Open Access Article
Open Access ArticleMicroRNAs and long noncoding RNAs: new regulators in cell fate determination of mesenchymal stem cells
Zixiang Wu
 abc,
Shujing Liang
abc,
Shujing Liang
 abc,
Wenyu Kuai
abc,
Lifang Hu
abc,
Wenyu Kuai
abc,
Lifang Hu
 *abc and
Airong Qian
*abc
*abc and
Airong Qian
*abc
aLaboratory for Bone Metabolism, Key Laboratory for Space Biosciences and Biotechnology, School of Life Sciences, Northwestern Polytechnical University, Xi'an 710072, China. E-mail: wuzx@mail.nwpu.edu.cn; liangsj@mail.nwpu.edu.cn; kuaiwenyu@mail.nwpu.edu.cn; hulifang@nwpu.edu.cn; qianair@nwpu.edu.cn
bResearch Center for Special Medicine and Health Systems Engineering, School of Life Sciences, Northwestern Polytechnical University, Xi'an 710072, China
cNPU-UAB Joint Laboratory for Bone Metabolism, School of Life Sciences, Northwestern Polytechnical University, Xi'an 710072, China
First published on 14th November 2019
Abstract
Mesenchymal stem cells (MSCs) are multipotent cells that are able to differentiate into numerous cell types, including well-known inherent osteoblasts, adipocytes, and chondrocytes, and other cell types, such as hepatocytes, cardiomyocytes and nerve cells. They have become a favorite source of cell-based therapy. Therefore, knowing the mechanism that determines the cell fate of MSCs is important not only for deep understanding of the MSC function but also for the manipulation of MSCs for clinical application. Recently, studies have demonstrated that microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), the two best studied noncoding RNAs, show key roles in cell fate determination of MSCs by functioning as vital regulators of their target gene expression or signaling transduction. Here, we summarize the characteristics of miRNAs and lncRNAs, and review the recent advances proving their profound involvement in determining the cell fate of MSCs to inherent osteoblast, adipocyte, and chondrocyte cells, and to several key cell types including hepatocytes, cardiomyocytes and nerve cells. This will provide researchers with a deep understanding of the role of miRNAs and lncRNAs in MSCs and provide guidance for future research.
1 Introduction
Mesenchymal stem cells (MSCs), with strong self-renewal ability and plasticity, have the potential to differentiate into multiple cell types, including well-known inherent osteoblasts, adipocytes, and chondrocytes, and other cell types such as hepatocytes, cardiomyocytes and nerve cells.1 Possessing multipotency, MSCs have been the favorite source of cell therapy and stem cell therapy and have attracted more and more researcher attention for treating various diseases, such as repairing ischemic heart, bone, and skeletal muscle.2–4 Therefore, uncovering the mechanism that regulates MSC differentiation or cell fate determination is essential for both understanding MSC function and their clinical application.MiRNAs and lncRNAs are two kinds of noncoding RNA, which are transcribed from DNA but do not encode proteins.5 MiRNAs and lncRNAs are not junk sequence but emerge as powerful regulators of gene expression. They are best studied and have been shown to play critical roles in diverse cellular processes, including cell proliferation, cell differentiation and apoptosis.6,7 Recently, miRNAs and lncRNAs have been demonstrated to play important regulatory roles in determining cell fate of MSCs.
In this review, we discuss characteristics of miRNAs and lncRNAs and review recent advances proving their profound involvement in determining cell fate of MSCs to inherent osteoblast, adipocyte, and chondrocyte, and to several key cell types including hepatocyte, cardiomyocyte and nerve cell. This review will provide researchers deep understanding of the role of miRNAs and lncRNAs in MSCs, and provide novel targets for disease treatment.
2 MicroRNAs (miRNAs)
2.1 Characteristics of miRNAs
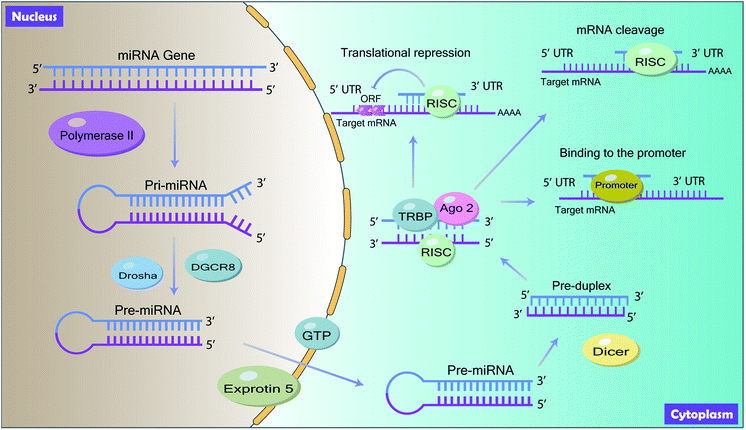
MiRNAs are noncoding, ∼22 nucleotides single-stranded RNAs that are involved in the regulation of gene expression to coordinate multiple biological processes.8,9 They are ubiquitous, ranging from virus10 to human tissues11 and emerge as key post-transcriptional regulators of gene expression by suppressing target messenger RNAs (mRNAs) translation efficiency or degrading target mRNAs.12,13MiRNAs are ubiquitous in animal genomes and they are mainly derived from the introns of the pre-mRNA or the genetic interval of the genome through transcription.14 Mature miRNAs are generated through a series of processes (Fig. 1). Firstly, the primary miRNA (pri-miRNA) with hundreds of nucleotides, which contains mature miRNA sequences, is first transcribed from the introns or the genetic interval of DNA by RNA polymerase II in the nucleus.15 Following, the pri-miRNA is trimmed twice by the RNase III Drosha with the double-stranded RNA binding protein DiGeorge Syndrome Critical Region 8 (DGCR8) to produce precursor miRNA (pre-miRNA) with approximately 60 to 100 nucleotides coil.16 Then, the pre-miRNA is exported from the nucleus to the cytoplasm by Ran-GTP dependent exportin-5 and is cleaved into mature miRNA by RNase III endonuclease enzyme Dicer. Finally, in the cytoplasm, mature miRNA is assembled into RNA-Induced Silencing Complex (RISC) with the interaction of TAR RNA-binding protein (TRBP) and Argonaute 2 (Ago 2) to target the 3′ untranslated regions (3′ UTR) of its target mRNA.17
2.2 Action mode of miRNAs
By interacting with target mRNA, miRNA regulates gene expression and induces functional changes. Generally, miRNAs inhibit the translation process of its target mRNA by binding to the 3′ terminal UTR of the target mRNA. The 5′ end of miRNA is known as a seed region and miRNA targets the 3′ UTR of mRNA by complementary base pairing to mediate post-transcriptional gene silencing18 (Fig. 1). According to the base pairing degree, mRNA is either degraded or inhibited by miRNAs.19 Besides, studies have shown that miRNAs can also bind to the promoter, coding region and 5′ UTR20 (Fig. 1).3 Long non-coding RNAs (lncRNAs)
3.1 Characteristics of lncRNAs
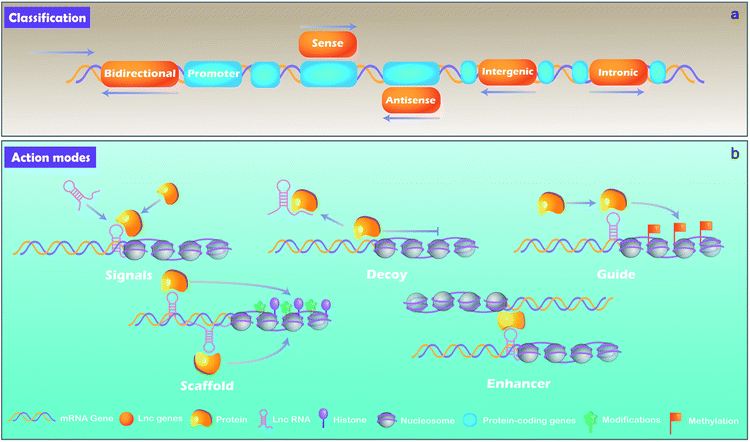
lncRNAs, which are distinguished from small RNA with more than 200 nucleotides, are another class of noncoding RNA.21 Rather than the “noise” considered previously, lncRNAs are critical regulators involved in almost a variety of biological processes, including X chromosome silencing, epigenetic and transcriptional activation, transcriptional interference and nuclear trafficking.22 They are divided into 5 main types depending on the position and orientation of the transcription site relative to adjacent gene.23,24 (1) Sense lncRNAs. The transcription direction of this type lncRNA is the same as the direction of transcription of its adjacent mRNA. (2) Antisense lncRNAs. They are endogenous RNAs of organisms and are often located at both ends of protein-coding genes. (3) Intergenic lncRNAs. They are discrete transcription units that intervene in protein-coding loci. (4) Intronic lncRNAs transcript completely derived from introns of protein-coding genes. (5) Bidirectional lncRNAs. The lncRNAs can be simultaneously transcribed from the same and opposite directions as the adjacent mRNA.lncRNA can be derived from promoters, enhancers, and intergenic regions with eukaryotic RNA processing.25 lncRNAs are generally produced through transcription mediated by RNA polymerase II and the transcriptional activators such as switching defective/sucrose non-fermenting (SWI/SNF).26 After transcription, lncRNAs are mostly spliced, capped and polyadenylated in a similar way as mRNA.25,27
3.2 Action mode of lncRNAs
By binding to protein partners, lncRNAs possess regulatory capacities. Although there are still limited knowledge of lncRNAs, studies have demonstrated several main action modes of lncRNAs, acting as molecular signal, decoy, guide, scaffold, and enhancer (Fig. 2).28 (1) Molecular signal: under different stimulation conditions and signaling pathways, lncRNAs are specifically transcribed and serve as signal molecules in the transmission of specific signaling pathways.29 Some lncRNAs possess regulatory functions while others are merely by-products of transcription.27 (2) Decoy: the action mode of lncRNA is molecular sink during cellular processes.7 After lncRNAs transcribed, they will bind to transcription factors directly and the function of such transcription factors is blocked, thereby regulating downstream gene transcription.30 In short, signals and decoys control the activation or suppression of gene.31 (3) Guides: lncRNAs can serve as a guide that recruits protein complex and nuclear proteins to chromatin sites to regulate downstream gene expression. Evidences demonstrate that lncRNAs can guide changes in gene expression either in cis (on neighboring genes) or in trans (distantly located genes). lncRNAs such as air and eRNAs (enhancer RNAs) play a role in cis by diffusing from transcriptionally controlled focus sequence elements such as enhancers or promoters, while lncRNAs such as HOTAIR and lincRNA-p21 function in trans requiring some interacting partners to be properly localized to the action sites.27 (4) Scaffold: as a “central scaffold”, scaffolds can bring multiple related transcription factors that can be bound to this lncRNA, and they can form lncRNA-ribonucleoprotein (RNPs) complexes. This form can realize the intersection and integration of information between different signal paths, which is beneficial for the body/cell to quickly feedback and adjust external signals,27,32 such as HOTAIR,33 7SL.34 (5) Enhancers: some lncRNAs act as enhancers to participate in the regulation of genes, and stabilize the chromatin loop.35 In addition, it can enhance the inflammatory response with a number of ways, such as increasing the transcription of pro-inflammatory cytokines by enhancing inflammatory signals, such as NF-κB signaling.364 MiRNAs and cell fate determination of MSCs
MSCs are multipotent stem cells with the capability of self-renewing and differentiating into multiple cell lineages, including osteoblasts, adipocytes and chondrocytes.37 Functioning as critical regulators, numerous miRNAs have been demonstrated to play important roles in regulating the differentiation of MSCs.38,394.1 MiRNAs and osteogenic differentiation of MSCs
Evidences have shown that miRNAs regulate the osteogenic differentiation of MSCs and multiple signaling pathways are involved in, such as Wnt signaling,40 BMP signaling,41,42 and Notch signaling.43 Wnt/β-catenin signaling pathway plays a critical role in cell fate determination of MSCs. Wnt ligands function by interacting 7-transmembrane domain-spanning frizzled (FZD) receptor and LRP5/6 coreceptors, thereby stabilizing β-catenin by preventing it from phosphorylation and degradation, which facilitates β-catenin translocate into the nucleus to regulate various target genes expression. Wnt/β-catenin signaling pathway has become a prominent channel during miRNA-mediated osteogenic differentiation of MSCs.44 Wang et al. found that increased miR-346 significantly promoted osteogenic differentiation of hMSCs by inhibiting the expression of glycogen synthase kinase-3 (GSK-3β).45 MiR-346 directly bound to the 3′-UTR of GSK-3β mRNA, which resulted in decreased level of GSK-3β, and increased osteogenic differentiation.45 Recent studies reveal that miRNAs also regulate osteogenic differentiation and bone formation through bone morphogenetic proteins (BMP)/Smads signaling pathway. In fact, BMPs are potent inducers of osteoblastogenesis during bone development.46 They undergo site-specific phosphorylation and activate Smad-1, Smad-5 and Smad-8, which are transcription factors and enter into the nucleus to regulate the expression of runt related transcription factor 2 (Runx2), osterix (OSX), osteocalcin (OCN) and osteopontin (OPN) to regulate osteoblast differentiation.47 Liu et al. have reported that miR-155 inhibits osteogenic differentiation induced by BMP9 both in mMSCs and hMSCs with reduced expression of osteogenesis-related genes.48 TGF-β acts as another important signaling pathway that inhibits osteoblast differentiation and works with BMP2 to form a TGF-β/BMP signaling pathway. Bhushan et al. have found that miR-181a binds directly to Rgs4 and Gata6 and promotes osteoblast differentiation by repressing the TGF-β signaling pathway.49 Besides, miR-21 promotes BMP9-induced osteogenic differentiation of MSCs by inhibiting the BMP/Smads signaling pathway under the interaction of Smad7.50 These findings suggest that different miRNAs play different roles in regulating MSCs differentiation. The difference may be due to the different targets of different miRNAs. Moreover, the role of miRNAs in osteogenic differentiation through the Notch signaling pathway is receiving increasing attention.51 MiR-34a inhibits hMSCs differentiation by siRNA-mediated reduction of the expression of Jagged1 (JAG1), a ligand of the Notch1 signaling pathway. The inhibition of miR-34a becomes a potential new therapeutic strategy for enhancing bone formation.52 The overexpression of miR-130a and miR-27b have been shown to promote osteogenic differentiation by targeting PPARγ and enhancing the expression of Runx2 and OSX genes.53 Recently, Jin et al. have demonstrated that miR-145 is able to decrease osteogenic differentiation of human jaw bone marrow mesenchymal stem cells by targeting semaphorin 3A.54 Moreover, miR-21,55 miR-148b-3p,56 Mmu-miR-185,57 and miR-381![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 58 have also show important roles in regulating osteogenic differentiation of hMSCs. Thus, numerous miRNAs play important role in regulating osteogenic differentiation of MSCs, showing either positive or negative regulatory role.
58 have also show important roles in regulating osteogenic differentiation of hMSCs. Thus, numerous miRNAs play important role in regulating osteogenic differentiation of MSCs, showing either positive or negative regulatory role.
4.2 MiRNAs and adipogenic differentiation of MSCs
Adipogenesis is another differentiation direction of MSCs and several key transcription factors are involved, including peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT/enhancer binding proteins (C/EBPs). MiRNAs also show key role in regulating adipogenic differentiation of MSCs. MiR-27a inhibits adipocyte differentiation by targeting 3′-UTR of PPARγ in a sequence-specific manner, and miR-223 regulates adipocyte differentiation via a C/EBPs/miR-223/FGFR2 regulatory feedback loop.59,60 Recent study has shown that miR-214-5p promotes adipogenic differentiation but inhibits osteogenic differentiation of hBMSCs. Overexpression of miR-214-5p promoted adipogenic differentiation, inhibited the mRNA expression of osteogenic genes ALP, Runx2, OCN and collagen α-1 (I) chain (COL1A1), and suppressed the level of transforming growth factor-β (TGF-β), phosphorylated-Smad2 (p-Smad2) and collagen type IV α1 chain (COL4A1) in hBMSC. These findings suggest that miR-214-5p promotes the adipogenic differentiation of hBMSCs by regulating TGF-β/Smad2/COL4A1 signaling pathway.61 Moreover, miRNAs also function through mitogen-activated protein kinase (MAPK) signaling to regulate adipogenic differentiation and commitment. Jin et al. have reported that MAPK7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) plays a major role in guiding miR-24 and miR-143 to promote 3T3-L1 adipogenesis differentiation and commitment.62 MAP2K5 (mitogen-activated protein kinase kinase 5), which is another novel member of the MAPK signaling pathway, has the positive effect of modulating adipose tissue-derived stromal cells (ADSC) differentiation by miRNA-143.63 In addition, miR-148a and miR-210 inhibit Wnt signaling pathway by targeting Wnt1 and Tcf7l2 (transcription factor 7 like 2) respectively, and ultimately promote adipogenic differentiation of ADSCs.64,65 Therefore, Wnt signaling pathway is also a crucial signaling pathway that targets miRNA-mediated adipogenic differentiation.
plays a major role in guiding miR-24 and miR-143 to promote 3T3-L1 adipogenesis differentiation and commitment.62 MAP2K5 (mitogen-activated protein kinase kinase 5), which is another novel member of the MAPK signaling pathway, has the positive effect of modulating adipose tissue-derived stromal cells (ADSC) differentiation by miRNA-143.63 In addition, miR-148a and miR-210 inhibit Wnt signaling pathway by targeting Wnt1 and Tcf7l2 (transcription factor 7 like 2) respectively, and ultimately promote adipogenic differentiation of ADSCs.64,65 Therefore, Wnt signaling pathway is also a crucial signaling pathway that targets miRNA-mediated adipogenic differentiation.
4.3 MiRNAs and chondrogenic differentiation of MSCs
Numerous evidence suggests that miRNAs are indispensable for chondrocyte differentiation and cartilage function.66 Yan et al. have revealed that 44 miRNAs are involved in cartilage differentiation by using microRNA array detection. Further, two miRNAs out of seven differentially-expressed miRNAs, called miR-29a and miR-29b, have been revealed to directly target 3′ UTR of col2α1 encoding type II collagen α1, suggesting them as key regulators for chondrogenic differentiation of mMSCs.67 Moreover, the expression of miR-29a and miR-29b is demonstrated to be regulated by SRY-related high mobility group-box gene 9 (Sox9), a key transcription factor for chondrocyte.67 Guerit et al. have also found that the expression of miRNA-574-3p is regulated by Sox9 and miRNA-574-3p inhibits chondrogenic differentiation of hMSCs.68 However, some studies show that Sox9 is a target of miRNAs to regulate chondrogenic differentiation of MSCs. MiR-495 suppresses chondrogenic differentiation of hMSCs by directly binding to 3′ UTR of Sox9 and reducing Sox9 expression.69 These findings reveal the complexity of miRNAs function. Furthermore, both miR-181a and miR-181b negatively regulate cartilage differentiation and function in maintaining cell integrity. Besides, their negative feedback regulation has gradually become an effective therapeutic target for cartilage-related diseases.70,71 Zhong et al. have reported that miR-377 is directly bound up with chondrogenesis through regulating TGFBR2 expression and enhancement of miR-377 modulates cartilage-specific genes expression, providing an important clue for research in arthritis pathogenesis and new treatment method for arthritis.72 Therefore, miRNAs are key regulators for chondrogenic differentiation of MSCs and can be novel targets for treatment of cartilage-related diseases.4.4 MiRNAs and other cell types differentiation of MSCs
Besides being involved in the regulation of the inherent multipotent differentiation potential of MSCs to osteoblasts, adipocytes and chondrocytes, miRNAs also show key role in regulating the differentiation of MSCs to other cell types, such as hepatocytes, cardiomyocytes and nerve cells. Cui et al. have screened six overexpressed miRNAs (miR-1246, miR-1290, miR-148a, miR-30a, miR-424 and miR-542-5p) during hepatic differentiation of human umbilical cord lining-derived mesenchymal stem cells (hMSCs). They found that downregulation of any one of the six miRNAs inhibited hepatocyte growth factor (HGF)-induced hepatic differentiation and ectopic overexpression of seven miRNAs (miR-1246, miR-1290, miR-148a, miR-30a, miR-424, miR-542-5p and miR-122) together stimulated hMSC conversion into functionally mature induced hepatocytes.73 Moreover, the transplantation of the induced hepatocytes into mice with liver injury not only improved liver function, but also restored injured livers.73 These findings indicate the capability of miRNAs converting hMSCs to hepatocytes. Chen et al. have also reported the therapeutic potential of miRNA in treating acute myocardial infarction by modulating MSC function.74 They found that overexpression of miR-133 decreased apoptosis of rat MSCs (rMSCs) and enhanced the therapeutic effect of rMSCs in a rat model with myocardial infarction.74 MicroRNA let-7f-5p has been shown to target partitioning defective 6 homologue alpha (Par6α), a component of the Par3/Par6/aPKC complex, which is essential for axon specification during neuronal development. Downregulation of let-7f-5p in rat BMSCs induces the cells into neuron-like cells by directly targeting Par6α.75 More recently, key miRNAs and related pathways involved in neural differentiation of rBMSCs have been revealed by Wei et al.76 They identified 83 significantly differentially expressed miRNAs during neural differentiation of rBMSCs and showed that Hippo, Wnt, TGF-β and Hedgehog signaling pathways were involved in the neural differentiation.76 All these findings demonstrate the key role of miRNAs in regulating differentiation of MSCs into other cell types rather than osteoblast, adipocyte, and chondrocyte and provide potential targets for MSCs-based cell therapy.5 LncRNAs and MSC differentiation
Recently, with the fast advances in lncRNAs research, evidences have shown that lncRNAs are widely involved in the growth and development by regulating the fate of MSCs.775.1 LncRNAs and osteogenic differentiation of MSCs
Possessing complicated characteristics and action modes, lncRNAs show a more complex regulatory role in osteogenic differentiation of MSCs compared with miRNAs. lncRNAs regulate osteogenic differentiation through regulating signaling pathways, acting as “miRNA sponge”, targeting miRNAs, or functioning in other ways. Jia et al. have reported that canonical Wnt signaling pathway is activated by ANCR-RNAi, and low level of ANCR promotes the process of osteogenic differentiation in periodontal ligament stem cells (PDLSCs).78 By acting as “miRNA sponge”, lncRNAs play important role in mediating osteogenic differentiation of MSCs. It has been demonstrated that the expression level of lncRNA-H19 gradually increases during osteogenic differentiation of both hBMSCs and mBMSCs. Further investigation shows that lncRNA-H19 promotes osteogenic differentiation by sponging various miRNAs, including miR-188,79 miR-138,80 and miR-675.81 Moreover, lncRNAs also function by targeting miRNAs. LncRNAs HULC and MALAT1 promote osteoblast differentiation of hBMSCs either by targeting miR-195 or by targeting miRNA-143.82,83 LncRNA-ANCR inhibits osteogenic differentiation of PDLSCs by targeting miRNA-758, which has a role in promoting Notch2 expression by targeting 3′-UTR of Notch2.84 Other findings show different functional ways of lncRNAs in regulating osteogenic differentiation of MSCs. LncRNA TUG1 promotes osteogenic differentiation of PDLSCs through interacting with lin-28 homolog A (Lin28A), which is a potential target for TUG1-mediated bone formation analyzed by bioinformatics.85 lncRNA AK141205 positively regulates osteogenic differentiation of mMSCs by promoting CXC chemokine ligand-13 (CXCL13) expression via acetylation of H4 histone, providing a potential therapeutic target of disease treatment using MSCs.86 Zhuang et al. have found that lncRNA MEG3 promotes osteogenic differentiation of hMSCs partly by activating BMP4 transcription,87 while Jin et al. have shown that knockdown of lncRNA myocardial infarction-associated transcript (MIAT) promotes osteogenic differentiation of human adipose stem cells (hASCs) both in vitro and in vivo, suggesting that lncRNA MIAT negatively regulates osteogenic differentiation.88 These findings provide reference for studying the mechanism by which lncRNAs mediate osteogenic differentiation of MSCs and provide new therapeutic targets for treating diseases.5.2 LncRNAs and adipogenic differentiation of MSCs
In most cases, lncRNAs act in two opposite processes including osteogenic differentiation and adipogenic differentiation. Therefore, some lncRNAs involved in the regulation of osteogenic differentiation of MSCs are also involved in the regulation of adipogenic differentiation of MSCs.89,90 LncRNA H19 regulates adipogenic differentiation by targeting miR-675 and is significantly down-regulated in human BMSCs differentiated into adipocytes through a CTCF/H19/miR-675/HDAC axis.91 Similarly, lnc-ADNCR has been shown to inhibit adipogenic differentiation by competing with miRNA-204 which is also called “miRNA sponge”. MiRNA-204 inhibits adipocyte differentiation by suppressing PPARγ activity.92 LncRNA Gm15290 facilitates PPARγ- and C/EBPa-induced fat deposition by targeting miR-27b and results in a significant increase in body weight in mice.93 Li et al. found that lnc-GAS5 overexpression negatively regulated the formation of fat cells. The mechanism is that lnc-GAS5 acts as an endogenous competitive RNA to inhibit the binding sites of miR-18a. This process reduces the expression of connective tissue growth factor (CTGF), thereby negatively modulating the adipogenic differentiation of MSCs.94 In addition, lincRNA-Para1 activates PPARγ to promote adipogenic differentiation by interacting with paraspeckle component and hnRNP-like RNA binding protein 14 (RBM14/NCoAA).95 Recently, Gernapudi and colleagues have identified a novel miR-140/lncRNA NEAT1 signaling network necessary for adipogenesis by using adipocyte-derived stem cells (ADSCs) from wild-type and miR-140 knockout mice.96 They found that miR-140 knockout ADSCs showed dramatically decreased adipogenic capabilities associated with down-regulation of NEAT1 expression. Further, they demonstrated that miR-140 promoted NEAT1 expression by physically binding to it, which promotes adipogenic differentiation of mADSCs.96 LncRNA HOXA11-AS1 and MIR31HG also show promotion effect on adipogenic differentiation of hADSCs.97,98 All these findings reveal the important role of lncRNAs in regulating adipogenic differentiation of MSCs and provide potential therapy targets for diseases, such as obesity.5.3 LncRNAs and chondrogenic differentiation of MSCs
Compared with a large number of lncRNAs involved in osteogenic differentiation and adipogenic differentiation of MSCs, there are few studies on the role of lncRNAs in the regulation of chondrogenic differentiation of MSCs. It has been reported that Sox4 promotes proliferation and chondrogenesis of human Synovium-derived MSCs (SMSCs) by up-regulating lncRNA DANCR.99 Further study has revealed that lncRNA DANCR promotes proliferation and chondrogenesis of human SMSCs by targeting myc, Smad3, and STAT3 to regulate their stability.100 As a transcription factor of the same family, SOX9, which plays a role in regulating cartilage formation under the mediation of lncRNA-ROCR. In the absence of ROCR, SOX9 induction was significantly abolished, while overexpression of SOX9 promoted differentiation of MSCs into chondrocytes. Therefore, the important role of lncRNA-ROCR in chondrocyte biology has been revealed.101 Apart from this, studies have found that lncRNA-ZBED3-AS1 can promote cartilage formation by up-regulating the expression of zbed3. This pathway acts primarily through the Wnt/β-catenin pathway, so the Wnt inhibitor Dickkopf-1 (DKK1) reverses the stimulatory effect of ZBED3-AS1 on cartilage formation.102 Recently, Pang et al. have revealed the role of lnRNA H19 in regulating chondrogenic differentiation of ADSCs.103 They found that overexpression of lnRNA H19 in ADSCs induced differentiation towards chondrocytes by competing miRNA regulation of STAT2.103 All these findings gradually uncover the role of lncRNAs in chondrogenic differentiation and provide potential targets for chondrocyte-related diseases.5.4 LncRNAs and other cell types differentiation of MSCs
There are emerging evidences illustrating the regulatory role of lncRNAs in differentiation of MSCs into other cell types, although they are still rare in comparison to the studies on the three well-known osteogenic, adipogenic and chondrogenic differentiation. Wang et al. have reported that lncRNA MIAT promotes rBMSCs differentiation into endothelial cells by regulating vascular endothelial growth factor (VEGF) via targeting miR-200a, which restores erectile dysfunction in rat.104 Sun and colleagues have demonstrated another lncRNA, lncRNA MEG3, involved in regulating the differentiation of rBMSCs to endothelial cells.105 They showed that down-regulation of lncRNA MEG3 promoted BMSCs differentiation into endothelial cells by decreasing VEGF expression via facilitating forkhead box protein M 1 (FOXM1) ubiquitination, resulting in the repair of erectile dysfunction.105 Recently, Tan et al. used HGF to induce rMSCs to differentiate into hepatocytes. They found that β-catenin-coordinated lncRNA MALAT1/miR-217 axis up-regulated the expression of zinc finger E-box-binding homeobox 1 (ZEB-1) and further enhanced the telomerase activity by regulating telomerase reverse transcriptase (TERT) in rMSCs differentiating into hepatocytes.106 Moreover, lncRNA Braveheart can facilitate mMSCs differentiation into cardiomyocytes by magnifying the expression of cardiac-specific transcription factors and epithelial–mesenchymal transition (EMT)-associated genes, providing new clue for cell-mediated cardiomyocyte regeneration using lncRNA.107 Moreover, lncRNA H19 regulates the differentiation of hMSCs into nerve cells through the miRNA-675/IGFR axis.108 Wu et al. have revealed the expression patterns of lncRNAs in rBMSCs during their differentiation into neural cells.109 All these findings show the key involvement of lncRNAs in differentiation of MSCs to other cell types besides osteoblast, adipocyte and chondrocyte.6 Conclusion and perspectives
MSCs have become a favorite source of cell-based therapy for their multipotency and plasticity to differentiate into multiple cell types, including well-known inherent osteoblasts, adipocytes, and chondrocytes, and other cell types such as hepatocytes, cardiomyocytes and nerve cells. Therefore, uncovering the mechanism that determining cell fate of MSCs is important for better understanding MSCs and manipulating them for clinical application. Here, we highlight the key role of miRNAs and lncRNAs in cell fate determination of MSCs, not only focusing on the well-known inherent osteoblast, adipocyte and chondrocyte differentiation, but also introducing the differentiation of MSCs to hepatocyte, cardiomyocyte and nerve cell, respectively. Based on the numerous recent advances, we know that miRNAs and lncRNAs are key regulators in determining cell fate of MSCs by modulating their target gene expression or signaling transduction. Many molecules and signaling pathways (e.g. Wnt/β-catenin signaling, BMP signaling, and Notch signaling) have been demonstrated to be involved in (Table 1). Besides, there are interactions between some miRNAs and lncRNAs in cell fate determination of MSCs. Thus, the role of miRNAs and lncRNAs in MSCs attracts more and more attention.| Type | Name | Cell fate determination of MSCs | Main mechanism | Key references |
|---|---|---|---|---|
| MiRNA | miR-346 | Promotes osteogenic differentiation | Suppresses the expression of GSK-3β protein and activates the Wnt/β-catenin pathway | 45 |
| miR-155 | Inhibits osteogenic differentiation | Targets Runx2 and BMPR2, and downregulates their expression | 48 | |
| miR-181a | Promotes osteoblast differentiation | Targets the negative regulator of Tgfbi and TβR-I/Alk5 to degrade it | 49 | |
| miR-34a | Inhibits osteogenic differentiation | Reduces the expression of JAG1 of the ligand for Notch1 | 52 | |
| miR-130a and miR-27b | Promote osteogenic differentiation | Promote the expression of Runx2 and Osterix and inhibit the expression of PPARγ | 53 | |
| miR-145 | Inhibits osteogenic differentiation | Targets semaphorin 3A | 54 | |
| miR-21 | Promotes osteogenic differentiation | Inhibits the BMP/Smads signaling pathway under the interaction of Smad7. Promotes maxillofacial bone regeneration via the PTEN/PI3K/Akt/HIF-1α pathway | 50 and 55 | |
| miR-148b-3p | Promotes osteogenic differentiation | Increases alkaline phosphatase and collagen type I | 56 | |
| Mmu-miR-185 | Promotes osteogenic differentiation | Via the Bgn-mediated BMP/Smad pathway | 57 | |
| miR-381 | Inhibits osteogenic differentiation | Upregulates PPARγ via suppressing Wnt signaling pathway | 58 | |
| miR-27a | Inhibits adipocyte differentiation | Targets 3′ UTR of PPARγ to degrade it | 52 | |
| miR-223 | Regulates adipocyte and osteoblast differentiation | Targets a novel C/EBPs/miR-223/FGFR2 regulatory feedback loop | 53 | |
| miR-214-5p | Promotes adipogenic differentiation; attenuates the osteogenic differentiation | Promotes adipogenic differentiation by regulating TGF-β/Smad2/COL4A1 signaling pathway; inhibits ALP, Runx2, OC and COL1 expression | 61 | |
| miR-143 | Promotes adipogenesis differentiation | Targets the MAPK7 to repress it | 62 | |
| miR-148a | Promotes adipogenic differentiation | Represents a CREB-modulated miRNA that acts to repress Wnt1 | 57 | |
| miR-210 | Promotes adipogenic differentiation | Promotes adipogenesis by repressing Wnt signaling through targeting Tcf7l2 | 58 | |
| miR-29a and miR 29b | Inhibits cartilage differentiation | Target 3′ UTR of Col2a1 encoding type II collagen | 67 | |
| miR-495 and miRNA-574-3p | Suppress chondrogenic differentiation | Target Sox9 | 68 and 69 | |
| miR-181a | Suppress cartilage differentiation | Suppresses the expression of two genes, CCN1 and a major cartilaginous proteoglycan, aggrecan; | 63 | |
| miR-181b | Suppress cartilage differentiation | Reduces MMP-13 expression while inducing type II collagen expression | 64 | |
| miR-377 | Promotes chondrogenic differentiation | Regulates TGFBR2 expression and modulates the expression of cartilage-specific genes such as AGC1 in C-28/I2 | 72 | |
| miR-1246, miR-1290, miR-148a, miR-30a, miR-424 and miR-542-5p | Promote differentiation into hepatocytes | Unknown | 73 | |
| miR-133 | Promotes differentiation into cardiomyocytes | Decreases cell apoptosis | 74 | |
| let-7f-5p | Promotes differentiation into nerve-like-cells | Directly targets Par6α | 75 | |
| LncRNA | lncRNA-ANCR | Inhibits osteogenic differentiation | Targets miR-758 and miR-758, regulates Notch2 expression by targeting 3′-UTR of Notch2, Wnt signaling pathway is activated by lncRNA-ANCR/RNAi | 78 and 84 |
| lncRNA H19 | Promotes osteogenic differentiation | Mediates LCoR through sponging miR-188 | 79 | |
| lncRNA-HULC and lncRNA-MALAT1 | Promote osteoblast differentiation | lncRNA-HULC acts through sponging miR-195 and enhances activation of Wnt/β-catenin and p38MAPK pathway; lncRNA – MALAT1 regulates Osx expression through targeting miR-143 | 82 and 83 | |
| TUG1 | Promotes osteogenic differentiation | Promotes osteogenic differentiation though suppressing Lin28A | 85 | |
| lncAK141205 | Facilitates osteogenic differentiation | LncAK141205 promotes CXCL13 expression by acetylation of H4 histone | 86 | |
| lncRNA MIAT | Inhibits osteogenic differentiation | Unknown | 88 | |
| lncRNA H19 | Inhibits adipogenic differentiation | Sponge microRNA-675 and miR-675 targetes the 3′ UTRs of the histone deacetylase (HDAC) 4–6 transcripts | 91 | |
| lnc-ADNCR | Inhibits adipogenic differentiation | Through sponging miR-204 and promotes the expression of SIRT1, then the NCoR and SMART were blocked to repress PPARγ activity | 92 | |
| lncRNA Gm15290 | Promotes adipogenic differentiation | Sponges miR-27b to promote PPARγ activity | 93 | |
| lnc-GAS5 | Inhibits adipogenic differentiation | Acts as a sponge for miR-18a and suppresses CTGF protein translation | 94 | |
| lincRNA-Para1 | Promote adipogenic differentiation | Activates the PPARγ through interacting with the RBM14/NCoAA | 95 | |
| lncRNA NEAT1 | Promotes adipogenic differentiation | Interacts with NEAT1 and lead to increase NEAT1 expression | 96 | |
| lncRNA HOXA11-AS1 | Promotes adipogenic differentiation | Suppresses the transcription genes of CEBP-α, DGAT2, CIDEC, and perilipin | 97 | |
| lncRNA MIR31HG | Inhibits adipocyte differentiation | Reduces the enrichment of active H3K4me3 and AcH3, FABP4 | 98 | |
| lncRNA-DANCR | Promotes proliferation and chondrogenesis | Upregulates the expression of Smad3 and STAT3 | 100 | |
| lncRNA-ROCR | Promotes proliferation and chondrogenesis | Upregulates SOX9 | 101 | |
| lncRNA-ZBED3-AS1 | Promotes cartilage formation | Increases zbed3 expression | 102 | |
| lncRNA H19 | Promotes chondrogenic differentiation![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
Competes miRNA regulation of STAT2 | 103 | |
| lncRNA MIAT | Promotes differentiation into endothelial cells | Targets miR-200a | 104 | |
| lncRNA MEG3 | Promotes differentiation into endothelial cells | Decreases VEGF expression via facilitating FOXM1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ubiquitination ubiquitination |
105 | |
| lncRNA MALAT1 | Promotes differentiation hepatocytes | β-Catenin-coordinated lncRNA MALAT1/miR-217 axis | 106 | |
| lncRNA Braveheart | Promotes differentiation into cardiomyocytes | Enhances the expression of cardiac-specific transcription factors and EMT-associated genes | 107 | |
| lncRNA H19 | Promotes differentiation into nerve cells | Function through miRNA-675/IGFR axis | 108 |
With the gradual uncovery of the critical regulatory role and underlying mechanism of miRNAs and lncRNAs in regulating cell fate determination of MSCs, more and more aspects of miRNAs and lncRNAs have been known, which make them as potential targets for modulating MSCs or treating disease. However, there are still lots of questions to be answered. How is the miRNA and lncRNA expression regulated during MSCs differentiation? Are there some main sets of miRNAs or lncRNAs involved in cell fate determination of MSCs? As there is interaction between miRNAs and lncRNAs, what is their relationship during MSCs differentiation? Future study should target these kinds of questions to clearly clarify the regulatory mechanism of some specific miRNAs and lncRNAs in differentiation of MSCs. Most important, as many evidences reveal that there are miRNAs networks, lncRNAs networks, or miRNAs-lncRNAs interaction networks involved in the regulation of cell function, physiological and pathological processes, these networks should be identified and their functions should be uncovered in cell fate determination of MSCs, which will be the future research targets.
In summary, miRNAs and lncRNAs are novel and critical regulators for the cell fate determination of MSCs. The uncovery of the molecular mechanisms of miRNAs and lncRNAs determining cell fate of MSCs and the modulation of MSCs by targeting miRNAs or lncRNAs to facilitate clinical application of MSCs are attractive research areas.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (81772017, 31570940), the China Postdoctoral Science Foundation (2018T111099, 2017M610653), Young Talent Fund of University Association for Science and Technology in Shaanxi, China (20170401), The Project Supported by Natural Science Basic Research Plan in Shaanxi Province of China (2018JM3040), the grant (BKJ17J004).References
- M. P. Alfaro, S. Saraswati and P. P. Young, Vitam. Horm., 2011, 87, 39–59 CrossRef CAS PubMed.
- R. Litwinowicz, B. Kapelak, J. Sadowski, A. Kedziora and K. Bartus, Polish Journal of Thoracic and Cardiovascular Surgery, 2018, 15, 196–199 CrossRef PubMed.
- Y. Z. Jin and J. H. Lee, Clinics in Orthopedic Surgery, 2018, 10, 271–278 CrossRef PubMed.
- S. Hamidian Jahromi and J. E. Davies, Stem Cells Transl. Med., 2019, 8, 456–465 CrossRef PubMed.
- J. X. Yang, R. H. Rastetter and D. Wilhelm, Adv. Exp. Med. Biol., 2016, 886, 13–32 CrossRef CAS PubMed.
- S. Crippa, M. Cassano and M. Sampaolesi, Curr. Pharm. Des., 2012, 18, 1718–1729 CrossRef CAS PubMed.
- J. Li, H. Tian, J. Yang and Z. Gong, DNA Cell Biol., 2016, 35, 459–470 CrossRef CAS PubMed.
- J. Chen, M. Qiu, C. Dou, Z. Cao and S. Dong, Drug Dev. Res., 2015, 76, 235–245 CrossRef CAS PubMed.
- T. Moghaddam and Z. Neshati, J. Cell. Biochem., 2019, 120, 14136–14155 CrossRef CAS PubMed.
- D. W. Trobaugh and W. B. Klimstra, Trends Mol. Med., 2017, 23, 80–93 CrossRef CAS PubMed.
- N. Ludwig, P. Leidinger, K. Becker, C. Backes, T. Fehlmann, C. Pallasch, S. Rheinheimer, B. Meder, C. Stahler, E. Meese and A. Keller, Nucleic Acids Res., 2016, 44, 3865–3877 CrossRef CAS PubMed.
- Q. Lin, L. Ma, Z. Liu, Z. Yang, J. Wang, J. Liu and G. Jiang, Oncotarget, 2017, 8, 15961–15970 Search PubMed.
- B. P. Towler, C. I. Jones and S. F. Newbury, Biochem. Soc. Trans., 2015, 43, 1208–1214 CrossRef CAS PubMed.
- N. Shomron and C. Levy, J. Biomed. Biotechnol., 2009, 2009, 594678 Search PubMed.
- Y. Lee, M. Kim, J. Han, K. H. Yeom, S. Lee, S. H. Baek and V. N. Kim, EMBO J., 2004, 23, 4051–4060 CrossRef CAS PubMed.
- V. N. Kim and J. W. Nam, Trends Genet., 2006, 22, 165–173 CrossRef CAS PubMed.
- V. M. King and G. M. Borchert, Methods Mol. Biol., 2017, 1617, 27–37 CrossRef CAS PubMed.
- C. Shin, J. W. Nam, K. K. Farh, H. R. Chiang, A. Shkumatava and D. P. Bartel, Mol. Cell, 2010, 38, 789–802 CrossRef CAS PubMed.
- S. Hosseinpour, Y. He, A. Nanda and Q. Ye, Calcif. Tissue Int., 2019, 105, 223–238 CrossRef CAS PubMed.
- J. O'Brien, H. Hayder, Y. Zayed and C. Peng, Front. Endocrinol., 2018, 9, 402 CrossRef PubMed.
- S. Dahariya, I. Paddibhatla, S. Kumar, S. Raghuwanshi, A. Pallepati and R. K. Gutti, Mol. Immunol., 2019, 112, 82–92 CrossRef CAS PubMed.
- B. K. Dey, A. C. Mueller and A. Dutta, Transcription, 2014, 5, e944014 CrossRef PubMed.
- T. R. Mercer, M. E. Dinger and J. S. Mattick, Nat. Rev. Genet., 2009, 10, 155–159 CrossRef CAS PubMed.
- V. J. Peschansky and C. Wahlestedt, Epigenetics, 2014, 9, 3–12 CrossRef CAS PubMed.
- H. Wu, L. Yang and L. L. Chen, Trends Genet., 2017, 33, 540–552 CrossRef CAS PubMed.
- Y. Hori, Y. Tanimoto, S. Takahashi, T. Furukawa, K. Koshiba-Takeuchi and J. K. Takeuchi, BMC Genomics, 2018, 19, 967 CrossRef CAS PubMed.
- K. C. Wang and H. Y. Chang, Mol. Cell, 2011, 43, 904–914 CrossRef CAS PubMed.
- F. Leti and J. K. DiStefano, Genes, 2017, 8, 207 CrossRef PubMed.
- M. Huarte, M. Guttman, D. Feldser, M. Garber, M. J. Koziol, D. Kenzelmann-Broz, A. M. Khalil, O. Zuk, I. Amit, M. Rabani, L. D. Attardi, A. Regev, E. S. Lander, T. Jacks and J. L. Rinn, Cell, 2010, 142, 409–419 CrossRef CAS PubMed.
- X. Li, Z. Wu, X. Fu and W. Han, Mutat. Res., Rev. Mutat. Res., 2014, 762, 1–21 CrossRef CAS PubMed.
- L. Da Sacco, A. Baldassarre and A. Masotti, Int. J. Mol. Sci., 2012, 13, 97–114 CrossRef CAS PubMed.
- L. Yang, H. Wang, Q. Shen, L. Feng and H. Jin, Cell Death Dis., 2017, 8, e3073 CrossRef PubMed.
- M. C. Tsai, O. Manor, Y. Wan, N. Mosammaparast, J. K. Wang, F. Lan, Y. Shi, E. Segal and H. Y. Chang, Science, 2010, 329, 689–693 CrossRef CAS PubMed.
- P. Walter and G. Blobel, Nature, 1982, 299, 691–698 CrossRef CAS PubMed.
- M. T. Lam, W. Li, M. G. Rosenfeld and C. K. Glass, Trends Biochem. Sci., 2014, 39, 170–182 CrossRef CAS PubMed.
- N. W. Mathy and X. M. Chen, J. Biol. Chem., 2017, 292, 12375–12382 CrossRef CAS PubMed.
- L. Hu, C. Yin, F. Zhao, A. Ali, J. Ma and A. Qian, Int. J. Mol. Sci., 2018, 19, 360 CrossRef PubMed.
- D. Hamam, D. Ali, M. Kassem, A. Aldahmash and N. M. Alajez, Stem Cells Dev., 2015, 24, 417–425 CrossRef CAS PubMed.
- C. Huang, J. Geng and S. Jiang, Cell Tissue Res., 2017, 368, 229–238 CrossRef CAS PubMed.
- I. D'Alimonte, A. Lannutti, C. Pipino, P. Di Tomo, L. Pierdomenico, E. Cianci, I. Antonucci, M. Marchisio, M. Romano, L. Stuppia, F. Caciagli, A. Pandolfi and R. Ciccarelli, Stem Cell Rev., 2013, 9, 642–654 CrossRef PubMed.
- J. D. Lamplot, J. Qin, G. Nan, J. Wang, X. Liu, L. Yin, J. Tomal, R. Li, W. Shui, H. Zhang, S. H. Kim, W. Zhang, J. Zhang, Y. Kong, S. Denduluri, M. R. Rogers, A. Pratt, R. C. Haydon, H. H. Luu, J. Angeles, L. L. Shi and T. C. He, Am. J. Stem Cells, 2013, 2, 1–21 CAS.
- Y. Peng, Q. Kang, H. Cheng, X. Li, M. H. Sun, W. Jiang, H. H. Luu, J. Y. Park, R. C. Haydon and T. C. He, J. Cell. Biochem., 2003, 90, 1149–1165 CrossRef CAS PubMed.
- F. Sun, M. Wan, X. Xu, B. Gao, Y. Zhou, J. Sun, L. Cheng, O. D. Klein, X. Zhou and L. Zheng, J. Dent. Res., 2014, 93, 589–595 CrossRef CAS PubMed.
- M. Zhang, Y. Q. Bian, H. M. Tao, X. F. Yang and W. D. Mu, Eur. Rev. Med. Pharmacol. Sci., 2018, 22, 2896–2905 CAS.
- Q. Wang, J. Cai, X. H. Cai and L. Chen, PLoS One, 2013, 8, e72266 CrossRef CAS PubMed.
- S. Mori, H. Yoshikawa, J. Hashimoto, T. Ueda, H. Funai, M. Kato and K. Takaoka, Bone, 1998, 22, 99–105 CrossRef CAS PubMed.
- A. Nohe, Cell. Signalling, 2004, 16, 291–299 CrossRef CAS PubMed.
- H. Liu, L. Zhong, T. Yuan, S. Chen, Y. Zhou, L. An, Y. Guo, M. Fan, Y. Li, Y. Sun, W. Li, Q. Shi and Y. Weng, Int. J. Mol. Med., 2018, 41, 3379–3393 CAS.
- R. Bhushan, J. Grunhagen, J. Becker, P. N. Robinson, C. E. Ott and P. Knaus, Int. J. Biochem. Cell Biol., 2013, 45, 696–705 CrossRef CAS PubMed.
- Q. Song, L. Zhong, C. Chen, Z. Tang, H. Liu, Y. Zhou, M. Tang, L. Zhou, G. Zuo, J. Luo, Y. Zhang, Q. Shi and Y. Weng, Int. J. Mol. Med., 2015, 36, 1497–1506 CrossRef CAS PubMed.
- J. Cao, Y. Wei, J. Lian, L. Yang, X. Zhang, J. Xie, Q. Liu, J. Luo, B. He and M. Tang, Int. J. Mol. Med., 2017, 40, 378–388 CrossRef CAS PubMed.
- L. Chen, K. HolmstrØm, W. Qiu, N. Ditzel, K. Shi, L. Hokland and M. Kassem, Stem Cells, 2014, 32, 902–912 CrossRef CAS PubMed.
- K. Seenprachawong, T. Tawornsawutruk, C. Nantasenamat, P. Nuchnoi, S. Hongeng and A. Supokawej, Front. Genet., 2018, 9, 543 CrossRef CAS PubMed.
- Y. Jin, F. Hong, Q. Bao, Q. Xu, R. Duan, Z. Zhu, W. Zhang and C. Ma, Connect. Tissue Res., 2019, 1–9 CrossRef PubMed.
- C. Yang, X. Liu, K. Zhao, Y. Zhu, B. Hu, Y. Zhou, M. Wang, Y. Wu, C. Zhang, J. Xu, Y. Ning and D. Zou, Stem Cell Res. Ther., 2019, 10, 65 CrossRef CAS PubMed.
- S. Mollazadeh, B. S. F. Bazzaz, V. Neshati, A. A. de Vries, H. Naderi-Meshkin, M. Mojarad, M. Mirahmadi, Z. Neshati and M. A. Kerachian, BMC Med. Genet., 2019, 20, 117 CrossRef PubMed.
- Q. Cui, J. Xing, M. Yu, Y. Wang, J. Xu, Y. Gu, X. Nan, W. Ma, H. Liu and H. Zhao, Cell Death Dis., 2019, 10, 172 CrossRef PubMed.
- H. Long, Y. Zhu, Z. Lin, J. Wan, L. Cheng, M. Zeng, Y. Tang and R. Zhao, Cell Death Dis., 2019, 10, 470 CrossRef PubMed.
- Y. H. Son, S. Ka, A. Y. Kim and J. B. Kim, Endocrinol. Metab., 2014, 29, 122–135 CrossRef PubMed.
- X. Guan, Y. Gao, J. Zhou, J. Wang, F. Zheng, F. Guo, A. Chang, X. Li and B. Wang, Stem Cells, 2015, 33, 1589–1600 CrossRef CAS PubMed.
- J. Qiu, G. Huang, N. Na and L. Chen, Mol. Med. Rep., 2018, 17, 6301–6310 CAS.
- M. Jin, Y. Wu, J. Wang, J. Chen, Y. Huang, J. Rao and C. Feng, Biochem. Biophys. Res. Commun., 2016, 474, 76–82 CrossRef CAS PubMed.
- L. Chen, J. Hou, L. Ye, Y. Chen, J. Cui, W. Tian, C. Li and L. Liu, Sci. Rep., 2014, 4, 3819 CrossRef PubMed.
- C. Shi, M. Zhang, M. Tong, L. Yang, L. Pang, L. Chen, G. Xu, X. Chi, Q. Hong, Y. Ni, C. Ji and X. Guo, Sci. Rep., 2015, 5, 9930 CrossRef PubMed.
- L. Qin, Y. Chen, Y. Niu, W. Chen, Q. Wang, S. Xiao, A. Li, Y. Xie, J. Li, X. Zhao, Z. He and D. Mo, BMC Genomics, 2010, 11, 320 CrossRef PubMed.
- E. Hong and A. H. Reddi, Tissue Eng., Part B, 2012, 18, 445–453 CrossRef CAS PubMed.
- C. Yan, Y. Wang, X. Y. Shen, G. Yang, J. Jian, H. S. Wang, G. Q. Chen and Q. Wu, Biomaterials, 2011, 32, 6435–6444 CrossRef CAS PubMed.
- D. Guerit, D. Philipot, P. Chuchana, K. Toupet, J. M. Brondello, M. Mathieu, C. Jorgensen and D. Noel, PLoS One, 2013, 8, e62582 CrossRef CAS PubMed.
- S. Lee, D. S. Yoon, S. Paik, K. M. Lee, Y. Jang and J. W. Lee, Stem Cells Dev., 2014, 23, 1798–1808 CrossRef CAS PubMed.
- K. Sumiyoshi, S. Kubota, T. Ohgawara, K. Kawata, T. Abd El Kader, T. Nishida, N. Ikeda, T. Shimo, T. Yamashiro and M. Takigawa, J. Cell. Biochem., 2013, 114, 2094–2100 CrossRef CAS PubMed.
- J. Song, M. Lee, D. Kim, J. Han, C. H. Chun and E. J. Jin, Biochem. Biophys. Res. Commun., 2013, 431, 210–214 CrossRef CAS PubMed.
- N. Zhong, J. Sun, Z. Min, W. Zhao, R. Zhang, W. Wang, J. Tian, L. Tian, J. Ma, D. Li, Y. Han and S. Lu, Osteoarthritis and Cartilage, 2012, 20, 593–602 CrossRef CAS PubMed.
- L. Cui, Y. Shi, X. Zhou, X. Wang, J. Wang, Y. Lan, M. Wang, L. Zheng, H. Li and Q. Wu, Cell Death Dis., 2013, 4, e918 CrossRef CAS PubMed.
- Y. Chen, Y. Zhao, W. Chen, L. Xie, Z.-A. Zhao, J. Yang, Y. Chen, W. Lei and Z. Shen, Stem Cell Res. Ther., 2017, 8, 268 CrossRef PubMed.
- L. Han, Y. Wang, L. Wang, B. Guo, S. Pei and Y. Jia, Biochem. Biophys. Res. Commun., 2018, 495, 1476–1481 CrossRef CAS PubMed.
- Z.-J. Wei, B.-Y. Fan, Y. Liu, H. Ding, H.-S. Tang, D.-Y. Pan, J.-X. Shi, P.-Y. Zheng, H.-Y. Shi, H. Wu, A. Li and S.-Q. Feng, Neural Regener. Res., 2019, 14, 1462–1469 CrossRef PubMed.
- Z. Y. Xie, P. Wang, Y. F. Wu and H. Y. Shen, World Journal of Stem Cells, 2019, 11, 167–179 CrossRef PubMed.
- Q. Jia, W. Jiang and L. Ni, Arch. Oral Biol., 2015, 60, 234–241 CrossRef CAS PubMed.
- Y. Wang, W. Liu, Y. Liu, J. Cui, Z. Zhao, H. Cao, Z. Fu and B. Liu, J. Cell. Physiol., 2018, 233, 7435–7446 CrossRef CAS PubMed.
- J. Wu, J. Zhao, L. Sun, Y. Pan, H. Wang and W. B. Zhang, Bone, 2018, 108, 62–70 CrossRef CAS PubMed.
- Y. Huang, Y. Zheng, L. Jia and W. Li, Stem Cells, 2015, 33, 3481–3492 CrossRef CAS PubMed.
- X. R. Jiang, N. Guo, X. Q. Li, H. Y. Yang, K. Wang, C. L. Zhang, G. S. Li and G. D. Li, Eur. Rev. Med. Pharmacol. Sci., 2018, 22, 2954–2965 Search PubMed.
- Y. Gao, F. Xiao, C. Wang, C. Wang, P. Cui, X. Zhang and X. Chen, J. Cell. Biochem., 2018, 119, 6986–6996 CrossRef CAS PubMed.
- W. Peng, W. Deng, J. Zhang, G. Pei, Q. Rong and S. Zhu, Biochem. Biophys. Res. Commun., 2018, 503, 815–821 CrossRef CAS PubMed.
- Q. He, S. Yang, X. Gu, M. Li, C. Wang and F. Wei, Cell Death Dis., 2018, 9, 455 CrossRef PubMed.
- H. Li, Z. Zhang, Z. Chen and D. Zhang, Biochem. Biophys. Res. Commun., 2015, 466, 82–88 CrossRef CAS PubMed.
- W. Zhuang, X. Ge, S. Yang, M. Huang, W. Zhuang, P. Chen, X. Zhang, J. Fu, J. Qu and B. Li, Stem Cells, 2015, 33, 1985–1997 CrossRef CAS PubMed.
- C. Jin, Y. Zheng, Y. Huang, Y. Liu, L. Jia and Y. Zhou, Cell Biol. Int., 2017, 41, 33–41 CrossRef CAS PubMed.
- Y. Yu, Y. Chen, X. Zhang, X. Lu, J. Hong, X. Guo and D. Zhou, Cell Cycle, 2018, 17, 2374–2385 CrossRef CAS PubMed.
- Q. Wang, Q. Yang, G. Chen, Z. Du, M. Ren, A. Wang, H. Zhao, Z. Li, G. Zhang and Y. Song, Sci. Rep., 2018, 8, 9127 CrossRef PubMed.
- Y. Huang, Y. Zheng, C. Jin, X. Li, L. Jia and W. Li, Sci. Rep., 2016, 6, 28897 CrossRef CAS PubMed.
- M. Li, X. Sun, H. Cai, Y. Sun, M. Plath, C. Li, X. Lan, C. Lei, F. Lin, Y. Bai and H. Chen, Biochim. Biophys. Acta, 2016, 1859, 871–882 CrossRef CAS PubMed.
- W. Liu, C. Ma, B. Yang, C. Yin, B. Zhang and Y. Xiao, Biochem. Biophys. Res. Commun., 2017, 493, 1168–1175 CrossRef CAS PubMed.
- M. Li, Z. Xie, P. Wang, J. Li, W. Liu, S. Tang, Z. Liu, X. Wu, Y. Wu and H. Shen, Cell Death Dis., 2018, 9, 554 CrossRef PubMed.
- F. F. Firmin, F. Oger, C. Gheeraert, J. Dubois-Chevalier, A. S. Vercoutter-Edouart, F. Alzaid, C. Mazuy, H. Dehondt, J. Alexandre, B. Derudas, Q. Dhalluin, M. Ploton, A. Berthier, E. Woitrain, T. Lefebvre, N. Venteclef, F. Pattou, B. Staels, J. Eeckhoute and P. Lefebvre, Sci. Rep., 2017, 7, 14087 CrossRef PubMed.
- R. Gernapudi, B. Wolfson, Y. Zhang, Y. Yao, P. Yang, H. Asahara and Q. Zhou, Mol. Cell. Biol., 2016, 36, 30–38 CAS.
- N. Nuermaimaiti, J. Liu, X. Liang, Y. Jiao, D. Zhang, L. Liu, X. Meng and Y. Guan, Biochem. Biophys. Res. Commun., 2018, 495, 1878–1884 CrossRef CAS PubMed.
- Y. Huang, C. Jin, Y. Zheng, X. Li, S. Zhang, Y. Zhang, L. Jia and W. Li, Sci. Rep., 2017, 7, 8080 CrossRef PubMed.
- L. Zhang, S. Chen, N. Bao, C. Yang, Y. Ti, L. Zhou and J. Zhao, J. Mol. Histol., 2015, 46, 467–473 CrossRef CAS PubMed.
- L. Zhang, C. Yang, S. Chen, G. Wang, B. Shi, X. Tao, L. Zhou and J. Zhao, DNA Cell Biol., 2017, 36, 136–142 CrossRef CAS PubMed.
- M. J. Barter, R. Gomez, S. Hyatt, K. Cheung, A. J. Skelton, Y. Xu, I. M. Clark and D. A. Young, Development, 2017, 144, 4510–4521 CrossRef CAS PubMed.
- F. Ou, K. Su, J. Sun, W. Liao, Y. Yao, Y. Zheng and Z. Zhang, Biochem. Biophys. Res. Commun., 2017, 487, 457–463 CrossRef CAS PubMed.
- H.-l. Pang, Q.-q. Zhao, Y. Ma, Y.-l. Song, J. Min, J.-r. Lu, H. Li and D.-q. Zhao, Stem Cells Int., 2019, 2019, 11 Search PubMed.
- H. Wang, X. G. Ding, J. J. Yang, S. W. Li, H. Zheng, C. H. Gu, Z. K. Jia and L. Li, Eur. J. Cell Biol., 2018, 97, 180–189 CrossRef CAS PubMed.
- X. Sun, L. H. Luo, L. Feng and D. S. Li, Life Sci., 2018, 208, 246–252 CrossRef CAS PubMed.
- Y.-F. Tan, L. Tang, W.-X. OuYang, T. Jiang, H. Zhang and S.-J. Li, Pathol., Res. Pract., 2019, 215, 546–554 CrossRef CAS PubMed.
- J. Hou, H. Long, C. Zhou, S. Zheng, H. Wu, T. Guo, Q. Wu, T. Zhong and T. Wang, Stem Cell Res. Ther., 2017, 8, 4 CrossRef PubMed.
- A. Farzi-Molan, S. Babashah, B. Bakhshinejad, A. Atashi and M. Fakhr Taha, Cell Biol. Int., 2018, 42, 940–948 CrossRef CAS PubMed.
- A.-M. Wu, W.-F. Ni, Z.-Y. Huang, Q.-L. Li, J.-B. Wu, H.-Z. Xu and L.-H. Yin, J. Neurol. Sci., 2015, 351, 160–167 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |