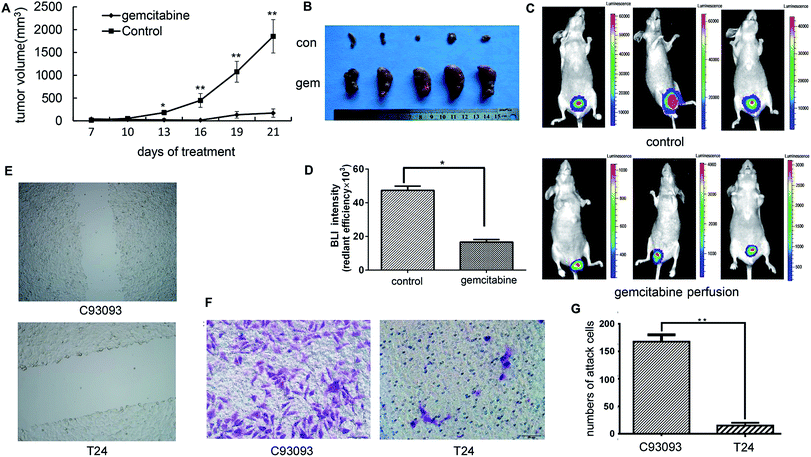
To study the therapeutic effect of gemcitabine on bladder cancer, 3 mm3 pieces of the tumor tissue from the C09303 PDX model were subcutaneously implanted into nude mice. The mice were then randomly divided into two groups, and one week later intraperitoneal injection with saline (control) or 2 mg kg−1 of gemcitabine.20 The tumor size was measured at 7, 10, 13, 16, 19, and 21 days after administration of gemcitabine. The growth of C09303 tumor was inhibited by gemcitabine (Fig. 3A and B).One week after the establishment of the orthotopic model, the mice were intravesically instilled gemcitabine at a dose of 2 mg per mice three times, and the therapeutic efficacy was evaluated by whole-body optical imaging. Unlike in the control group, the BLI intensity in the tumor site of the gemcitabine group was significantly weakened (Fig. 3C and D), indicating that intravesical gemcitabine effectively controlled the growth of bladder tumors.
Invasiveness and migration behavior of C09303 cell
To investigate the invasiveness and migration ability of C09303 tumor cells, we conducted a transwell assay and scratch test using the human bladder cancer cell line T24 as a control. The scratch test demonstrated that C09303 cells have strong migration and invasion ability (Fig. 3E). The number of C09303 tumor cells invading the transwell upper chamber was significantly higher than that of T24 cells (Fig. 3F and G; p < 0.001), indicating the high metastasis potential of C09303 cells after implantation in the bladder.Screening and analysis of metastasis-related genes of C09303
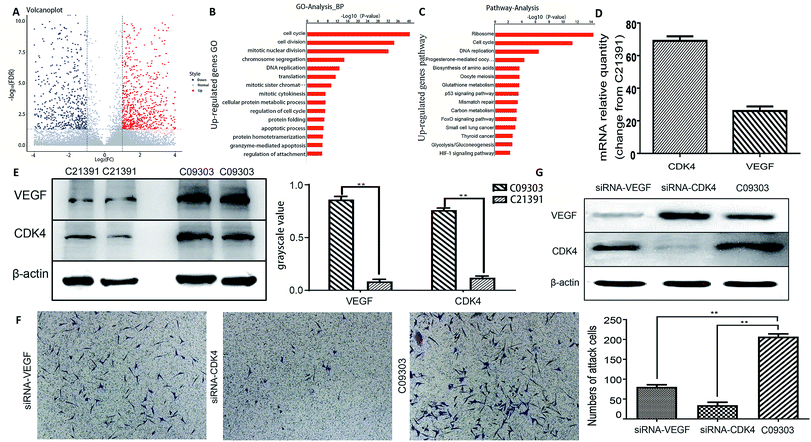
To further investigate the metastatic potential of C09303 and C21391 tumors, we determined their differentially expressed genes (p < 0.01, false discovery rate < 0.01) using RNA-sequencing, and then analyzed their characteristics. Among the 9280 genes compared, 985 were upregulated, and 941 were downregulated in C09303 (Fig. 4A). To screen for genes involved in tumor growth and metastasis, the significantly upregulated genes were analyzed for gene ontology and pathway enrichment. Both ribosome and cell cycle-related genes were quite different (Fig. 4B and C; ESI Fig. 1 and 2†). Reference to previous reports,22–24 we screened two representative genes, VEGF (ribosome relative gene) and CDK4 (cell cycle relative gene) for the metastasis characteristics analysis of C09303 based on p-value values and enrichment.RT-PCR results showed that VEGF and CDK4 levels were significantly higher in C09303 than C21391 tissues (p < 0.01; Fig. 4D), consistent with the results of pathway enrichment of differentially expressed genes. This up-regulation was further confirmed at the protein level by western blotting (p < 0.01; Fig. 4E).
To examine whether VEGF or CDK4 expression is associated with C09303 cell metastasis, stable knockdown of VEGF or CDK4 using a targeting RNAi system respectively led to less C09303 tumor cells invading the transwell upper chamber and inhibit C09303 cell proliferation significantly (Fig. 4F and G; ESI Fig. 3†). It indicated that both VEGF and CDK4 are essential factors related to the metastatic potential of C09303 bladder cancer.
Discussion
PDX model has become an effective tool to evaluate anti-tumor drugs and conduct research on personalized cancer treatment.25,26 In this study, we established two bladder cancer PDX models, one of which has multi-organ metastasis potential. H&E staining and IHC analysis confirmed that the xenograft tumor well retained the pathological features of the primary tumor. The occurrence of tumor metastasis depends mainly on the metastatic potential of tumor cells and the microenvironment of tumor growth. Conventional PDX models are mainly transplanted subcutaneously, and thus rarely form clinically similar metastatic features. By contrast, the present orthotopic xenograft developed a microenvironment similar to that of the primary tumor, indicating that it could simulate the evolutionary process of the tumor in vivo, and thus may serve as a useful individualized metastasis model. Although subcutaneous implantation of tumors in immunodeficient mice can increase the success rate of a PDX model, further orthotopic implantation of the tumor tissue or primary culture cells from a PDX model is more conducive to achieving tumor metastasis. In order to well keep the heterogeneity of the primary tumor, we selected tumor tissue from the third-generation PDX model to culture, and obtained the primary tumor cell to be labeled with luciferase by lentiviral transfection. These tumor cells for transplantation were cultured in vitro no more than three generations.Although a few PDX bladder models have been previously established by orthotopic transplantation, metastasis occurrence was rare owing to the lack of the physiological environment of the bladder.27 In this study, we cultured primary tumor cells derived from the C09303 PDX model to conduct orthotopic transplantation by bladder instillation. In this process, the tumor cells were not serially passaged, and the biological characteristics of the patient's tumor were retained. A strong BLI signal was detected in the lung and kidney, indicating metastasis formation. H&E staining of the sections showing these signals demonstrated a prominent cancer tissue structure, consistent with the results obtained by surgical exploration and in vivo imaging. Furthermore, the transwell analysis and the scratch test confirmed the metastatic potential of the C09303 tumor cells, which was significantly higher than that of the bladder cancer cells T24. In order to test the application of the PDX model in drug screening, we tested the response of C09303 model to chemotherapeutic drugs. BCG is the most effective adjuvant agent for treating non-muscle invasive bladder cancer (NMIBC),28 but approximately 30–40% of NMIBC patients do not respond to BCG treatment.29 Gemcitabine is used as neoadjuvant chemotherapy in patients with transitional cell carcinoma. Intravesical instillation of gemcitabine is effective against BCG-refractory NMIBC as well as advanced bladder cancer.30 In this study, C09303 PDX subcutaneous transplantation and the orthotopic transplantation model showed good reactivity to gemcitabine therapy.
Radical cystectomy and CDDP -based chemotherapy has demonstrated perfect results in some invasive muscle disease patients. However, part of these has distant metastases, resulting in a fatal disease. Thus, it is vital to explore a novel approach to treat advanced bladder cancer through molecularly targeted therapies. Individualized metastasis models of bladder cancer provide a useful tool to screen therapeutic drugs and to understand the metastasis-related mechanisms. RNA sequencing analysis is a widely used technique to compare related genes in tumor tissues and screen for differentially expressed genes. In this study, RNA sequencing of tissues from two patients with bladder cancer, C09303 and C21391, was performed to screen for metastasis-associated genes. We found that both VEGF and CDK4 were significantly upregulated in C09303, and their increased expression levels were confirmed in metastatic bladder cancer tissues from the model at the mRNA and protein levels. VEGF-knockdown or CDK4-knockdown C09303 cell showed a lower ability of invasiveness and migration respectively. All these indicated that the metastatic potential of the C09303 is closely related to the up-regulation of VEGF and CDK4 expression. Previous studies have found that both VEGF and CDK4 are closely related to tumor metastasis.22–24 VEGF encodes a transcription factor that is required for vascular endothelial growth,31 and CDK4 can help the tumor pass through the R point of the cell cycle, a point of no return, after which cells become committed to a new round of replication, and thus have positive effects on the malignant proliferation of bladder cancer.32 As one of the primary angiogenic factors for bladder cancer, VEGF can enhance tumorigenesis and tumor invasiveness, and can induce tumor angiogenesis to accelerate growth.33 This patient-derived bladder cancer xenograft model revealed that VEGF and CDK4 enhanced tumor metastasis behavior.
Based on these, drug targeting above protein may be an attractive therapeutic tools. FOXM1, a transcriptional activator with multiple oncogenic roles, exerts major roles in bladder cancer,34 which was identified as a critical phosphorylation target in a proteomic analysis of potential substrates of CDK4 kinase.35 Increased activity and expression of FOXM1 in NMIBC tumors at high risk of recurrence and these tumors display invasive characteristics in the recurrences. Treatment with CDK4/6 inhibitor or knockdown of CDK4 led to reduced FOXM1 phosphorylation in vitro and in vivo, allowing a significant tumor regression. Angiogenesis plays a significant role in the pathogenesis of bladder cancer. Many studies demonstrated that targeted agents directed against the molecular signaling pathways involved in angiogenesis such as VEGF pathway.36 However, clinical trials suggested that a single agent targeting the VEGF pathway did not show a significant improvement in outcomes.37 Conceptually promising concepts are the sequential and/or combination therapy, including agents targeting different pro-angiogenic factors and pathway, such as VEGF and CDK4.
In conclusion, the bladder cancer PDX model C09303 showed highly metastatic behavior. The up-regulated expression of VEGF and CDK4 were closely related to the metastatic potential. We demonstrated the feasibility of creating a bladder cancer PDX model which may more closely resemble the true tumor microenvironment when compared with cell line-derived xenograft models. A PDX model, such as the one described here, could be potentially utilized to inform patient-specific treatment recommendations.
Conflicts of interest
There are no conflicts to declare.Abbreviations
| PDX | Patient-derived xenograft |
| H&E | Hematoxylin and eosin |
| IHC | Immunohistochemistry |
| CEA | Carcinoembryonic antigen |
| BLI | Bioluminescent imaging |
| RT-PCR | Reverse transcription-polymerase chain reaction |
| VEGF | Vascular endothelial growth factor |
| CDK4 | Cyclin-dependent kinase4 |
| NMIBC | Non-muscle invasive bladder cancer |
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 31772546, No. 31572340 and No. 81372606).References
- S. Antoni, J. Ferlay, I. Soerjomataram, A. Znaor, A. Jemal and F. Bray, Eur. Urol., 2017, 71, 96–108 CrossRef PubMed.
- C. Rubio, M. Martínez-Fernández, C. Segovia, I. Lodewijk, C. Suarez-Cabrera, C. Segrelles, F. López-Calderón, E. Munera-Maravilla, M. Santos and A. Bernardini, Clin. Cancer Res., 2019, 25, 390–402 CrossRef PubMed.
- W. J. Alfred, C. Eva, N. C. Cowan, D. S. Maria, G. Georgios, L. Thierry, M. J. Ribal, A. G. Heijden Van Der and S. Amir, Eur. Urol., 2014, 65, 778–792 CrossRef PubMed.
- T. Inoue, N. Terada, T. Kobayashi and O. Ogawa, Nat. Rev. Urol., 2017, 14, 267–283 CrossRef PubMed.
- G. Fluegen, A. Avivar-Valderas, Y. Wang, M. R. Padgen, J. K. Williams, A. R. Nobre, V. Calvo, J. F. Cheung, J. J. Bravo-Cordero, D. Entenberg, J. Castracane, V. Verkhusha, P. J. Keely, J. Condeelis and J. A. Aguirre-Ghiso, Nat. Cell Biol., 2017, 19, 120–132 CrossRef CAS PubMed.
- K. A. Lodhia, A. M. Hadley, P. Haluska and C. L. Scott, Biochim. Biophys. Acta, 2015, 1855, 223–234 CAS.
- S. Aparicio, M. Hidalgo and A. L. Kung, Nat. Rev. Cancer, 2015, 15, 311–316 CrossRef CAS PubMed.
- M. Hidalgo, F. Amant, A. V. Biankin, E. Budinská, A. T. Byrne, C. Caldas, R. B. Clarke, J. S. De, J. Jonkers and G. M. Mælandsmo, Cancer Discovery, 2014, 4, 998–1013 CrossRef CAS PubMed.
- T. Voskoglounomikos, J. L. Pater and L. Seymour, Clin. Cancer Res., 2003, 9, 4227–4239 Search PubMed.
- J. Bellmunt, W. R. De, D. J. Vaughn, Y. Fradet, J. L. Lee, L. Fong, N. J. Vogelzang, M. A. Climent, D. P. Petrylak and T. K. Choueiri, N. Engl. J. Med., 2017, 376, 1015–1026 CrossRef CAS PubMed.
- R. Akbani, H. Al-Ahmadie, M. Albert, I. Alexopoulou, A. Ally, T. Antic, M. Aron, M. Balasundaram, J. Bartlett and S. B. Baylin, Cell, 2017, 171, 540–556 CrossRef PubMed.
- N. Zhao, C. Zhang, Y. Zhao, B. Bai, J. An, H. Zhang, J. B. Wu and C. Shi, Oncotarget, 2016, 7, 57277–57289 Search PubMed.
- Z. Cheng, D. Garvin, A. Paguio, P. Stecha, K. Wood and F. Fan, Curr. Chem. Genomics, 2010, 4, 84–91 CrossRef CAS PubMed.
- F. Wang, L. Ma, Z. Zhang, X. Liu, H. Gao, Y. Zhuang, P. Yang, M. Kornmann, X. Tian and Y. Yang, J. Cancer, 2016, 7, 408–417 CrossRef CAS PubMed.
- C. Zhang, M. P. Barrios, R. M. Alani, M. Cabodi and J. Y. Wong, Exp. Cell Res., 2016, 342, 159–165 CrossRef CAS PubMed.
- G. Redelman-Sidi, M. S. Glickman and B. H. Bochner, Nat. Rev. Urol., 2014, 11, 153–162 CrossRef CAS PubMed.
- C. J. Nemeth, R. M. Khan, P. Kirchner and R. Adams, Invest. Urol., 1977, 15, 149–150 CAS.
- C. Zhang, Y. Zhao, H. Zhang, X. Chen, N. Zhao, D. Tan, H. Zhang and C. Shi, Int. J. Mol. Sci., 2017, 18, e1332 CrossRef PubMed.
- C. Shi, J. B. Wu, G. C. Chu, Q. Li, R. Wang, C. Zhang, Y. Zhang, H. L. Kim, J. Wang and H. E. Zhau, Oncotarget, 2014, 5, 10114–10126 Search PubMed.
- H. K. Seo, W. A. Kwon, W. S. Dong, J. H. Son, W. S. Park, K. H. Lee, S. J. Lee and K. C. Jeong, J. Urol., 2014, 191, e231 Search PubMed.
- M. Horinaga, R. Fukuyama, M. Iida, H. Yanaihara, Y. Nakahira, S. Nonaka, N. Deguchi and H. Asakura, Urology, 2010, 76, 1267 CrossRef PubMed.
- S. Chen, X. Guan, L. L. Wang, B. Li, X. B. Sang, Y. Liu and Y. Zhao, Gene, 2017, 635, 3–8 CrossRef CAS PubMed.
- D. Huang, S. Song, Z. Z. Wu, W. Wu, X. Cui, J. N. Chen, M. S. Zeng and S. Su, Cancer Res., 2017, 77, 3591–3604 CrossRef CAS PubMed.
- T. Liu, J. Yu, M. Deng, Y. Yin, H. Zhang, K. Luo, B. Qin, Y. Li, C. Wu and T. Ren, Nat. Commun., 2017, 8, 13923 CrossRef CAS PubMed.
- F. R. Harris, P. Zhang, L. Yang, X. Hou, K. Leventakos, S. J. Weroha, G. Vasmatzis and I. V. Kovtun, Mol. Oncol., 2019, 13, 132–152 CrossRef CAS PubMed.
- C. Bernardo, C. Costa, N. Sousa, F. Amado and L. Santos, Transl. Res., 2015, 166, 324–331 CrossRef PubMed.
- L. J. Williams, S. K. Klein, D. Greenawalt, D. Trueman, H. Musgrove, H. Jones, L. Ruston and B. R. Davies, Cancer Res., 2015, 75, 5146 Search PubMed.
- H. W. Herr and M. Alvaro, J. Urol., 2008, 179, 53–56 CrossRef PubMed.
- G. Klement, S. Baruchel, J. Rak, S. Man, K. Clark, D. J. Hicklin, P. Bohlen and R. S. Kerbel, J. Clin. Invest., 2000, 105, R15 CrossRef CAS PubMed.
- L. Li and T. Yang, Colloids Surf., B, 2003, 30, 315–322 CrossRef CAS.
- G. Ding, S. Yu, S. Cheng, G. Li and Y. Yu, Am. J. Transl. Res., 2016, 8, 578–587 CAS.
- L. Li, T. Yang and X. Lian, Cancer Invest., 2005, 23, 309–315 CrossRef CAS PubMed.
- G. Di Lorenzo, S. Perdona, R. Damiano, A. Faiella, F. Cantiello, S. Pignata, P. Ascierto, E. Simeone, M. De Sio and R. Autorino, Cancer, 2010, 116, 1893–1900 CrossRef CAS PubMed.
- L. Li, W. Dang, Q. Yu, L. Li and P. Wu, Oncotarget, 2017, 8, 32298–32308 Search PubMed.
- L. Anders, N. Ke, P. Hydbring, Y. Choi, H. Widlund, J. Chick, H. Zhai, M. Vidal, S. Gygi and P. Braun, Cancer Cell, 2011, 20, 620–634 CrossRef CAS PubMed.
- A. A. Elfiky and J. E. Rosenberg, Curr. Oncol. Rep., 2009, 11, 244–249 CrossRef CAS PubMed.
- C. R. Mazzola and J. Chin, Expert Opin. Invest. Drugs, 2015, 24, 913–927 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra02362c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |