Open Access Article
Open Access ArticleWhite light emission produced by CTMA-DNA nanolayers embedded with a mixture of organic light-emitting molecules
Prathamesh
Chopade
 a,
Sreekantha Reddy
Dugasani
a,
Sreekantha Reddy
Dugasani
 a,
Sohee
Jeon
b,
Jun-Ho
Jeong
a,
Sohee
Jeon
b,
Jun-Ho
Jeong
 *bc and
Sung Ha
Park
*bc and
Sung Ha
Park
 *a
*a
aSKKU Advanced Institute of Nano Technology (SAINT), Department of Physics, Sungkyunkwan University, Suwon 16419, South Korea. E-mail: sunghapark@skku.edu
bNanomechanical Systems Research Division, Korea Institute of Machinery and Materials (KIMM), Daejeon 34103, Republic of Korea. E-mail: jhjeong@kimm.re.kr
cDepartment of Nanomechatronics, Korea University of Science and Technology (UST), Daejeon 34113, Republic of Korea
First published on 4th October 2019
Abstract
Researchers have started to recognize that biomaterial-based devices and sensors can be used in the development of high-performing environmentally-friendly technologies. In this regard, DNA can be utilized as a competent scaffold for hosting functional nanomaterials to develop a designated platform in the field of bionanotechnology. Here, we introduce a novel methodology to construct CTMA-modified DNA nanolayers (CDNA NLs) embedded with single (e.g., red, green, and blue), double (violet, yellow, and orange), and triple (white) iridium-based organic light-emitting materials (OLEMs, including Ir(piq)2(acac) for red, Ir(ppy)2(acac) for green, FIrpic for blue) that can serve as active light-emitting layers. The OLEM-embedded CDNA NLs were fabricated using simple solution processes, and their spectral properties were investigated via Fourier-transform infrared (FTIR), X-ray photoelectron (XPS), UV-Vis, and photoluminescence (PL) spectroscopies. FTIR analysis of OLEM-embedded CDNA NLs suggested that the complexes are stable and chemically inert. XPS revealed the various modes of interaction between OLEMs and CDNA. The evidence of interactions between blue OLEM and CDNA was demonstrated by peak shifts. The wide band gap characteristics (∼4.76 eV) and relatively high optical quality (no absorption in the visible region) of OLEM-embedded CDNA NLs were observed in UV-Vis absorption measurements. We observed PL emission in OLEM-embedded CDNA NLs, which was caused by the energy transfer from CDNA to OLEMs (ligand-centered and metal to ligand charge transfer). Lastly, a white light-emitting OLEM-embedded CDNA thin film was constructed using a combination of appropriate concentrations of red, green, and blue OLEMs. Its characteristic was demonstrated through spectral measurements. In addition, colour coordinates were plotted in the International Commission on Illumination (CIE) colour space, which confirmed the colour identity for the developed colours (including white). Consequently, the OLEM-embedded CDNA NLs can likely be used as a functional material in bio-imaging and bio-photonics.
Introduction
Electronic devices based on biomaterials are an emerging concept aimed at achieving high performance and generating environment-friendly technologies. Efforts in the field of bionanotechnology have resulted in a unique opportunity to interface biological systems and electronic devices. Biomaterials are renewable, biodegradable and present a wide range of properties such as self-assembly, scaffold characteristics, non-toxic nature, and diverse functionalities, which are not easily obtained from conventional nanomaterials.1,2Researchers have realized that biomolecules can be incorporated in optically engineered light-emitting diodes (LED) to fabricate efficient light-emitting devices. Organic light-emitting diodes (OLED) are devices that rely on active organic light-emitting materials (OLEM) sandwiched between metal electrodes. With OLEDs already making a large impact in the market of flat panel displays, overcoming the challenges that negatively affect their performance is key. OLEDs are energy efficient host–dopant devices that incorporate fluorescent or phosphorescent OLEMs in the host material to emit light under operating conditions. In this regard, cyclometalated heavy metal complexes are being developed as dopants for harvesting the forbidden emissive triplet states formed in OLEDs, which can help realize an internal quantum efficiency as high as 100%. Among them, iridium (Ir) complexes emitting red, green and blue colours have been developed with good device efficiency.3–5
The host material needs to meet some criteria related to transport, stability, and energy levels to classify as an efficient host material. Excitons are formed on the host and then are rapidly transferred by a nonradiative process to the chromophore in an OLED device that employs a host–dopant strategy as an active light-emitting material. A host material is supposed to have (i) energy levels (i.e., HOMO and LUMO) close to the dopant energy levels for efficient charge transfer, (ii) a higher triplet energy than the dopant (emitter) to avoid inefficient recombination, (iii) the ability to promote ambipolar charge transport, (iv) a high glass transition temperature to maintain structural stability, and (v) inherent chemical stability to avoid negative interactions with the adjacent layers. These characteristics play an important role in developing effective host materials that provide efficient charge injection, charge transport, and exciton transfer to ensure the stability of the emitting layer.6
DNA has unique scaffolding characteristics that can be utilized as an efficient template for hosting OLEMs to develop an active component in optical devices. Cetyltrimethylammonium chloride-modified DNA (CDNA) (which is a complex of DNA conjugated with a cationic surfactant prepared by an ion-exchange reaction) can be soluble in organic solvents (such as alcohols, chloroform, and chlorobenzene). Thin-layers made of CDNA show significant optoelectronic characteristics (such as a transmission of ∼100%, a refractive index around 1.5, a wide band gap of ∼4.7 eV, a high triplet energy of 2.75 eV, a HOMO level of ∼5.6 eV and a shallow LUMO level of ∼0.9 eV), which result in good hole transport and excellent electron blocking ability.7,8 In addition, a CDNA thin-layer shows considerable thermal stability (up to 250 °C).9–12 Researchers started to use DNA as electron-blocking layers in OLEDs due to efficient electron–hole recombination in the emitting layers, with emitting layers made of OLEMs and quantum-dots to enhance the device performance.13 Thermally deposited nucleic acid thin-films and a water-soluble DNA-Pan complex containing Ru(bpy)32+ were used in OLEDs to enhance the photoemission efficiency.14,15 Dye molecule embedded-DNA nanofibers prepared by electrospinning were used to fabricate white light emitting LEDs.16 The use of metalloporphyrins and vitamin-derivatives as optoelectronic emitters resulted in the incorporation of biomolecules in OLEDs.17–19 Although DNA has been heavily used, researchers mostly used DNA as electron-blocking layers in devices.
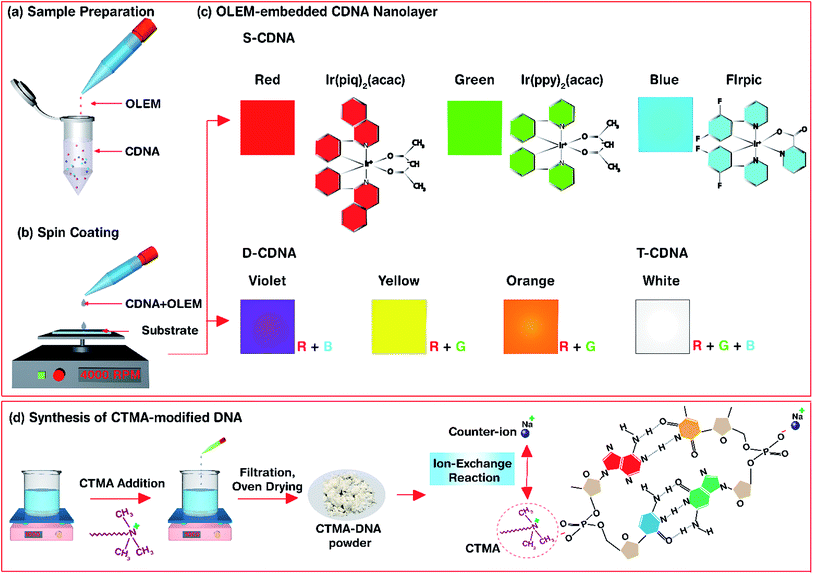
Here, we describe a method to develop CDNA nanolayers (NLs) embedded with single (unmixed), double (mixed), and triple (mixed) Ir-based OLEMs (i.e., R, G, and B-colour emitting), which can serve as active light emitting layers. Single, double, and triple OLEM-embedded CDNA NLs were labelled as S-CDNA, D-CDNA, and T-CDNA NLs, respectively. D-CDNA (e.g., violet (V), yellow (Y), and orange (O)-coloured NLs) and T-CDNA (white (W)-coloured) NLs were fabricated by mixing R, G, and B at appropriate concentrations. OLEM-embedded CDNA NLs formed by spin-coating on fused silica substrates were characterized by Fourier transform infrared (FTIR), UV-Vis, photoluminescence (PL), and X-ray photoelectron spectroscopy (XPS) for understanding the important characteristics of samples. We believe our approach demonstrates that CDNA is an efficient scaffold to host OLEMs, and it can be used in various devices and sensors.
Experimental section
Preparation of OLEM-embedded CTMA-DNA nanolayers
A solution of DNA extracted from salmon (GEN Corporation, Shiga, Japan) was prepared by dissolving 0.1 g of DNA in 10 mL of deionized (DI) water. This was stirred overnight to achieve a homogeneous mixture of DNA. An appropriate amount of cetyltrimethylammonium chloride (CTMA) was added to the DNA solution, which resulted in a white precipitate. The precipitate was collected by centrifuging and washing with DI water and was dried overnight. Eventually, CTMA modified DNA (CDNA) powder was obtained. CDNA powder was ready to be dissolved in an organic solvent. We used a mixture of chlorobenzene (for OLEMs) and butanol (for CDNA) with a ratio of 1 to 5 to achieve a homogenous solution of CDNA. Stock solutions (1 mM) of Iridium-based red, green and blue emitter molecules, viz., bis[2-(1-isoquinolinyl-N)phenyl-C](2,4-pentanedionato-O2,O4)iridium(III) (Ir(piq)2(acac)), bis[2-(2-pyridinyl-N)phenyl-C](2,4-pentanedionato-O2,O4)iridium(III) (Ir(ppy)2(acac)), and bis[2-(4,6-difluorophenyl)pyridine-C2,N](picolinato)iridium(III) (FIrpic) were prepared by mixing in chlorobenzene.Aliquots of CDNA solution were pipetted into a new test-tube along with a single type of proper [OLEM]. The solution was vortexed for 2 min, followed by incubation for 24 h at room temperature. This process resulted in single (red, green and blue light emitting) OLEM-embedded CDNA solutions (i.e., R-CDNA, G-CDNA, and B-CDNA). The final concentration of each OLEM, i.e., [R], [G], and [B], was 30 μM in a fixed [CDNA] of 1.5 wt% for S-CDNA. For D-CDNA, the final concentrations of OLEMs in double combinations, i.e., [R, B] for violet, [R, G] for yellow, and [R, G] for orange were 15 and 20 μM, 20 and 30 μM, and 15 and 10 μM in a fixed [CDNA] of 1.5 wt%. Similarly, for T-CDNA, final concentrations of the OLEMs in the triple combination, i.e., [R, G, B] for white were 20, 5, and 20 μM in a fixed [CDNA] of 1.5 wt%.
An S-CDNA, D-CDNA, or T-CDNA solution (40 μL) was spin-coated at 4000 rpm for 180 s on a 5 mm × 5 mm fused-silica substrate to obtain a NL with an average thickness of ∼80 nm. An average thickness of 3.5 μm was achieved by drop-casting 20 μL of either S-CDNA, D-CDNA, or T-CDNA solution (Fig. 1 and 2).
FTIR measurement
FTIR spectra of pristine CDNA and S/D/T-CDNA NLs formed on fused-silica substrates were obtained using an IFS-66/S TENSOR27 spectrometer (Bruker Inc., MA, USA) in the range of 3600–600 cm−1 at a resolution of 4 cm−1 (Fig. 3 and 6).XPS measurement
XPS of pristine CDNA and S-CDNA NLs were carried out using an ESCALAB 250Xi spectrometer (Thermo Fisher Scientific, Loughborough, UK) radiated with a monochromatic Al Kα X-ray source (energy of 1450 eV) with a single spot (diameter of 650 μm) on the sample surface (Fig. 4).UV-Vis absorbance measurement
The optical absorbances of pristine CDNA and S/D/T-CDNA NLs were measured using a Cary 5G spectrophotometer (Varian, CA, USA). The spectrophotometer was equipped with two light sources (a deuterium arc lamp and a quartz W – halogen lamp) and two detectors (a cooled PbS detector and a photomultiplier tube) (Fig. 5 and 6).PL measurement
The PL emission spectra of S/D/T-CDNA NLs were captured using a FS-2 fluorimeter (Scinco, Seoul, Korea) equipped with a Xe-arc lamp at a power of 25 W. The emission spectra were acquired by exciting the samples at a wavelength of 350 nm (Fig. 5 and 6).Results and discussion
A high optical quality NL with a smooth surface made of CDNA can be constructed by spin-coating. This NL can act as a charge transfer medium between adjacent layers in an OLED device. Transition metal OLEMs, i.e., cyclometaled Ir(III) complexes, were chosen as dopants primarily due to the strong spin–orbit coupling effect, which can integrate the singlet and triplet excited states, leading to bright emission covering the complete visible spectrum. In addition, these OLEMs are charge neutral, and thermally and chemically stable, which make them effective materials for use in OLEDs.
Fig. 1 shows the experimental procedure of OLEM-embedded CDNA NLs and synthesis of CDNA. Ir-based phosphorescent light emitting molecules (i.e., red, green and blue emitters) with appropriate concentrations were mixed with a CDNA solubilized in a mixture of chlorobenzene and butanol with ratios of 1 to 5. The as-prepared sample solution was spin-coated on a fused silica substrate to yield OLEM-embedded CDNA NLs emitting colours e.g., red-CDNA (R-CDNA) NLs for S-CDNA NLs, yellow-CDNA (Y-CDNA formed by mixing red and green OLEMs at a ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) for D-CDNA, and white-CDNA (W-CDNA formed by mixing red, green and blue OLEMs with a ratio of 4
3) for D-CDNA, and white-CDNA (W-CDNA formed by mixing red, green and blue OLEMs with a ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) for T-CDNA. We addressed OLEM-embedded CDNA NLs by FTIR and XPS spectra (to understand the interactions between OLEMs and CDNA), UV-Vis absorption (to determine the electronic transitions in given samples), and PL (to study the emission characteristics produced from S-, D- and T-CDNA NLs).
4) for T-CDNA. We addressed OLEM-embedded CDNA NLs by FTIR and XPS spectra (to understand the interactions between OLEMs and CDNA), UV-Vis absorption (to determine the electronic transitions in given samples), and PL (to study the emission characteristics produced from S-, D- and T-CDNA NLs).
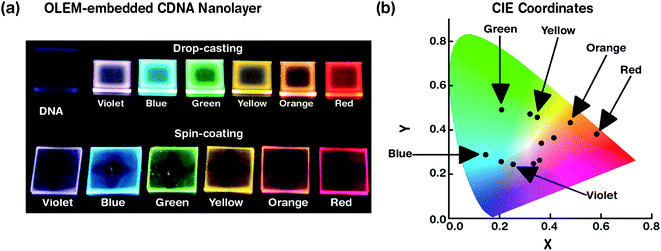
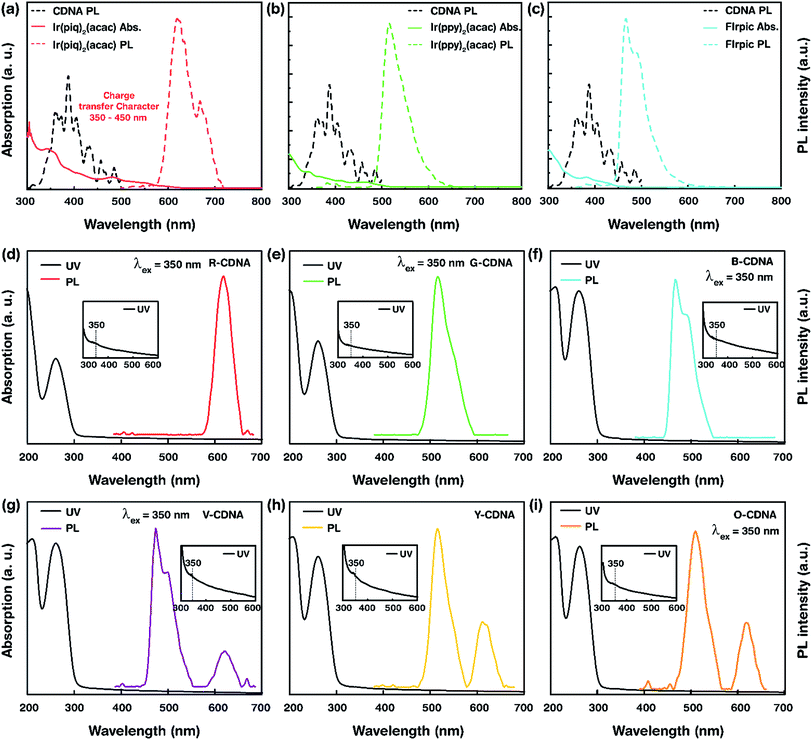
Photographs of S- and D-CDNA NLs under UV light illumination with a wavelength of 365 nm are shown in Fig. 2a. To compare the visibilities of S- and D-CDNA NLs with different thicknesses, we prepared drop-cast (average thickness of ∼3.5 μm) as well as spin-coated (∼80 nm) samples. As we expected, enhanced fluorescence was observed with NLs prepared by drop-casting compared to spin-coating. For S-CDNA NLs, the final concentration of each OLEM, i.e., [R], [G], and [B] was 30 μM in a fixed [CDNA]. For D-CDNA, the final concentrations of two OLEMs were as follows: [R, B] of 15 and 20 μM for violet, [R, G] of 20 and 30 μM for yellow, and [R, G] of 15 and 10 μM for orange in a fixed [CDNA]. The International Commission on Illumination (CIE) colour space provides information related to the wavelengths in the visible spectrum and colours observed by the human eye. It serves as a critical factor for colour management to determine the true colour. Fig. 2b displays the colour coordinates of 3 different S- (R, G and B) and 9 different D-CDNA NLs on the CIE 1931 RGB colour space. Various light emitting colours (except white) can be easily developed by mixing two OLEMs with different concentrations.
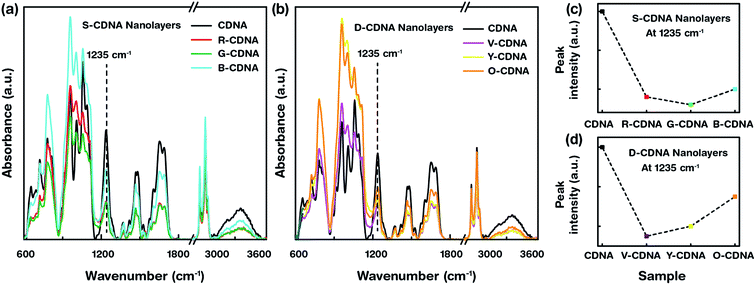
Attenuated total reflectance FTIR spectra of pristine CDNA and OLEM-embedded CDNA NLs were obtained to probe the interactions between OLEM and CDNA (Fig. 3). When excited by IR radiation, specific wavelengths are absorbed, causing chemical bonds to undergo vibrations such as stretching and bending in the wavelength range of 750–4000 cm−1, giving rise to a molecular fingerprint of the sample. Although no shifts in peak positions were noticed (indicating that minute chemical interactions occurred between OLEMs and CDNA), changes in peak intensities for S- and D-CDNA NLs were observed compared to pristine CDNA. This indicates the existence of OLEMs with different concentrations. The band range in 750–1300 cm−1 was sensitive to sugar (representative peak placed at 1055 cm−1) and phosphate backbone (1235 cm−1) vibrations. Within the 1300–1800 cm−1 range, bands were produced due to stretching vibrations of double bonds in DNA bases. Decreased peak intensities of S- and D-CDNA NLs compared to pristine CDNA were observed in the phosphate backbone vibration at 1235 cm−1 due to the presence of dopants, which caused a decrease in the relative amount of DNA (Fig. 3c and d).
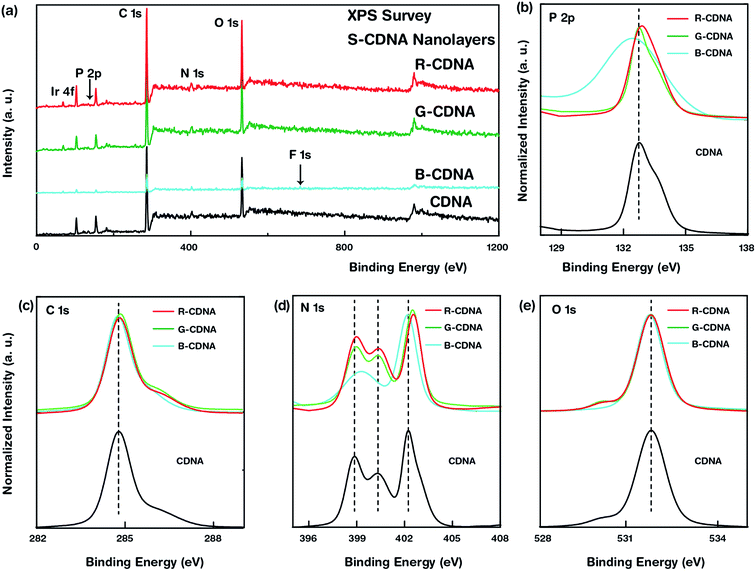
XPS measurements were conducted to investigate the interactions between OLEMs and CDNAs. Survey spectra of pristine CDNA and S-CDNA (i.e., R-, G-, and B-CDNA) NLs were acquired in the binding energy range from 0 to 1200 eV followed by high-resolution XPS spectra of core elements such as P 2p, C 1s, N 1s, and O 1s as illustrated in Fig. 4. Any shifts in binding energies of the core elements were the result of electron-transfer occurring through the interactions between dopant and host. Therefore, to understand the interaction between OLEMs and CDNA, we considered the shift in the peak positions in S-CDNA NLs, while the peak intensities were normalized for clarity. The presence of a single chemical state was observed in P 2p, C 1s, and O 1s, whereas three chemical states were observed in the high-resolution N 1s (two states in B-CDNA) spectra of S-CDNA NLs. R-CDNA and G-CDNA showed relatively negligible peak shifts in all elemental scans, while noticeable peak shifts in P 2p (negative, towards lower binding energy) and N 1s (positive/negative) of B-CDNA were observed. This meant that R and G had negligible effects on CDNA, whereas perceptible interactions between B and CDNA were evident.
Among many OLEM emitters, fluorine (F) dictates the emission characteristics in small-molecule B-OLEMs (i.e., FIrpic). The addition of F in FIrpic increases the energy gap between their HOMO and LUMO levels, thus leading to the emission of blue light. Although this strategy results in efficient emission, the presence of F makes these small-molecules prone to degradation, resulting in suppression of emission characteristics. We suspected the formation of a FIrpic isomer due to defluorination of FIrpic when mixed with CDNA. The effect of isomer formation on the core elements of CDNA was seen as a shift in peak position for P 2p and N 1s in B-CDNA compared to CDNA. Interestingly, the emission characteristics of B-CDNA were retained after mixing the blue emitter with CDNA. The mechanism by which such a phenomenon would occur remains to be explored in another study.20–22
Fig. 5 depicts the UV absorption of pristine OLEMs, S- and D-CDNA NLs. Based on UV absorption, characteristic peaks were observed at ∼260 nm from DNA molecules, and weak humps were present at ∼350 nm (shown in insets) due to the electronic transitions of OLEMs. The wide optical band gap associated with the π to π* electronic transition (at ∼260 nm) was estimated to be ∼4.76 eV, which indicated the corresponding HOMO–LUMO transitions in OLEM-embedded CDNA molecules. The minimum amount of energy (corresponding to the optical band gap) was required to form excitons upon relaxation by emitting light as dissipated energy. Thus, the wide band gap of CDNA allowed a CDNA NL to serve as an efficient host for energy transfer. Additionally, the strong absorption band observed at ∼190 nm (electronic transition of high energy) could also promote recombination i.e., formation of exciton that would lead to photoluminescence.23–25 UV absorption of OLEMs was associated with electronic transition from the ground state to the excited singlet state, which led to two major electronic transitions viz., ligand centered transition (LC) and metal to ligand charge transfer transition (MLCT). Usually, the singlet LC transition occurred at relatively higher energy than singlet and triplet MLCT bands, which were observed between singlet MLCT and triplet LC absorption bands within the range of 300–500 nm on the energy spectrum. Ir-based OLEMs showed characteristic bands at 300–360 nm, which can be attributed to the singlet LC transition (π to π*), while the bands appearing between 400–500 nm (charge transfer character) were associated with inter-ligand charge transfer (ILCT) and MLCT transitions.26–28
The PL spectra of pristine OLEMs, CDNA, S- and D-CDNA NLs are shown in Fig. 5. The maximum PL emission was observed at an excitation wavelength (λex) of 350 nm that yielded the characteristic emission peaks of R, G, and B OLEMs at 620, 520 and 469 nm. As we expected, two characteristic emission peaks corresponding to the mixing of OLEMs with CDNA were observed in the case of D-CDNA NLs. For instance, V-CDNA NLs formed by mixing R and B OLEMs clearly showed emission peaks at 620 and 469 nm (Fig. 5d). Consequently, violet, yellow, and orange colours from V-CDNA, Y-CDNA, and O-CDNA NLs can be perceived by the naked eye (as shown in Fig. 2). Excitation of S- and D-CDNA NLs at 350 nm promoted the population of excited LC and MLCT transition states located primarily on the ligands of the OLEMs. Consequently, the emission bands observed were a result of the radiative relaxation of photoexcited charge carriers that moved from the relatively higher energy ligands to the lower energy metal (Ir) center (π* to Ir, ligand to metal charge transfer) as well as between the ligands (π* to π, ILCT). In addition, the intermolecular interactions between the electron donor (CDNA) and electron acceptor (OLEMs) should give rise to a phenomenon known as exciplex, which can be exploited to develop highly efficient OLEDs. As displayed in Fig. 5a–c, the energy transfer between OLEMs and CDNA was noticed by the spectral overlap between the emission of CDNA (at ∼390 nm) and absorption of OLEMs (within the range of 350–450 nm). The energy transfer suggested that the excited charge carriers emerging from CDNA could be transferred onto OLEMs, which in turn were excited and underwent radiative relaxation emitting light with specific colours.29–32
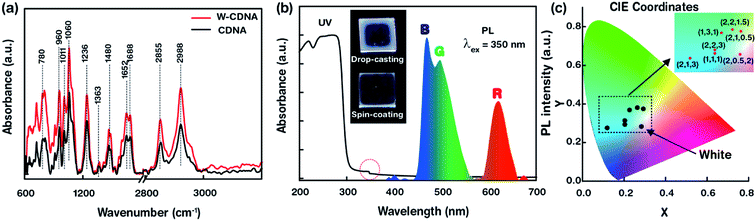
The attenuated FTIR, UV absorption, PL spectra, and CIE colour coordinates for white light producing T-CDNA NLs (e.g., W-CDNA) is shown in Fig. 6. The T-CDNA NLs were synthesized by mixing of R, G and B OLEMs in different concentrations at a fixed [CDNA]. We used R, G and B OLEM concentrations of 20, 5, and 20 μM to obtain W-CDNA NLs. In contrast to S- and D-CDNA NLs, FTIR spectra of W-CDNA NLs shown in Fig. 6a displayed hyperchromism (increasing intensity) in most of the FTIR absorbance bands upon addition of R, G, and B OLEMs in CDNA NLs. Fig. 6b showed the UV absorption (solid line) and PL (dotted line) spectra of W-CDNA NLs. The UV absorption spectrum showed the characteristic absorption band at 260 nm (attributed to CDNA) and a weak hump at 350 nm associated with the electronic transition of mixed OLEMs. The PL spectra obtained at a λex of 350 nm showed emission bands which corresponded to R (620 nm), G (490 nm, blue-shifted), and B (469 nm) OLEMs in CDNA. As mentioned above, the photoexcitation of OLEM-embedded CDNA NLs led to the generation of photoexcited electrons by a cascade of electronic transitions of LC and MLCT characteristics of the Ir-based OLEMs. These electrons relax to the ground state by the emission of light, which relates to R, G and B OLEM concentration in the CDNA NLs. Mixing of the R, G, and B lights led to the observed white light arising from W-CDNA NLs (as shown in the photograph displayed in Fig. 6b). Lastly, we plotted CIE colour coordinates of T-CDNA NLs in a colour gamut. T-CDNA NLs were produced by mixing different amounts of R, G, and B OLEMs at a fixed CDNA concentration (amount ratios of R, G, and B were marked in Fig. 6c). White light produced by a W-CDNA NL (formed by mixing R, G, and B OLEMs with the ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) were identified by the CIE colour coordinates, X = 0.33 and Y = 0.33 (Fig. 6c).
4) were identified by the CIE colour coordinates, X = 0.33 and Y = 0.33 (Fig. 6c).
Conclusions
We developed and characterized various colour (including white) light emitting CDNA NLs by mixing appropriate concentrations of OLEMs. CDNA proved to be an efficient template for designated nanomaterials. Spectral measurements (such as FTIR, XPS, UV-Vis, and PL) of OLEM-embedded CDNA NLs provided insight into the interactions between OLEMs (i.e., Ir(piq)2(acac) for R, Ir(ppy)2(acac) for G, FIrpic for B) and CDNA. FTIR analysis revealed that OLEM-embedded CDNA NLs are chemically stable. XPS showed evidence of OLEM interactions with CDNA. A FIrpic OLEM interacted noticeably with CDNA compared to Ir(piq)2(acac) and Ir(ppy)2(acac). The peak shifts of elemental states such as P 2p and N 1s in OLEM-embedded CDNA NLs supported this observation. UV-Vis absorption confirmed the presence of CDNA and provided information essential for efficient exciton formation on OLEMs via ligand-centered (LC) and metal to ligand-charge transfer (MLCT) electronic transitions. PL emission spectra represented emission bands corresponding to R, G, and B OLEMs via inter-ligand charge transfer (ILCT) and MLCT electronic relaxation of excited charge carriers to ground states. Further, a white light-emitting OLEM-embedded CDNA NL was fabricated by a combination of appropriate concentrations of R, G, and B OLEMs and was characterized using FTIR, UV-Vis and PL spectroscopies. In addition, CIE colour coordinates were placed in a colour gamut to confirm the colour identity of various types of OLEM-embedded CDNA NLs emitting white light as well as violet, blue, green, yellow, orange, and red light. Our results suggest that CDNA can be used as an efficient host matrix (that is physically robust and chemically stable) for OLEMs and as an active layer in OLEDs. OLEM-embedded CDNA NLs are promising candidates to be applied as a platform for developing multi-functional devices and highly sensitive sensors for bionanotechnology.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the Center for Advanced Meta-Materials (CAMM) funded by the Ministry of Science, ICT and Future Planning as the Global Frontier Project (CAMM-No. 2014M3A6B3063707 and CAMM-No. 2019M3A6B3032062) and by the National Research Foundation (NRF) of Korea (2016R1D1A1B03933768 and 2018R1A2B6008094).Notes and references
- I. V. Mihai, Chem. Soc. Rev., 2014, 43, 588–610 RSC.
- J. B. Christopher and Z. Bao, Polym. Int., 2010, 59, 563–567 Search PubMed.
- Y. Kawamura, K. Goushi, J. Brooks, J. J. Brown, H. Sasabe and C. Adachi, Appl. Phys. Lett., 2005, 86, 071104 CrossRef.
- P. Tao, W. Li, J. Zhang, S. Guo, Q. Zhao, H. Wang, B. Wei, S. Liu, X. Zhou, Q. Yu, B. Xu and W. Huang, Adv. Funct. Mater., 2016, 26, 881–884 CrossRef CAS.
- W. Li, J. Li, F. Wang, Z. Gao and S. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 26206–26216 CrossRef CAS.
- W. S. Jeon, T. J. Park, S. Y. Kim, R. Pode, J. Jang and H. J. Kwon, Org. Electron., 2009, 10, 240–246 CrossRef CAS.
- J. A. Hagen, W. Li, A. J. Steckl and J. G. Grote, Appl. Phys. Lett., 2006, 88, 171109 CrossRef.
- A. J. Steckl, Nat. Photonics, 2007, 1, 3–6 CrossRef CAS.
- S. Gong, C. Yang and J. Qin, Chem. Soc. Rev., 2012, 41, 4797–4807 RSC.
- Q. Sun, D. W. Chang, L. Dai, J. Grote and R. Naik, Appl. Phys. Lett., 2008, 92, 251108 CrossRef.
- F. W. Wang, D. Liu and J. Li, J. Photochem. Photobiol., A, 2018, 355, 152–157 CrossRef CAS.
- R. Bersohn and I. Isenberg, Biochem. Biophys. Res. Commun., 1963, 13, 205–208 CrossRef.
- Q. Sun, G. Subramanyam, L. Dai, M. Check, A. Campbell, R. Naik, J. G. Grote and Y. Wang, ACS Nano, 2009, 3, 737–743 CrossRef CAS.
- E. F. Gomez, V. Venkatraman, J. G. Grote and A. J. Steckl, Sci. Rep., 2014, 4, 7105 CrossRef CAS.
- N. Kobayashi, S. Uemura, K. Kusabuka, T. Nakahirac and H. Takahashi, J. Mater. Chem., 2001, 11, 1766–1768 RSC.
- Y. Ner, J. G. Grote, J. A. Stuart and G. A. Sotzing, Angew. Chem., Int. Ed., 2009, 48, 5134–5138 CrossRef CAS.
- H. Tajima, S. Ikeda, K. Shimatani, M. Matsuda, Y. Ando and J. O. H. Akiyama, Synth. Met., 2005, 153, 29–32 CrossRef CAS.
- H. Tajima, K. Shimatani, T. Komino, S. Ikeda, M. Matsuda, Y. Ando and H. Akiyama, Colloids Surf., A, 2006, 284–285, 61–65 CrossRef.
- N. Jürgensen, M. Ackermann, T. Marszalek, J. Zimmermann, A. J. Morfa, W. Pisula, U. H. F. Bunz, F. Hinkel and G. H. Sosa, ACS Sustainable Chem. Eng., 2017, 5, 5368–5372 CrossRef.
- A. Hany, D. P. Zoran, X. H. Nan, M. H. Ah and X. Gu, Science, 1999, 283, 1900–1902 CrossRef PubMed.
- V. Sivasubramaniam, F. Brodkorb, S. Hanning, H. P. Loebl, V. Elsbergen, H. Boerner, U. Scherf and M. Kreyenschmidt, J. Fluorine Chem., 2009, 130, 640–649 CrossRef CAS.
- E. Baranoff, B. F. E. Curchod, J. Frey, R. Scopelliti, F. Kessler, I. Tavernelli, U. Rothlisberger, M. Gratzel and Md. K. Nazeeruddin, Inorg. Chem., 2012, 51, 215–224 CrossRef CAS.
- C. E. Crespo-Hernandez, K. De La Harpe and B. Kohler, J. Am. Chem. Soc., 2008, 130, 10844–10845 CrossRef CAS.
- T. Takaya, C. Su, K. De La Harpe, C. E. Crespo-Hernandez and B. Kohler, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 10285–10290 CrossRef CAS PubMed.
- E. M. Conwell, P. M. McLaughlin and S. M. Bloch, J. Phys. Chem. B, 2008, 112, 2268–2272 CrossRef CAS.
- Y. You and W. Nam, Chem. Soc. Rev., 2012, 41, 7061–7084 RSC.
- M. X. Song, G. F. Wang, J. Wang, Y. H. Wanga, F. Q. Bai and Z. K. Qin, Spectrochim. Acta, Part A, 2015, 134, 406–412 CrossRef CAS.
- Y. You, K. S. Kim, T. K. Ahn, D. Kim and S. Y. Park, J. Phys. Chem. C, 2007, 111, 4052–4060 CrossRef CAS.
- V. Chandrasekhar, B. Mahanti, M. D. Pandey and R. S. Narayanan, ACS Omega, 2018, 3, 2786–2792 CrossRef CAS.
- S. Lamansky, P. Djurovich, D. Murphy, F. Abdel-Razzaq, R. Kwong, I. Tsyba, M. Bortz, B. Mui, R. Bau and M. E. Thompson, Inorg. Chem., 2001, 40, 1704–1711 CrossRef CAS.
- Y. Seino, H. Sasabe, Y. J. Pu and J. Kido, Adv. Mater., 2014, 26, 1612–1616 CrossRef CAS.
- V. S. Gorelika, G. I. Dovbeshkoc and A. Y. Pyatyshev, Bull. Lebedev Phys. Inst., 2016, 43, 69–73 CrossRef.
| This journal is © The Royal Society of Chemistry 2019 |