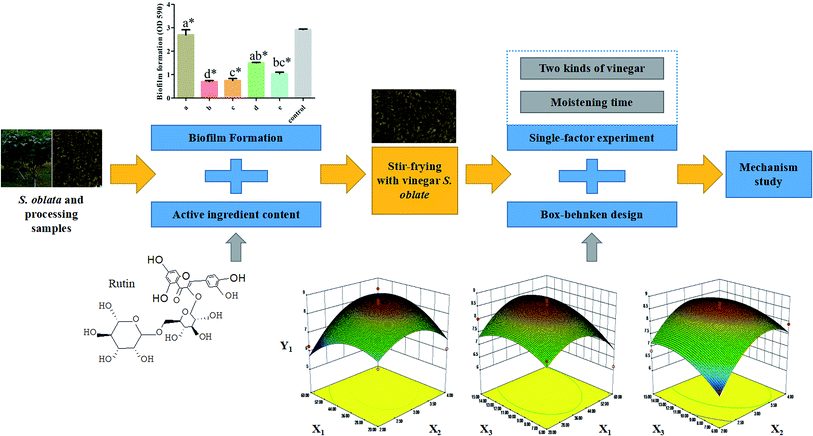
Open Access Article
Open Access ArticleProcess optimization of Syringa oblata Lindl. by response surface methodology and its effect on Staphylococcus xylosus biofilm†
Xing-Ru Chen‡
ab,
Yan-Yan Liu‡ab,
Yong-Hui Zhouab,
Xiao-Xu Xingab,
Qian-Wei Quab,
Xue-Ying Chenab,
Wen-Ya Dingab,
Guang-Long Chengab,
Ai-Juan Weiab,
Xi-Wen Fengab,
Bello-Onaghise God'spowerab,
Nsabimana Eliphazab and
Yan-Hua Li *ab
*ab
aCollege of Veterinary Medicine, Northeast Agricultural University, 600 Changjiang Road, Xiangfang, Harbin, Heilongjiang 150030, P. R. China. E-mail: liyanhua1970@163.com; Tel: +86 451 55191881
bHeilongjiang Key Laboratory for Animal Disease Control and Pharmaceutical Development, Harbin, China
First published on 5th November 2019
Abstract
Syringa oblata Lindl. (S. oblata) is a medicinal plant with effective broad-spectrum antibacterial activity, which can also inhibit Streptococcus suis biofilm formation. The processing of herbal medicine can purify medicinal materials, provide acceptable taste, reduce toxicity, enhance efficacy, influence performance and facilitate preparation. Thus, the aim of this study was to enhance the biofilm inhibition activity of S. oblata toward Staphylococcus xylosus (S. xylosus) using the best processing method. The content of rutin and flavonoids and the ability to inhibit the biofilm formation by S. oblata were examined using four processing methods. One of the best methods, the process of stir-frying S. oblata with vinegar, was optimized based on the best rutin content by response surface methodology. The histidine content and hisB gene expression of S. xylosus biofilm in vitro, resulting from stir-frying S. oblata with vinegar, were evaluated and were found to be significantly decreased and down-regulated, respectively. The results show that S. oblata stir-fried with vinegar can be used to effectively treat diseases resulting from S. xylosus infection. This is because it significantly inhibited S. xylosus biofilm formation by interfering with the biosynthesis of histidine; thus, its mechanism of action is decreasing histidine synthesis.
1. Introduction
The processing of herbal medicines by different methods, including parching, stir-baking with liquid, stir-frying with liquid, calcination, roasting in fresh cinders, steaming and boiling,1 plays an important role2 in achieving unique functions such as toxicity reduction, improving the pharmacological properties,3 stabilizing active ingredients,4 and flavor improvement or correction and content purification before clinical application.3 The safety, effectiveness, and quality of processed herbal medicines have attracted worldwide attention.2 Generally, the main mechanisms underlying herbal processing have been found to be related to the changes in the composition and/or activity of the components in the herbs.3Syringa oblata Lindl. (S. oblata) belongs to the Oleaceae plant family. The leaves and bark have a bitter taste and have important medicinal and industrial application purposes.5 S. oblata has promising biological effects such as anti-tumor, anti-hypertensive, anti-oxidant, anti-inflammatory activities.5 One of the major constituents associated with the anti-bacterial activity of S. oblata is rutin,6 which has been reported as the main bioactive ingredient in S. oblata that inhibits the formation of biofilm in Streptococcus suis (S. suis).7 However, it is unknown whether the concentration of rutin in S. oblata is affected by different processing methods and the effect of S. oblata against the formation of biofilm by Staphylococcus xylosus (S. xylosus) remains unexplored. The application of traditional Chinese medicine processing has a long history, and modern research shows that it has a certain scientific foundation. In order to improve the applications of S. oblata, it is important to influence its composition and pharmacological effects.
S. xylosus, a member of the coagulase-negative staphylococci (CoNS)8 is the most frequently isolated species from cows with mastitis9 and has the ability to form a biofilm.10,11 L-histidine biosynthesis is an ancient metabolic pathway present in bacteria and plants12 and is involved in the formation of biofilm in S. xylosus13 and Saccharomyces cerevisiae.14 However, there are no relevant pharmacological studies that evaluate the antibacterial activity of raw and processed S. oblata. Thus, the selection and optimization of processing methods may be able to effectively improve the association between the main components in herbal medicines and anti-bacterial activity.
The conventional optimization of a processing method in analytical chemistry involves monitoring the influence of one factor at a time on an experimental response, while other parameters are kept at a constant level. This, however, has certain disadvantages in that it does not depict all the effects of the parameter on the response, and the increase in the number of experiments necessary to conduct the research leads to an increase in the amount of time and expenses as well as an increase in the consumption of reagents and materials.15,16 Thus, this process response is a direct function of the single varied parameter.16 In order to overcome this problem, statistical optimization procedures allow one to take the interaction of variables into consideration by using multivariate statistical techniques.16 Currently, response surface methodology (RSM) is one of the most conventional and potent approaches to multivariate optimization analytical methodologies.17,18 This statistical tool tests the applicability and reliability of optimization to improve the process,19,20 reduce the number of experiments required to result in a less laborious task,21 and it indicates the possible influences of some variables on others.18 RSM is faster and more informative than the classical one-variable-at-a-time approach or the use of full factorial designs.22 Furthermore, RSM has been successfully used for developing, improving and optimizing processes.16
In this study, the changes in the rutin and flavonoid content and the effect of S. oblata against S. xylosus biofilm formation in vitro were tested under different S. oblata processing methods. The best method of processing S. oblata was then optimized using RSM. The histidine content and hisB gene expression of S. xylosus biofilm in vitro were evaluated after stir-frying S. oblata with vinegar. This study provides an explanation of how the methods of processing S. oblata affect S. xylosus biofilm formation and lays the foundation for further developing and optimizing the use of processed S. oblata. The detailed screening workflow is shown in Fig. 1.
2. Materials and methods
2.1. Plant materials
S. oblata, identified by Professor Xiuju Wu (College of Life Sciences, Northeast Agricultural University, Harbin, China), was grown under natural sunlight on the campus of the Northeast Agricultural University (N 45°44′33.64′′, E 126°43′22.07′′) in Harbin, Heilongjiang Province of China. The samples, collected in September 2018, were dried, pulverized, and sifted through the 24 mesh sieve.2.2. Preparation of Syringa oblata Lindl. for processing
The four kinds of processed S. oblata (leaves, 10 g each) were prepared in accordance with the Veterinary Pharmacopoeia of the People's Republic of China (volume II, 2015 edition) as follows.In the process of stir-frying with vinegar, salt-water and ginger juice, the leaves of S. oblata were mixed well with 7 mL of diluted rice vinegar, salt-water and ginger juice in a pot, respectively, and moistened for 15 min. They were then fried for 3 min and heated with a mild flame for 3 min at 400 °C until the vinegar, salt-water and ginger juice were completely absorbed. The ratios of 7 mL diluted rice vinegar, salt-water and ginger juice were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, 0.2
6, 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 and 1.25
7 and 1.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7, respectively. In the process of stir-frying with honey, S. oblata was prepared by mixing it well with 7 mL of 50% refined honey in a pot, moistening for 3 h, frying for 3 min and heating with a mild fire for 3 min at 400 °C until the refined honey was completely absorbed. The refined honey was prepared by placing 20 g of honey in a pot, heating to slight boiling at 120 °C until the surface was light yellow and shiny uniform bubbles appeared, then filtering through cotton gauze. Finally, all of the processed S. oblata was removed and allowed to cool.
7, respectively. In the process of stir-frying with honey, S. oblata was prepared by mixing it well with 7 mL of 50% refined honey in a pot, moistening for 3 h, frying for 3 min and heating with a mild fire for 3 min at 400 °C until the refined honey was completely absorbed. The refined honey was prepared by placing 20 g of honey in a pot, heating to slight boiling at 120 °C until the surface was light yellow and shiny uniform bubbles appeared, then filtering through cotton gauze. Finally, all of the processed S. oblata was removed and allowed to cool.
2.3. Determination of rutin and flavonoid content
A total of 20 to 50 mg of each type of processed S. oblata was used to extract flavonoids by adding 2 mL of 50% (v/v) methanol (HPLC grade) in H2O. The mixture was placed in an ultrasonic cleaner for 20 min and centrifuged for 10 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm.23 The supernatant was filtered through a 0.45 mm membrane filter and analyzed using a Waters Alliance HPLC system (Shimadzu, Corporation, Kyoto, Japan) and UV-1700 spectrophotometer (Shimadzu Corporation, Japan). Rutin and total flavonoids in normal and four kinds of processed S. oblata were determined using the chromatographic fingerprint analysis6 and aluminum chloride colorimetric method, respectively, as described previously.24 Among them, the chromatogram and colorimetric values were monitored at a wavelength of 355 nm and 510 nm, respectively during the experiment.
000 rpm.23 The supernatant was filtered through a 0.45 mm membrane filter and analyzed using a Waters Alliance HPLC system (Shimadzu, Corporation, Kyoto, Japan) and UV-1700 spectrophotometer (Shimadzu Corporation, Japan). Rutin and total flavonoids in normal and four kinds of processed S. oblata were determined using the chromatographic fingerprint analysis6 and aluminum chloride colorimetric method, respectively, as described previously.24 Among them, the chromatogram and colorimetric values were monitored at a wavelength of 355 nm and 510 nm, respectively during the experiment.
2.4. Biofilm formation
In this study, the S. xylosus ATCC 700404 strain was cultured in trypticase soy broth (TSB: Summus Ltd., Harbin, Heilongjiang, China) at 37 °C for 12 h with constant shaking. The methanolic extracts of processed S. oblata were used for minimum inhibitory concentration (MIC) assays and crystal violet staining using the protocol described previously.25 Rutin was used as a positive control.2.5. Experimental design of one of the Syringa oblata Lindl. processes
In order to improve the rutin content in one of the best processed S. oblata samples, two parameters, including two kinds of vinegar (rice vinegar and vinegar) and moistening times (15 min, 30 min and 40 min), were screened using a single-factor experiment. On the basis of the single-factor experiment, the BBD is a useful statistical technique26 that can be used to assess how the processing conditions influence the rutin and total flavonoid content. In this study, a three-level BBD experimental design required 17 experimental runs with three central points, which were employed to determine the optimal processing conditions. The selected independent variables were X1 (moistening time), X2 (frying time) and X3 (the dosage of vinegar) (Table 1). Three levels were coded 1, 0 and −1 for high, intermediate and low level, respectively (Table 1). The actual experimental design matrix is given in Table 2. Experimental data were analyzed by using a second-order polynomial model that correlated the interaction between the independent variables and the response variable.27 The second-order polynomial equation is as follows:where Y1 (μg g−1) and Y2 (mg g−1) are the predicted responses (the content of rutin and total flavonoids), Xi and Xj are the input variables (moistening time, frying time and the dosage of vinegar), b0, bi, bii, and bij represent the regression coefficients of constant, linear, quadratic, and interactions terms, respectively, and k represents the number of variables.28 According to the analysis of variance (ANOVA) with 95% confidence level, the regression coefficients, coefficient of determination, and the lack of fit for each studied response were employed to evaluate the fitness of the regression model. Design-expert software (version 8.0) was used to analyze the experimental data. The p-values < 0.05 were considered to be statistically significant.
| Independent variable/unit | Coded symbol | Level | ||
|---|---|---|---|---|
| −1 | 0 | 1 | ||
| Moistening time/min | X1 | 20 | 40 | 60 |
| Frying time/min | X2 | 2 | 3 | 4 |
| The dosage of vinegar/mL | X3 | 6 | 10.5 | 15 |
| Run | X1 | X2 | X3 | Y1 (μg g−1) | Y2 (mg g−1) | ||
|---|---|---|---|---|---|---|---|
| Predicted value (%) | Experiment value (%) | Predicted value (%) | Experiment value (%) | ||||
| 1 | 1 | 1 | 0 | 8.37 | 8.00 | 306.06 | 287.84 |
| 2 | 2 | 0 | −1 | 6.13 | 6.83 | 249.31 | 260.20 |
| 3 | 0 | 0 | 0 | 8.57 | 8.55 | 311.98 | 310.02 |
| 4 | −1 | −1 | 0 | 6.56 | 6.92 | 242.52 | 260.74 |
| 5 | 0 | 1 | 1 | 7.64 | 7.80 | 278.47 | 281.01 |
| 6 | 0 | 0 | 0 | 8.76 | 8.55 | 311.98 | 310.02 |
| 7 | −1 | 0 | 1 | 8.01 | 7.32 | 288.35 | 277.46 |
| 8 | 0 | 1 | −1 | 7.90 | 7.58 | 263.97 | 271.29 |
| 9 | 3 | 0 | 1 | 7.71 | 7.91 | 257.01 | 272.69 |
| 10 | −1 | 1 | 0 | 6.13 | 6.66 | 270.16 | 278.51 |
| 11 | 0 | 0 | 0 | 8.68 | 8.55 | 311.98 | 310.02 |
| 12 | 0 | −1 | 1 | 6.82 | 7.15 | 270.75 | 263.43 |
| 13 | 4 | −1 | 0 | 6.27 | 5.74 | 261.74 | 253.39 |
| 14 | 0 | 0 | 0 | 8.42 | 8.55 | 311.98 | 310.02 |
| 15 | 0 | −1 | −1 | 6.39 | 6.23 | 239.18 | 236.65 |
| 16 | −1 | 0 | −1 | 7.46 | 7.26 | 269.14 | 253.46 |
| 17 | 0 | 0 | 0 | 8.34 | 8.55 | 302.15 | 310.02 |
2.6. Determination of the histidine content
Overnight cultures of S. xylosus were diluted with TSB (corresponding to 1 × 105 CFU mL−1) and treated with sub-MICs processing S. oblata. This mixture was incubated at 37 °C for 24 h. Untreated S. xylosus served as the control. At the same time, the standard solution of 15 mg histidine (99% pure histidine was purchased from Beijing Solarbio Technology Co. Ltd.) was prepared by dissolving it in 250 mL (0.1 mol L−1) hydrochloric acid. The above assay was conducted using HPLC external standard analysis.25 The chromatographic separation was carried out on a Diamosil C18 column (4.6 mm × 250 mm, 5 μm) with a gradient solvent A (10 mmol L−1 diammonium hydrogen phosphate buffer containing 10 mmol L−1 sodium 1-octanesulfonate and obtaining pH 2.0 by phosphoric acid) and solvent B (acetonitrile) as the mobile phase at a flow rate of 1 mL min−1. The gradient conditions were 0 to 5 min, 95% solvent A; 5 to 6 min, 95% to 86% solvent A; 6 to 15 min, 86% solvent A; 15 to 16 min, 86% to 87% solvent A; 16 to 25 min, 87% solvent A. The detection wavelength was set at 205 nm, and the injection volume was 100 μL. Three independent sample analyses were performed for each sample.2.7. Verification of hisB expression in processing Syringa oblata Lindl. by quantitative real-time PCR
In order to investigate the effect of sub-MICs of processed S. oblata on the expression of the IGPD synthetic gene (hisB) of S. xylosus, the S. xylosus media culture (mid-log growth phase) was supplemented with sub-MICs of processed S. oblata and incubated with shaking at 37 °C for 24 h. Cells without sub-MICs of processed S. oblata served as the control. The supplemented solution was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 5 min and treated with an RNASE REMOVER I (Huayueyang Ltd., Beijing, China). The E.Z.N.ATM bacterial RNA isolation kit was used to determine the total RNA levels. The 16S RNA gene was selected as an internal control. The specific primers were obtained from Sangon Biotech (Shanghai) and listed in Table S1.† Quantitative real-time PCR was performed as described in our previous study.29
000 × g for 5 min and treated with an RNASE REMOVER I (Huayueyang Ltd., Beijing, China). The E.Z.N.ATM bacterial RNA isolation kit was used to determine the total RNA levels. The 16S RNA gene was selected as an internal control. The specific primers were obtained from Sangon Biotech (Shanghai) and listed in Table S1.† Quantitative real-time PCR was performed as described in our previous study.29
2.8. Statistical analysis
All the experiments were performed in triplicate. Data analysis and calculation of the standard deviation were done using SPSS 11.0.0 (IBM, USA) and p < 0.05 was considered indicative of statistically significant differences.3. Results
3.1. Rutin detection in processed Syringa oblata Lindl.
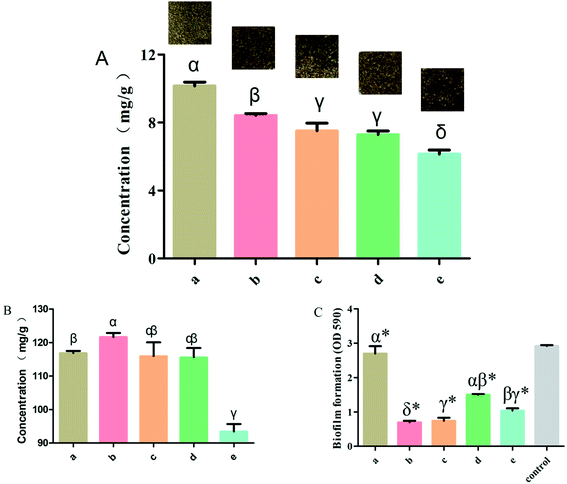
To investigate the effect of rutin in the normal and the four processed S. oblata samples, an investigation was conducted as described in the Materials and Methods section above. Previous studies have shown that at any given temperature, the longer the herb was subjected to stir-frying, the less rutin was extracted.30 When the heating temperature was 180 °C to 200 °C, rutin was partially decomposed to quercetin and was entirely damaged at temperatures above 210 °C. This is also in tandem with our study that when the heated temperature exceeded 200 °C, the rutin content in all the processed S. oblata was significantly lower (p < 0.05) than the normal S. oblata (Fig. 2A). At the same time, it may be beneficial to extract rutin with acid at a suitable pH (pH 2.0), which is consistent with the pH of rice vinegar.31 Thus, this study showed that the content of rutin in the stir-frying with vinegar was the highest among all the processed S. oblata. Chromatograms of rutin and S. oblata in the different processing groups are shown in Fig. S1.†3.2. Flavonoid detection in processed Syringa oblata Lindl.
To investigate the effect of rutin in normal and the four processed S. oblata samples, an investigation was done as described in the Materials and Methods section above. This study showed that the total flavonoid contents of the S. oblata samples stir-fried with salt-water, ginger juice and honey were 115.92 ± 5.88, 115.59 ± 4.84 and 93.41 ± 3.91 mg g−1, respectively. This indicated that the concentration of total flavonoids in S. oblata improved and was slightly lower than the normal S. oblata (116.86 ± 1.14 mg g−1) (p < 0.05) (Fig. 2B). This is in consonance with the previous study wherein the total flavonoid content decreased after stir-frying.32 In addition to the above-mentioned processing method, it has been reported that the total flavonoids content in S. oblata processed with vinegar increased by 21% in comparison with unprocessed S. oblata.33 This study has proven that the stir-frying S. oblata with vinegar had the strongest effect and the concentration of total flavonoids was 121.67 ± 2.08 mg g−1 and significantly higher than the normal S. oblata (p < 0.05).3.3. Biofilm inhibition properties of processed Syringa oblata Lindl.
The normal and the four processed S. oblata samples were evaluated against S. xylosus biofilm formation with the same MICs (31.25 mg mL−1). The results revealed that compared with the control, 1/2 MIC of the normal and four processed S. oblata had the significant ability to inhibit S. xylosus biofilm formation (p < 0.05) (Fig. 2C). Most recent studies have shown that S. oblata can inhibit Streptococcus suis (S. suis) biofilm formation and rutin is the active ingredient.6 Interestingly, we have shown here that the S. oblata stir-fried with vinegar had the greatest ability to inhibit the formation of S. xylosus biofilm (Fig. 2C). At the same time, the rutin and total flavonoid contents in the S. oblata stir-fried with vinegar were the highest among all processed S. oblata. Thus, in order to improve the ability of S. xylosus to form biofilm, the rutin and flavonoid content needed to be improved by optimizing the S. oblata stir-fried with vinegar.3.4. Box–Behnken design and response surface methodology
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
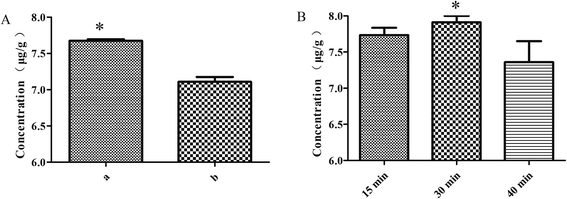
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 ratio), moistening for 15 min and frying for 2 min, the rutin of the S. oblata stir-fried with rice vinegar was significantly higher than the S. oblata stir-fried with vinegar (p < 0.05) (Fig. 3A). As the moistening time increased from 15 min to 40 min, the rutin of the S. oblata stir-fried with rice vinegar increased first and then decreased (Fig. 3B). Thus, the rice vinegar and 30 min moistening time were chosen as the central point of the BBD experiment.
6 ratio), moistening for 15 min and frying for 2 min, the rutin of the S. oblata stir-fried with rice vinegar was significantly higher than the S. oblata stir-fried with vinegar (p < 0.05) (Fig. 3A). As the moistening time increased from 15 min to 40 min, the rutin of the S. oblata stir-fried with rice vinegar increased first and then decreased (Fig. 3B). Thus, the rice vinegar and 30 min moistening time were chosen as the central point of the BBD experiment.
By applying multiple regression analysis to the experimental data, the relationship between the response variable and the test variables was described by the following second-order polynomial equation:
| Y1 = 8.55 + 0.5X1 + 0.63X1X2 − 0.93X12 − 0.79X22 | (1) |
| Y2 = 310.02 + 13.06X1 − 21.38X12 − 18.52X22 − 25.54X32 | (2) |
The results from the ANOVA for the adjusted model for Y1 and Y2 are presented in Table 3. The model F-values of 4.28 and 4.06, both with p-value < 0.05, and the lack of fit F-values of 23.22 and 28.48, imply that the model was significant and well adapted to the response. The results showed that the determination coefficient (R2) was 0.8464 and 0.8392, which were in good agreement with the adjusted Radj2 of 0.6488 and 0.6323. It is also indicated that the model was highly significant. A relatively lower value of the coefficient of variation (CV = 7.54% and 5.63%) indicated the better precision and reliability of the experiments carried out and the selected quadratic models exhibited high accuracy and significant reproducibility.35
| Source | Sum of squares | df | Mean square | F value | p-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| a | b | a | b | a | b | a | b | a | b | |
| a *Significant (p < 0.05); a indicated rutin content. b indicated total flavonoid content. | ||||||||||
| Model | 12.45 | 9024.24 | 9 | 9 | 1.38 | 1002.69 | 4.28 | 4.06 | 0.034* | 0.039* |
| X1 | 2.01 | 1363.95 | 1 | 1 | 2.01 | 1363.95 | 6.22 | 5.52 | 0.041* | 0.051 |
| X2 | 0.01 | 1.96 | 1 | 1 | 0.01 | 1.96 | 0.04 | 7.92 × 10−3 | 0.851 | 0.932 |
| X3 | 0.66 | 665.85 | 1 | 1 | 0.66 | 665.85 | 2.03 | 2.69 | 0.197 | 0.145 |
| X1X2 | 1.59 | 69.60 | 1 | 1 | 1.59 | 69.60 | 4.94 | 0.28 | 0.062 | 0.612 |
| X1X3 | 0.12 | 72.77 | 1 | 1 | 0.12 | 72.77 | 0.37 | 0.29 | 0.561 | 0.604 |
| X2X3 | 0.26 | 33.12 | 1 | 1 | 0.26 | 33.12 | 0.82 | 0.13 | 0.396 | 0.725 |
| X12 | 3.64 | 1924.09 | 1 | 1 | 3.64 | 1924.09 | 11.28 | 7.79 | 0.012* | 0.027* |
| X22 | 2.63 | 1443.72 | 1 | 1 | 2.63 | 1443.72 | 8.13 | 5.84 | 0.025* | 0.046* |
| X32 | 0.79 | 2747.37 | 1 | 1 | 0.79 | 2747.37 | 2.45 | 11.12 | 0.162 | 0.013* |
| Residual | 2.26 | 1729.78 | 7 | 7 | 0.32 | 247.11 | ||||
| Lack of fit | 2.14 | 1652.42 | 3 | 3 | 0.71 | 550.81 | 23.22 | 28.48 | 0.005* | 0.004* |
| Pure error | 0.12 | 77.36 | 4 | 4 | 0.03 | 19.34 | ||||
| Cor total | 14.71 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 754.02 754.02 |
16 | 16 | ||||||
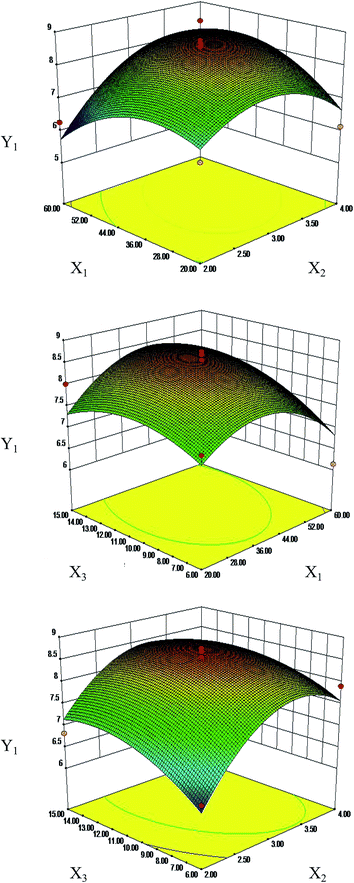
Response surface plots for each response condition and composite desirability functions were also analyzed to optimize the formulation. The optimized conditions were 44.03 and 40.68 moisturizing time (min), 3.3 and 3.29 frying time (min) and 11.97 and 11.19 vinegar dosage (mL) with 8.68 μg mg−1 and 312.64 mg g−1 calculated as the predicted values of Y1 (Fig. 4) and Y2 (Fig. S2†), respectively. This similarity indicated that the processing regression model was highly significant and could be used to predict the concentration of S. oblata.
3.5. Verification of the predictive model by biofilm formation
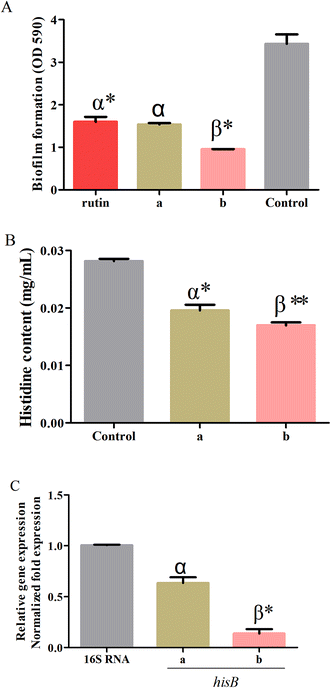
The optimum conditions obtained by RSM were used to validate the predictive model for the processing conditions of the S. oblata stir-fried with rice vinegar as compared with normal S. oblata by inhibiting biofilm formation. The results revealed that compared with the control, 1/2 MIC of the S. oblata stir-fried with vinegar had the highest ability to inhibit the formation of S. xylosus biofilm as compared to 1/2 MIC of normal S. oblata and a positive control rutin (Fig. 5A). Therefore, the response model was adequate for reflecting the expected optimization, and the extraction conditions and the ability to inhibit biofilm formation achieved by RSM were reliable and practical.3.6. Determination of histidine content
The histidine content of S. xylosus was determined in sub-MICs processed S. oblata in the normal and the stir-fried with vinegar group. When supplemented with sub-MICs of processed S. oblata in the culture medium, the histidine content of S. xylosus showed significant depletion (p < 0.05) in comparison with the control (Fig. 5B). Additionally, the histidine content of S. xylosus in the S. oblata stir-fried with vinegar S. oblata was significantly lower than in normal S. oblata (p < 0.05) (Fig. 5B). At the same time, the chromatograms of histidine, the control group without S. oblata, normal S. oblata and the S. oblata stir-fried with vinegar are shown in Fig. S3.†3.7. Verification of hisB expression in processed Syringa oblata Lindl. by quantitative real-time PCR
To further investigate the expression of hisB during the interference of the normal S. oblata and the S. oblata stir-fried with vinegar with the formations of S. xylosus biofilms, the sub-MICs were determined by quantitative real-time PCR. The results showed that the sub-MICs of the normal and the S. oblata stir-fried with vinegar interfered with the formation of biofilms by S. xylosus, and the expression of hisB gene was significantly down-regulated (p < 0.05 and p < 0.01) (Fig. 5C). Compared with the sub-MICs of the normal S. oblata, the expression of hisB gene was significantly down-regulated with the sub-MICs of the S. oblata stir-fried with vinegar.4. Discussion
Herbal medicine processing is a traditional herbal pharmaceutical technology.36 There are complex chemical changes during herbal medicine processing, and these chemical constituents that change during processing may be the basis for relieving drug irritation, enhancing therapeutic effect, biological efficacy, clinical applicability37 and clinical efficacy changes.36In recent years, there have been many studies on the chemical and molecular mechanisms involved in the processing of herbal medicine.36 For example, in stir-fried-treated Adinandra nitida tea, a well-known resource of functional foods, the main compounds such as total water-soluble solids, phenolics and flavonoids content were higher than in the corresponding untreated samples.38 Bai-Zhu possesses significantly different therapeutic effects as compared to processed Bai-Zhu since the processing increases the content of atractylenolide, which could strengthen the effect on gastrointestinal function.3 There are no studies on the processing of S. oblata at home and abroad. This study has demonstrated that stir-frying S. oblata with vinegar led to the highest content of rutin and total flavonoids among all processed S. oblata.
At present, the RSM experimental designs most commonly applied in pharmaceutical experiments are the central composite design (CCD) and Box–Behnken design (BBD).39 Compared to CCD, BBD employs a reduced number of experimental runs, leading to higher efficiency39 that is obtained with the best conditions of resources21 for quadratic models,21 and it is extensively used in RSM for three-level factors.20 BBD has been used to optimize the extraction of bioactive and antibacterial compounds from different processing samples. Thus, it is necessary to improve the content of rutin and biofilm formation in the S. oblata stir-fried with vinegar by the RSM method. This study has confirmed that the optimized conditions were rice vinegar, 44.03 min moisturizing time, 3.3 min frying time and 11.97 mL vinegar dosage, with rutin content calculated as 8.68 μg mg−1 for the single-factor and BBD experiments with S. oblata stir-fried with vinegar. When the main factors were the dosage of rice vinegar, vinegar infiltration, and processing temperature, with values of 20%, 3 h and 130, the contents of effective components were increased.40
We have demonstrated that the S. oblata stir-fried with vinegar had the greatest ability to inhibit the formation of S. xylosus biofilm as compared to the normal S. oblata. A study has found that the nitrogen metabolic limitation has a key role in the formation of biofilm, namely, histidine metabolism.13 Therefore, it was necessary to investigate the mechanisms underlying the effects of S. oblata stir-fried with vinegar on histidine metabolism and biofilm formation. The results showed that compared with normal S. oblata, the histidine content and hisB expression of S. xylosus in the S. oblata stir-fried with vinegar were significantly decreased and down-regulated. This is consistent with previous research where L-histidine dramatically decreased the biofilm formation by Saccharomyces cerevisiae flor yeasts.14 This may prove that the S. oblata stir-fried with vinegar may indirectly interfere with S. xylosus biofilm formation by decreasing histidine content and hisb gene expression.
5. Conclusion
In conclusion, we successfully developed a method for processing S. oblata, namely, stir-frying the S. oblata with vinegar, which was sufficient to inhibit biofilm formation. The mechanism related to the biofilm formation of S. xylosus involved the decrease and down-regulation of the histidine content and hisB gene expression, respectively, upon treatment with S. oblata stir-fried with vinegar as compared with normal S. oblata. These results provide an interesting template for designing new and more effective processing methods.Author contributions
Y-HL designed the whole experiment; Y-YL and X-RC directed the completion of the experiment; Y-HZ, X-XX, Q-WQ, X-YC, W-YD, G-LC, A-JW, X-WF, GB, EN were supportive during the experiment.Funding
This work was sponsored by the earmarked fund for China Agriculture Research System-35 and National Science, Technology support plan of China (grant number 2015BAD11B03-07).Conflicts of interest
The authors declare no competing financial interest.References
- T. Yi, J.-Y. Fang, L. Zhu, Y.-N. Tang, H. Ji, Y.-Z. Zhang, J.-C. Yu, X.-J. Zhang, Z.-L. Yu, Z.-Z. Zhao and H.-B. Chen, Chin. Med., 2016, 11, 1–8 CrossRef.
- W. Wang, J. Xue, K. Li, D. Hu, G. Huang and L. Ye, Int. J. Food Prop., 2017, 20, S644–S653 CrossRef CAS.
- G.-S. Shan, L.-X. Zhang, Q.-M. Zhao, H.-B. Xiao, R.-J. Zhuo, G. Xu, H. Jiang, X.-M. You and T.-Z. Jia, J. Pharm. Biomed. Anal., 2014, 98, 74–84 CrossRef CAS.
- J.-Y. Fang, L. Zhu, T. Yi, J.-Y. Zhang, L. Yi, Z.-T. Liang, L. Xia, J.-F. Feng, J. Xu, Y.-N. Tang, Z.-Z. Zhao and H.-B. Chen, Chin. Med., 2015, 10, 1–7 CrossRef.
- S. Guozhu, C. Yuan, L. Chun, Y. Xuelong, G. Xiaoli, T. Pengfei and C. Xingyun, Chem. Cent. J., 2015, 9, 1–12 CrossRef.
- Y.-Y. Liu, X.-R. Chen, L.-F. Gao, M. Chen, W.-Q. Cui, W.-Y. Ding, X.-Y. Chen, B.-O. God'spower and Y.-H. Li, Front. Pharmacol., 2018, 9, 1–10 CrossRef.
- J. Bai, Y. Yang, S. Wang, L. Gao, J. Chen, Y. Ren, W. Ding, I. Muhammad and Y. Li, Front. Pharmacol., 2017, 8, 1–9 CAS.
- K. M. Osman, K. A. Abd El–Razik, H. S. H. Marie and A. Arafa, J. Food Saf., 2016, 36, 89–99 CrossRef CAS.
- B. Kot, M. Piechota, M. Antos-Bielska, E. Zdunek, K. M. Wolska, T. Binek, J. Olszewska, P. Gulinski and E. A. Trafny, Pol. J. Vet. Sci., 2012, 15, 741–749 CAS.
- S. Planchon, M. Desvaux, I. Chafsey, C. Chambon, S. Leroy, M. Hébraud and R. Talon, J. Proteome Res., 2009, 8, 1797 CrossRef CAS.
- S. Planchon, B. Gaillardmartinie, E. Dordetfrisoni, M. N. Bellonfontaine, S. Leroy, J. Labadie, M. Hébraud and R. Talon, Int. J. Food Microbiol., 2006, 109, 88–96 CrossRef CAS.
- R. K. Kulis-Horn, M. Persicke and J. Kalinowski, Microb. Biotechnol., 2014, 7, 5–25 CrossRef CAS.
- C. G. Xu, Y. B. Yang, Y. H. Zhou, M. Q. Hao, Y. Z. Ren, X. T. Wang, J. Q. Chen, I. Muhammad, S. Wang and D. Liu, Front. Pharmacol., 2017, 8, 543 CrossRef PubMed.
- M. B. Zeidan, G. Zara, C. Viti, F. Decorosi, I. Mannazzu and M. Budroni, PLoS One, 2015, 10, 1–10 Search PubMed.
- M. A. Bezerra, R. E. Santelli, E. P. Oliveira, L. S. Villar and L. A. Escaleira, Talanta, 2008, 76, 965–977 CrossRef CAS.
- C. Liyana-Pathirana and F. Shahidi, Food Chem., 2005, 93, 47–56 CrossRef CAS.
- J. Carlos Martinez-Patino, B. Gullon, I. Romero, E. Ruiz, M. Brncic, J. S. Zlabur and E. Castro, Ultrason. Sonochem., 2019, 51, 487–495 CrossRef.
- S. V. F. Gomes, L. A. Portugal, J. P. dos Anjos, O. N. de Jesus, E. J. de Oliveira, J. P. David and J. M. David, Microchem. J., 2017, 132, 28–35 CrossRef CAS.
- P. Naderi, M. Shirani, A. Semnani and A. Goli, Ecotoxicol. Environ. Saf., 2018, 163, 372–381 CrossRef CAS.
- V. Londhe and R. Shirsat, AAPS PharmSciTech, 2018, 19, 1392–1400 CrossRef CAS.
- F. G. B. Los, A. A. F. Zielinski, J. P. Wojeicchowski, A. Nogueira and I. M. Demiate, Food Anal. Methods, 2019, 12, 148–159 CrossRef.
- B. Singh, H. K. Sharma and B. C. Sarkar, J. Food Sci. Technol., 2012, 49, 294–308 CrossRef CAS.
- S. Czemmel, J. Hoell, R. Loyola, P. Arce-Johnson, J. Antonio Alcalde, J. Tomas Matus and J. Bogs, Front. Plant Sci., 2017, 8, 1–15 CrossRef.
- Z.-L. Sheng, P.-F. Wan, C.-L. Dong and Y.-H. Li, Ind. Crops Prod., 2013, 43, 778–786 CrossRef CAS.
- Y.-h. Zhou, C.-g. Xu, Y.-b. Yang, X.-x. Xing, X. Liu, Q.-w. Qu, W.-y. Ding, G. s. Bello-Onaghise and Y.-h. Li, Front. Microbiol., 2018, 9, 1–10 CrossRef.
- S. Teixeira, C. Delerue-Matos and L. Santos, Sci. Total Environ., 2019, 646, 168–176 CrossRef CAS.
- P. Sharma, L. Singh and N. Dilbaghi, J. Hazard. Mater., 2009, 164, 1024–1029 CrossRef CAS.
- D. C. Montgomery, Environ. Prog. Sustainable Energy, 2013, 32, 8–10 CrossRef.
- W. Ding, Y. Zhou, Q. Qu, W. Cui, B. O. God'spower, Y. Liu, X. Chen, M. Chen, Y. Yang and Y. Li, Front. Pharmacol., 2018, 9, 1–11 CrossRef.
- I. M. Liu and S. J. Sheu, Am. J. Chin. Med., 1989, 17, 179–187 CrossRef CAS.
- A. Tang, L. Ning and N. Cheng, Acta Med. Sin., 1994, 1–4 Search PubMed.
- Q. Zhang, Y.-L. Wang, D. Gao, L. Cai, Y.-Y. Yang, Y.-J. Hu, F.-Q. Yang, H. Chen and Z.-N. Xia, Pharm. Biol., 2018, 56, 67–75 CrossRef CAS PubMed.
- K. Tang, W. Q. Wang and L. I. Biao, Chin. J. Exp. Tradit. Med. Formulae, 2013, 1–3 Search PubMed.
- N. U. o. T. C. M. N. L. Tulin, C. Mao and D. Ye, Optimization of Processing Technique of Sparganium stoloniferum by Orthogonal Experimental Design, Chin. Tradit. Herb. Drugs, 2000, 1–3 Search PubMed.
- G. D. Vyavahare, R. G. Gurav, P. P. Jadhav, R. R. Patil, C. B. Aware and J. P. Jadhav, Chemosphere, 2018, 194, 306–315 CrossRef CAS.
- Y. Q. Xiao, L. Li, Y. Liu, Y. L. Ma and D. R. Yu, China J. Chin. Mater. Med., 2016, 41, 24–27 Search PubMed.
- Y. Y. Xu, H. Cai, G. Cao, Y. Duan, K. Pei, S. C. Tu, J. Zhou, L. Xie, D. D. Sun, J. Y. Zhao, J. Liu, X. Q. Wang and L. Shen, J. Chromatogr., B, 2018, 1083, 110–123 CrossRef CAS PubMed.
- Y. S. Chen, Y. B. Shen, X. Fu, A. M. Abbasi and R. Yan, Int. J. Food Sci. Technol., 2017, 52, 1820–1827 CrossRef CAS.
- J. W. Tak, B. Gupta, R. K. Thapa, K. B. Woo, S. Y. Kim, T. G. Go, Y. Choi, J. Y. Choi, J.-H. Jeong, H.-G. Choi, C. S. Yong and J. O. Kim, AAPS PharmSciTech, 2017, 18, 1125–1134 CrossRef CAS.
- J. Lei, Y. Cui and L. Fang, Shaanxi J. Tradit. Chin. Med., 2015, 1–5 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06224f |
| ‡ These authors have contributed equally to this study and share first authorship. |
| This journal is © The Royal Society of Chemistry 2019 |