Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
UPLC-QTOF-MS-guided isolation of anti-COPD ginsenosides from wild ginseng†
Hailin Zhua,
Junli Liua,
Hongqiang Lina,
Ying Zhangac,
Na Yanga,
Baisong Zhoua,
Zhongyao Wanga,
Alan Chen-Yu Hsud,
Jinping Liu *ab and
Pingya Li*a
*ab and
Pingya Li*a
aSchool of Pharmaceutical Sciences, Jilin University, Fujin Road 1266, Changchun 130021, Jilin, China. E-mail: liujp@jlu.edu.cn; lipy@jlu.edu.cn; Tel: +86-431-85619803
bResearch Center of Natural Drug, Jilin University, Changchun 130021, Jilin, China
cThe First Hospital of Jilin University, Changchun 130021, Jilin, China
dPriority Research Centre for Healthy Lungs, Faculty of Health and Medicine, The University of Newcastle, Newcastle, NSW, Australia
First published on 26th November 2019
Abstract
Four previously undescribed ginsenosides, along with five known analogues were isolated from wild ginseng by a UPLC-QTOF-MS-guided fractionation procedure. Their structures were elucidated on the basis of spectroscopic and spectrometric data (1D and 2D NMR, HR-ESI-MS). The isolated compounds could significantly inhibit the cigarette smoke extract (CSE)-induced inflammatory reaction in A549 cells. The HDAC2 pathway might be involved in the protective effect against the CSE-mediated inflammatory response in A549 cells.
1 Introduction
Panax ginseng has received much attention not only as a medicinal herb but also as a functional food.1 Ginseng has a wide range of biological activities such as anti-inflammatory, anti-oxidant and immunomodulatory functions. It is the traditional belief that wild ginseng is more medicinally efficacious and more valuable than cultivated ginseng. In the past, natural wild ginseng has been used to refer to the original ecological ginseng. But it is hard to find the natural wild ginseng because it is nearly extinct now. Currently, according to the “Identification and Grade Quality Standards of Wild Ginseng” (National Standards of People's Republic of China, GB/T 18765-2015), wild ginseng refers to the ginseng cultivated in mountains and grown for more than 15 years in a natural environment without any human intervention. This kind of wild ginseng is a replacement for natural wild ginseng grown within the original ecological environment. It has been reported that there were differences in immunomodulating effects between wild ginseng and cultivated ginseng.2 In our previous study, it was also found that the wild ginseng (20 year-old mountain-cultivated ginseng) was much differentiated from cultivated ginseng. And based on UPLC-QTOF/MS, a total of 126 chemical compositions were tentatively identified or characterized from wild ginseng.3Chronic Obstructive Pulmonary Disease (COPD), the fourth leading cause of death in the world currently, is usually caused by significant exposure to noxious gases or particles. The airflow limitation and persistent respiratory symptoms are the typical symptoms. Cigarette smokers have a higher prevalence of respiratory symptoms and higher COPD mortality rate, and the cigarette smoking is the leading environmental risk factor for COPD around the world.4 Along with the progressive lung inflammation, some pro-inflammatory mediators such as TNF-α, IL-1β and IL-6 participated in the occurrence and development of COPD.5,6 Ginsenosides are regarded as the main bioactive components in ginseng.7,8 Ginseng extract9,10 and monomer saponins such as Rg1 (ref. 11), Rh2 (ref. 12) and CK13 had been reported to significantly inhibit COPD-related inflammation in vivo and in vitro. Reduced HDAC2 activity and expression was found in COPD, resulting in amplification of the inflammatory response. Meanwhile, HDAC2 dysfunction is thought to play an important role in the development of corticosteroid resistance in COPD.14–16 Thus, increasing HDAC2 activity may be a promising strategy to overcome corticosteroid resistance in COPD. Existing treatments such as theophylline, nortriptyline, macrolides and selective phosphatidylinositol-3-kinase-δ inhibitors have been reported to increase HDAC2 activity effectively.17
As part of our continuing efforts to elucidate the chemical composition of wild ginseng furtherly and search for anti-COPD compounds from wild ginseng, a UPLC-QTOF-MS-guided fractionation procedure was performed to target ginsenosides from the anti-COPD fractions of wild ginseng, leading to the isolation of 4 previously unreported ginsenosides (1–4) and five known analogues (5–9). Herein, the isolation and structural elucidation of the isolated compounds as well as their anti-COPD activity on the CSE-stimulated A549 cells were discussed. The potential mechanism against the CSE stimulation was also preliminarily investigated in the paper.
2 Experimental
2.1 Plant materials
The 20 years-old mountain-cultivated ginsengs were collected from cultivation area in Fusong, Jilin Province, the main source of ginseng in China. The ginseng was identified by our group according to Chinese Pharmacopoeia (2015 version). The voucher specimen (no. MCG170931) was deposited at the Research Center of Natural Drug, Jilin University, Changchun, China.2.2 Apparatus and chemicals
The HR-ESI-MS spectra were performed on Waters Xevo G2-XS QTOF mass spectrometer (Waters, Milford, MA, USA). NMR spectra were measured on a Bruker Avance-600 spectrometer (Bruker, Karlsruhe, Germany) in DMSO-D6 operating at 600 MHz for 1H NMR, HMBC and HMQC, 150 MHz for 13C NMR, respectively. Tetramethylsilane (TMS) was used as internal standard. Column chromatography was performed with silica gel (200–300 mesh) purchased from Qingdao Ocean Chemical Group Co. Ltd (Qingdao, China). Thin-layer chromatography was conducted on silica gel G plates (Qingdao Marine Chemical Inc.). Semi-preparative HPLC with 1525 binary pump combined Waters 2998 UV detector (Waters Co., Milford, MA, USA.) and YMC C18 column (5 μm, 20 mm × 10 mm; YMC Co., Ltd., Japan) were also used to isolate the compounds. Methanol (MeOH) and acetonitrile (ACN) were HPLC grade (Fisher, USA). Purified water was purchased from YiBao Co, Ltd (Shenzhen, China). Other solvents used were analytical grade (Beijing Chemical Works, Beijing, China).2.3 Extraction and isolation
The fresh wild ginseng (1.0 kg) were crushed and extracted with 95% EtOH for three times. The extract was combined and then concentrated under vacuum to obtain the concentrated aqueous solution (0.5 L), and successively partitioned with petroleum ether, EtOAc and n-butanol, to afford petroleum ether (29.8 g), EtOAc (35.2 g) and n-butanol (133.5 g) soluble extracts, respectively. The n-butanol extract (inhibition of the release of pro-inflammatory mediators in CSE-induced A549 cells at 200 μM, Fig. S10A†) was subjected to rapid medium pressure purification chromatography on silica gel (200–300 mesh) eluted with a gradient of chloroform–methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0–0
0–0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give 10 fractions (Fr. A–J). Fraction G showed significant inhibition of the release of pro-inflammatory mediators in CSE-induced A549 cells at 200 μM (Fig. S10B†). Subsequent UPLC-QTOF-MS-guided separation and purification targeted the subfractions of fraction G containing compounds with m/z > 500. Fraction G was further fractionated on silica gel column chromatography eluting with gradient elution of chloroform–methanol (10
1) to give 10 fractions (Fr. A–J). Fraction G showed significant inhibition of the release of pro-inflammatory mediators in CSE-induced A549 cells at 200 μM (Fig. S10B†). Subsequent UPLC-QTOF-MS-guided separation and purification targeted the subfractions of fraction G containing compounds with m/z > 500. Fraction G was further fractionated on silica gel column chromatography eluting with gradient elution of chloroform–methanol (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to yield six subfractions (G1–G6, Fig. S10C†). Subfraction G5 (0.21 g) was further separated by semi-preparative RP-HPLC with ACN/H2O as mobile phase to obtain compound 1 (ACN–H2O: 19
1) to yield six subfractions (G1–G6, Fig. S10C†). Subfraction G5 (0.21 g) was further separated by semi-preparative RP-HPLC with ACN/H2O as mobile phase to obtain compound 1 (ACN–H2O: 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 81, 2 mL min−1, 19 min, 6 mg, 0.0006%), compound 2 (ACN–H2O: 19
81, 2 mL min−1, 19 min, 6 mg, 0.0006%), compound 2 (ACN–H2O: 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 81, 2 mL min−1, 22 min, 3 mg, 0.0003%), compound 8 (ACN–H2O: 21
81, 2 mL min−1, 22 min, 3 mg, 0.0003%), compound 8 (ACN–H2O: 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 79, 2 mL min−1, 5 min, 4 mg, 0.0003%), compound 9 (ACN–H2O: 42
79, 2 mL min−1, 5 min, 4 mg, 0.0003%), compound 9 (ACN–H2O: 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 58, 2 mL min−1, 36 min, 3 mg, 0.0003%). G4 (0.14 g) was also further separated by semi-preparative RP-HPLC with ACN/H2O as mobile phase to obtain compound 3 (ACN–H2O: 15
58, 2 mL min−1, 36 min, 3 mg, 0.0003%). G4 (0.14 g) was also further separated by semi-preparative RP-HPLC with ACN/H2O as mobile phase to obtain compound 3 (ACN–H2O: 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 85, 2 mL min−1, 15 min, 4 mg, 0.0004%), compound 5 (ACN–H2O: 18
85, 2 mL min−1, 15 min, 4 mg, 0.0004%), compound 5 (ACN–H2O: 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 82, 2 mL min−1, 32 min, 3 mg, 0.0003%) and compound 6 (ACN–H2O: 21
82, 2 mL min−1, 32 min, 3 mg, 0.0003%) and compound 6 (ACN–H2O: 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 79, 2 mL min−1, 13 min, 3 mg, 0.0003%). G2 (0.18 g) was also further separated by semi-preparative RP-HPLC with ACN/H2O as mobile phase to obtain compound 4 (ACN–H2O: 21
79, 2 mL min−1, 13 min, 3 mg, 0.0003%). G2 (0.18 g) was also further separated by semi-preparative RP-HPLC with ACN/H2O as mobile phase to obtain compound 4 (ACN–H2O: 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 79, 2 mL min−1, 10 min, 2 mg, 0.0002%) and compound 7 (ACN–H2O: 28
79, 2 mL min−1, 10 min, 2 mg, 0.0002%) and compound 7 (ACN–H2O: 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 72, 2 mL min−1, 10 min, 3 mg, 0.0003%).
72, 2 mL min−1, 10 min, 3 mg, 0.0003%).
2.4 Spectroscopic data
6-O-[α-L-rhamnopyranosyl-(1→2)-β-D-glucopyranosyl]-20-O-β-D-glucopyranosyl-dammar-23,25-diene-3β,6α,12β,20S-tetraol. White amorphous powder; Libermann–Burchard reaction was positive; Molisch reaction was positive; suggesting the existence of triterpenoid structure; 1H and 13C NMR: see Table 1; HR-ESI-MS m/z 943.5270 [M − H]− (calc. for C48H79O18 is 943.5266); HR-MS, 1H and 13C NMR, HMQC, HMBC spectra see in the ESI (Fig. S1-1–S1-5).†
| Position | 1 | 2 | 4 | |||
|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | |
| 1 | 38.73 | 1.44(1H, m), 0.91(1H, m) | 38.88 | 1.41(1H, m), 0.99(1H, m) | 38.76 | 1.32(1H, m), 0.92(1H, m) |
| 2 | 26.80 | 1.51(1H, m), 1.50(1H, m) | 26.94 | 1.60(1H, m), 1.47(1H, m) | 26.79 | 1.82(1H, m), 1.75(1H, m) |
| 3 | 76.90 | 3.54(1H, m) | 77.37 | 3.51(1H, m) | 77.21 | 3.65(1H, m) |
| 4 | 39.98 | — | 39.97 | — | 39.91 | — |
| 5 | 59.86 | 1.40(1H, m) | 60.01 | 1.45(1H, m) | 59.84 | 1.07(1H, m) |
| 6 | 73.84 | 4.66(1H, m) | 72.73 | 4.63(1H, m) | 72.63 | 4.53(1H, m) |
| 7 | 44.75 | 2.17(1H, m), 1.76(1H, m) | 44.90 | 2.14(1H, m), 1.89(1H, m) | 44.75 | 2.24(1H, m), 1.90(1H, m) |
| 8 | 40.16 | — | 40.31 | — | 40.16 | — |
| 9 | 48.64 | 1.42(1H, m) | 48.79 | 1.39(1H, m) | 48.72 | 1.17(1H, m) |
| 10 | 38.59 | — | 38.73 | — | 38.63 | — |
| 11 | 30.27 | 1.92(1H, m), 1.41(1H, m) | 30.43 | 2.01(1H, m), 1.41(1H, m) | 30.91 | 2.15(1H, m), 1.07(1H, m) |
| 12 | 69.08 | 3.86(1H, m) | 70.32 | 3.87(1H, m) | 69.81 | 3.90(1H, m) |
| 13 | 48.06 | 1.69(1H, m) | 48.61 | 1.73(1H, m) | 47.03 | 1.96(1H, m) |
| 14 | 50.80 | — | 50.95 | — | 50.94 | — |
| 15 | 30.09 | 1.40(1H, m), 0.91(1H, m) | 30.24 | 1.37(1H, m), 0.88(1H, m) | 30.73 | 1.37(1H, m), 0.86(1H, m) |
| 16 | 25.90 | 1.48(1H, m), 1.26(1H, m) | 26.94 | 1.45(1H, m), 1.26(1H, m) | 26.03 | 1.45(1H, m), 1.07(1H, m) |
| 17 | 51.05 | 2.50(1H, m) | 51.20 | 2.47(1H, m) | 53.49 | 2.30(1H, m) |
| 18 | 16.74 | 0.82(3H, s) | 16.80 | 0.91(3H, s) | 16.97 | 0.83(3H, s) |
| 19 | 16.84 | 0.81(3H, s) | 16.89 | 0.78(3H, s) | 17.56 | 0.83(3H, s) |
| 20 | 82.02 | — | 82.17 | — | 75.24 | — |
| 21 | 22.37 | 1.22(3H, s) | 22.10 | 1.19(3H, s) | 26.38 | 0.97(3H, s) |
| 22 | 40.01 | 2.13(1H, m), 1.88(1H, m) | 141.31 | 6.05(1H, d, J = 11.5 Hz) | 34.911 | 2.01(1H, m), 1.32(1H, m) |
| 23 | 127.283 | 5.53(1H, dt, J = 12.5, 7.5 Hz) | 127.43 | 6.25(1H, m) | 21.973 | 2.60(1H, m), 2.25(1H, m) |
| 24 | 134.62 | 6.39(1H, d, J = 11.0 Hz) | 121.77 | 6.02(1H, d, J = 11.5 Hz) | 125.517 | 5.48(1H, m) |
| 25 | 141.72 | — | 134.77 | — | 130.16 | — |
| 26 | 114.63 | 4.98(1H, m), 4.89(1H, m) | 31.12 | 2.25(3H, s) | 25.56 | 1.56(3H, s) |
| 27 | 17.79 | 1.83(3H, s) | 22.52 | 1.91(3H, s) | 16.57 | 1.22(3H, s) |
| 28 | 30.97 | 1.62(3H, s) | 31.12 | 1.92(3H, s) | 31.00 | 1.63(3H, s) |
| 29 | 17.79 | 0.94(3H, s) | 16.99 | 0.99(3H, s) | 16.74 | 0.96(3H, s) |
| 30 | 16.65 | 0.81(3H, s) | 16.80 | 0.78(3H, s) | 16.46 | 0.82(3H, s) |
| 6-Glc-1′ | 99.91 | 5.11(1H, d, J = 7.0 Hz) | 100.06 | 5.09(1H, d, J = 7.5 Hz) | 99.94 | 5.35(1H, m) |
| 6-Glc-2′ | 77.22 | 4.48(1H, m) | 77.37 | 4.40(1H, m) | 77.53 | 4.19(1H, m) |
| 6-Glc-3′ | 77.51 | 4.34(1H, m) | 78.43 | 4.32(1H, m) | 77.21 | 4.32(1H, m) |
| 6-Glc-4′ | 70.82 | 4.14(1H, m) | 70.96 | 4.12(1H, m) | 70.85 | 4.12(1H, m) |
| 6-Glc-5′ | 76.78 | 3.90(1H, brs) | 77.37 | 3.66(1H, brs) | 77.21 | 4.06(1H, brs) |
| 6-Glc-6′ | 61.47 | 4.62(1H, m), 4.43(1H, m) | 61.62 | 4.45(1H, m), 4.59(1H, m) | 61.49 | 4.37(1H, m), 4.24(1H, m) |
| 2′-Rham-1′′ | 99.91 | 6.32(1H, m) | 100.06 | 6.42(1H, m) | 99.87 | 6.59(1H, m) |
| 2′-Rham-2′′ | 73.65 | 5.06(1H, m) | 70.63 | 4.85(1H, m) | 70.50 | 4.60(1H, m) |
| 2′-Rham-3′′ | 70.48 | 4.87(1H, m) | 70.32 | 4.84(1H, m) | 70.20 | 4.47(1H, m) |
| 2′-Rham-4′′ | 72.05 | 4.35(1H, m) | 72.73 | 4.31(1H, m) | 72.06 | 4.12(1H, m) |
| 2′-Rham-5′′ | 67.84 | 4.88(1H, m) | 67.99 | 4.84(1H, m) | 67.88 | 4.94(1H, m) |
| 2′-Rham-6′′ | 16.74 | 1.58(3H, d, J = 6.5 Hz) | 17.94 | 1.60(3H, d, J = 6.5 Hz) | 17.80 | 1.34(3H, d, J = 6.5 Hz) |
| 20-Glc-1′′′ | 96.62 | 4.91(1H, d, J = 7.5 Hz) | 96.77 | 5.03(1H, d, J = 7.5 Hz) | — | — |
| 20-Glc-2′′′ | 73.84 | 3.66(1H, m) | 74.00 | 3.59(1H, m) | — | — |
| 20-Glc-3′′′ | 77.36 | 4.18(1H, m) | 77.66 | 4.15(1H, m) | — | — |
| 20-Glc-4′′′ | 69.28 | 3.84(1H, m) | 69.72 | 3.87(1H, m) | — | — |
| 20-Glc-5′′′ | 76.68 | 3.62(1H, m) | 77.37 | 3.51(1H, m) | — | — |
| 20-Glc-6′′′ | 61.23 | 4.56(1H, m), 4.26(1H, m) | 61.38 | 4.53(1H, m), 4.23(1H, m) | — | — |
| –O–CO– | — | — | — | — | 170.90 | — |
| –CH2 | — | — | — | — | 38.76 | 3.77(2H, m) |
| –CO–OH | — | — | — | — | 171.20 | — |
6-O-[α-L-rhamnopyranosyl-(1→2)-β-D-glucopyranosyl]-20-O-β-D-glucopyranosyl-dammar-22(23),24-diene-3β,6α,12β,20S-tetraol. White amorphous powder; Libermann–Burchard reaction was positive; Molisch reaction was positive; suggesting the existence of triterpenoid structure; 1H and 13C NMR: see Table 1; HR-ESI-MS m/z 943.5272 [M − H]− (calc. for C48H79O18 is 943.5266); HR-MS, 1H and 13C NMR, HMQC, HMBC spectra see in the ESI (Fig. S2-1–S2-5).†
3-O-[β-D-glucopyranosyl-(1→2)-β-D-glucopyranosyl]-3β,12β,20S,25-tetrahydroxydammar-23-ene. White amorphous powder; Libermann–Burchard reaction was positive; Molisch reaction was positive; suggesting the existence of triterpenoid structure; 1H and 13C NMR: see Table 2; HR-ESI-MS m/z 799.4897 [M − H]− (calc. for C42H71O14 is 799.4844); HR-MS, 1H and 13C NMR, HMQC, HMBC spectra see in the ESI (Fig. S3-1–S3-5).†
| Position | 3 | Position | 3 | ||
|---|---|---|---|---|---|
| δC | δH | δC | δH | ||
| a Delta is ppm of chemical shifts which are reported in parts per million (δ), and coupling constants (J) are expressed in hertz. | |||||
| 1 | 39.90 | 1.40(1H, m), 0.91(1H, m) | 22 | 136.16 | 6.07(1H, d, J = 16.0 Hz) |
| 2 | 25.79 | 2.25(1H, m), 1.55(1H, m) | 23 | 126.20 | 6.31(1H, m) |
| 3 | 88.22 | 3.15(1H, m) | 24 | 39.50 | 2.76(1H, m), 2.44(1H, m) |
| 4 | 39.60 | — | 25 | 80.34 | — |
| 5 | 55.61 | 0.69(1H, m) | 26 | 29.94 | 1.64(3H, s) |
| 6 | 17.74 | 1.48(1H, m), 1.21(1H, m) | 27 | 29.96 | 1.56(3H, s) |
| 7 | 34.42 | 1.36(1H, m), 1.33(1H, m) | 28 | 27.48 | 0.98(3H, s) |
| 8 | 40.02 | — | 29 | 15.99 | 0.90(3H, s) |
| 9 | 49.44 | 1.31(1H, m) | 30 | 16.48 | 0.81(3H, s) |
| 10 | 36.29 | — | 3-Glc-1′ | 103.66 | 4.96(1H, d, J = 7.5 Hz) |
| 11 | 30.98 | 1.92(1H, m), 1.63(1H, m) | 3-Glc-2′ | 81.35 | 4.02(1H, m) |
| 12 | 70.02 | 3.30(1H, m) | 3-Glc-3′ | 76.16 | 3.99(1H, m) |
| 13 | 48.53 | 1.92(1H, m) | 3-Glc-4′ | 69.88 | 3.98(1H, m) |
| 14 | 50.98 | — | 3-Glc-5′ | 76.43 | 3.16(1H, brs) |
| 15 | 30.44 | 1.47(1H, m), 0.94(1H, m) | 3-Glc-6′ | 61.06 | 4.59(1H, m), 4.42(1H, m) |
| 16 | 25.79 | 1.58(1H, m), 1.36(1H, m) | 2′-Glc-1′′ | 103.87 | 5.10(1H, d, J = 7.5 Hz) |
| 17 | 52.97 | 2.30(1H, m) | 2′-Glc-2′′ | 75.30 | 3.42(1H, m) |
| 18 | 15.37 | 0.82(3H, s) | 2′-Glc-3′′ | 76.88 | 4.14(1H, m) |
| 19 | 15.93 | 0.74(3H, s) | 2′-Glc-4′′ | 70.02 | 4.25(1H, m) |
| 20 | 72.29 | — | 2′-Glc-5′′ | 76.55 | 3.42(1H, m) |
| 21 | 27.48 | 1.02(3H, s) | 2′-Glc-6′′ | 60.89 | 4.42(1H, m), 4.26(1H, m) |
6-O-[α-L-rhamnopyranosyl-(1→2)-β-D-glucopyranosyl]-20-O-malonyl-dammar-24-ene-3β,6α,12β,20S-tetraol. White amorphous powder; Libermann–Burchard reaction was positive; Molisch reaction was positive; suggesting the existence of triterpenoid structure; 1H and 13C NMR: see Table 1; HR-ESI-MS m/z 869.4890 [M − H]− (calc. for C45H73O16 is 869.4899); HR-MS, 1H and 13C NMR, HMQC, HMBC spectra see in the ESI (Fig. S4-1–S4-5).†
Compound 5–9. White amorphous powder; Libermann–Burchard reaction was positive; Molisch reaction was positive; suggesting the existence of triterpenoid structure; 1H and 13C NMR, spectra see in the ESI (Fig. S5-1–S9-2).†
2.5 Bioassay
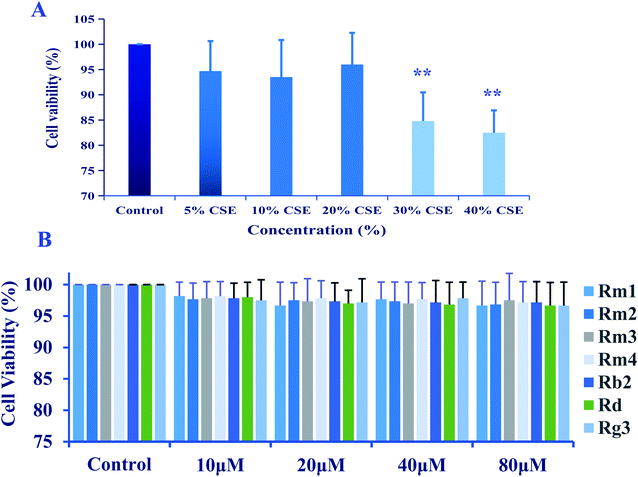
A549 cells were cultured in 96-well plates at a density of 5 × 105 cells per well treated with the presence of CSE (5%, 10%, 20%, 30% and 40%) for 18 h, or treated with ginsenosides at the concentrations of 0.0, 10.0, 20.0, 40.0, 80.0 μM for 24 h.
![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ±SD. Statistical significance was calculated with two tailed test or a one-way analysis of variance (ANOVA), and p-value < 0.05 was considered as statistically significant.
±SD. Statistical significance was calculated with two tailed test or a one-way analysis of variance (ANOVA), and p-value < 0.05 was considered as statistically significant.3 Results and discussion
3.1 Structure elucidation
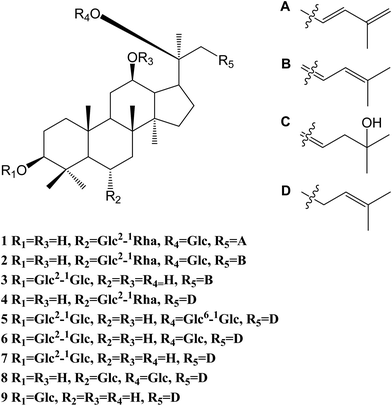
The 95% EtOH extract of wild ginseng was suspended in aqueous solution and partitioned with petroleum ether, EtOAc and n-butanol. UPLC-QTOF-MS-guided isolation of bioactive n-butanol soluble extracts (inhibition of the release of pro-inflammatory mediators in CSE-induced A549 cells at 200 μM) led to identify nine ginsenosides (Fig. S10A†). Five known compounds were identified as ginsenoside Rb2 (5),24 Rd (6),24 Rg3 (7),25 Rg1 (8)25 and Rh2 (9)26 by comparing spectroscopic data with those of reported values.Compound 1 was a white amorphous powder (MeOH). The molecular formula was established as C48H80O18 by combining NMR spectra and HR-ESI-MS at m/z 943.5270 [M − H]− (calc. for 943.5266). The 1H NMR spectrum of 1 displayed eight methyl signals at δ 0.81 (3H, s), 0.81 (3H, s), 0.82 (3H, s), 0.94 (3H, s), 1.22 (3H, s), 1.58 (3H, d, J = 6.5 Hz), 1.62 (3H, s) and 1.83 (3H, s). The 13C NMR spectrum of 1 showed 48 signals, including ten methylene [two of them bearing an oxygen atom respectively (δ 61.23, 61.47), and one of them was olefinic signal at δ 114.63], twenty-four methine [eighteen of them bearing an oxygen atom respectively (δ 67.84, 69.08, 69.28, 70.48, 70.82, 72.05, 73.65, 73.84, 73.84, 76.68, 76.78, 76.90, 77.22, 77.36, 77.51, 96.62, 99.91 and 99.91), and two of them were olefinic signals at δ 127.28 and 134.62], six quaternary carbon [δ 38.59, 39.98, 40.16, 50.80, 82.02 and an olefinic signal at δ 141.72], eight methyl carbons (δ 16.65, 16.74, 16.74, 16.84, 17.79, 17.79, 22.37 and 30.97). There is a resemblance with the chemical shifts of 1 along with ginsenoside Re27 except the signals of the side chain. The main differences were the replacement of two olefinic carbons by four olefinic carbons (δ 141.72, 134.62, 127.28 and 114.63), and only two methyl signals (δ 22.37 and 17.79) in 1. Furthermore, comparison of the NMR spectroscopic data of 1 with those of cylindrictone A,28 whose structure was 12β,20β-dihydroxy-23,25-diene-3-oxo-dammarane, a 23,25-diene group was deduced to exist in compound 1. The location of this group was confirmed by the heteronuclear multiple bond coherence (HMBC) spectrum of 1. The correlations between H-22 with C-21 and C-24, H-23 with C-20 and C-25, H-24 with C-22, C-26 and C-27, H-26 with C-24 and C-27, H-27 with C-24 and C-26, were observed in the HMBC spectrum of 1. The structure of 1 were deduced as 6-O-[α-L-rhamnopyranosyl-(1→2)-β-D-glucopyranosyl]-20-O-β-D-glucopyranosyl-dammar-23,25-diene-3β,6α,12β,20S-tetraol (ginsenoside Rm1, Fig. 1). Compound 1 was a minor glucoside in wild ginseng. The carbon and proton signals of 1 were fully assigned on the basis of HMQC and HMBC spectra (Table 1 and Fig. 2).
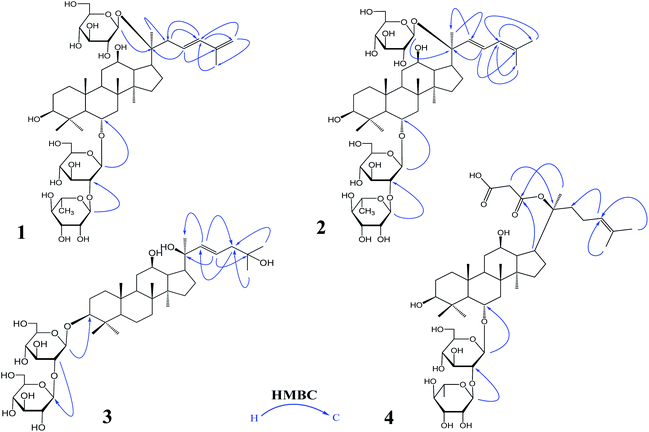 | ||
| Fig. 1 The important HMBC correlations of 1–4: ginsenoside Rm1 (1); ginsenoside Rm2 (2); ginsenoside Rm3 (3); ginsenoside Rm4 (4) HMBC: heteronuclear multiple bond correlation. | ||
Compound 2 was a white amorphous powder (MeOH). The molecular formula was established as C48H80O18 by combining the NMR spectra and HR-ESI-MS at m/z 943.5272 [M − H]− (calc. for 943.5266). The 1H NMR spectrum of 2 displayed nine methyl signals at δ 0.78 (3H, s), 0.78 (3H, s), 0.91 (3H, s), 0.99 (3H, s), 1.19 (3H, s), 1.60 (3H, d, J = 6.5 Hz), 1.91 (3H, s), 1.92 (3H, s) and 2.25 (3H, s). The 13C NMR spectrum of 2 revealed 48 signals, including eight methylene [two of them bearing an oxygen atom respectively (δ 61.38, 61.62)], twenty-five methine [eighteen of them bearing an oxygen atom respectively (δ 67.99, 69.72, 70.32, 70.32, 70.63, 70.96, 72.73, 72.73, 74.00, 77.37, 77.37, 77.37, 77.37, 77.66, 78.43, 96.77, 100.06 and 100.06), and three of them were olefinic signals at δ 121.77, 127.43 and 141.31], six quaternary carbon [δ 39.97, 40.31, 38.73, 50.95, 82.17 and an olefinic signal at δ 134.77], nine methyl carbons (δ 16.80, 16.80, 16.89, 16.99, 17.94, 22.10, 22.52, 31.12 and 31.12). There is a resemblance with the chemical shifts of 2 along with ginsenoside Re27 or compound 1 except the signals of the side chain. The main differences were the four olefinic carbons (δ 121.77, 127.43, 134.77 and 141.31), and three methyl signals (δ 22.10, 22.52 and 31.12) in 2. Furthermore, comparison of the NMR spectroscopic data of 2 with those of notoginsenoside ST14,29 whose structure was (3β,6α,12β,20S,22E)-3,6,12,20-tetrahydroxydammar-22(23),24-diene-6-O-β-D-glucopyranoside, a 22(23),24-diene group was deduced to exist in compound 2. The location of this group was confirmed by HMBC of 2. In HMBC spectrum, the correlations between H-22 with C-21 and C-24, H-23 with C-20 and C-25, H-24 with C-22, C-26 and C-27, H-26 with C-24 and C-27, H-27 with C-24 and C-26, were observed. Thus, the structure of compound 2 was identified as 6-O-[α-L-rhamnopyranosyl-(1→2)-β-D-glucopyranosyl]-20-O-β-D-glucopyranosyl-dammar-22(23), 24-diene-3β,6α,12β,20S-tetraol (ginsenoside Rm2, Fig. 1). Compound 2 was also a minor glucoside in wild ginseng. The complete proton and carbon signal assignments were shown in Table 1.
Compound 3, a white amorphous powder (MeOH), possessed a molecular formula of C42H72O14, as elucidated by combining the NMR spectra and HR-ESI-MS at m/z 799.4897 [M − H]− (calc. for 799.4844). The 1H NMR spectrum of 3 displayed eight methyl signals at δ 0.74 (3H, s), 0.81 (3H, s), 0.82 (3H, s), 0.90 (3H, s), 0.98 (3H, s), 1.02 (3H, s), 1.56 (3H, s) and 1.64 (3H, s). The 13C NMR spectrum of 3 revealed 42 signals, including ten methylene [two of them bearing an oxygen atom respectively (δ 60.89, 61.06)], twenty methine [twelve of them bearing an oxygen atom respectively (δ 69.88, 70.02, 70.02, 75.30, 76.55, 76.16, 76.43, 76.88, 81.35, 88.22, 103.6 and 103.87), and two of them were olefinic signals at δ 126.20 and 136.16], six quaternary carbon [δ 36.29, 39.60, 40.02, 50.98, 72.29 and 80.34], eight methyl carbons (δ 15.37, 15.93, 15.99, 16.48, 27.48, 27.48, 29.94 and 29.96). The NMR data of 3 were similar to those of quinquenoside A,29,30 whose structure was 3-O-[β-D-glucopyranosyl-(1→2)-β-D-glucopyranosyl]-20-O-[β-D-glu-copyranosyl-(1→6)-β-D-glucopyranosyl]-3β,12β,20S,25-tetrahydroxydammar-23-ene, except the absence of β-D-glucopyranosyl-(1→6)-β-D-glucopyranosyl group signals in 3. And the chemical shift of C-20 was changed to δ 72.29, indicated that C-20 was linked to a free hydroxyl group instead of being glycosylated. The location of hydroxyl group was determined by HMBC of 3. In HMBC spectrum, the correlations between H-22 with C-21 and C-24, H-23 with C-20 and C-25, were observed. Thus, the structure of compound 3 was identified as 3-O-[β-D-glucopyranosyl-(1→2)-β-D-glucopyranosyl]-3β,12β,20S,25-tetrahydroxydammar-23-ene (ginsenoside Rm3, Fig. 1). Compound 3 was also a minor glucoside in wild ginseng. The complete signal assignments on basis of HMQC and HMBC were shown in Table 1.
Compound 4, a white amorphous powder (MeOH), possessed a molecular formula of C45H74O16, as elucidated by the NMR spectra and HR-ESI-MS at m/z 869.4890 [M − H]− (calc. for 869.4899). The 1H NMR spectrum of 4 displayed eight methyl signals at δ 0.82 (3H, s),0.83 (3H, s), 0.83 (3H, s), 0.96 (3H, s), 0.97 (3H, s), 1.22 (3H, s), 1.34 (3H, d, J = 6.5 Hz), 1.56 (3H, s) and 1.63(3H, d, J = 4.5 Hz). The 13C NMR spectrum of 4 revealed 45 signals, including ten methylene [one of them bearing an oxygen atom (δ 61.49)], eighteen methine, eight quaternary carbons [δ 38.63, 39.91, 40.16, 50.94, 75.24, 130.16, 170.90 and 171.20], eight methyl carbons (δ 16.46, 16.57, 16.74, 17.56, 17.80, 25.56, 26.38 and 31.00). The NMR data of 4 were similar to those of ginsenoside Rg2,31 except a malonyl group was appeared in 4. The location of the malonyl group was determined by HMBC of 4. The following correlations were found: H-21 with C-17 and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, H-17 with C-21 and C
O, H-17 with C-21 and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, which indicated that the malonyl group was linked to C-20. Thus, the structure of 4 was identified as 6-O-[α-L-rhamnopyranosyl-(1→2)-β-D-glucopyranosyl]-20-O-malonyl-dammar-24-ene-3β,6α,12β,20S-tetraol (ginsenoside Rm4, Fig. 1). Compound 4 was also a minor glucoside in wild ginseng. The proton and carbon signals of 4 were fully assigned on the basis of HMQC and HMBC spectra (Table 1).
O, which indicated that the malonyl group was linked to C-20. Thus, the structure of 4 was identified as 6-O-[α-L-rhamnopyranosyl-(1→2)-β-D-glucopyranosyl]-20-O-malonyl-dammar-24-ene-3β,6α,12β,20S-tetraol (ginsenoside Rm4, Fig. 1). Compound 4 was also a minor glucoside in wild ginseng. The proton and carbon signals of 4 were fully assigned on the basis of HMQC and HMBC spectra (Table 1).
3.2 Bioactivity evaluation
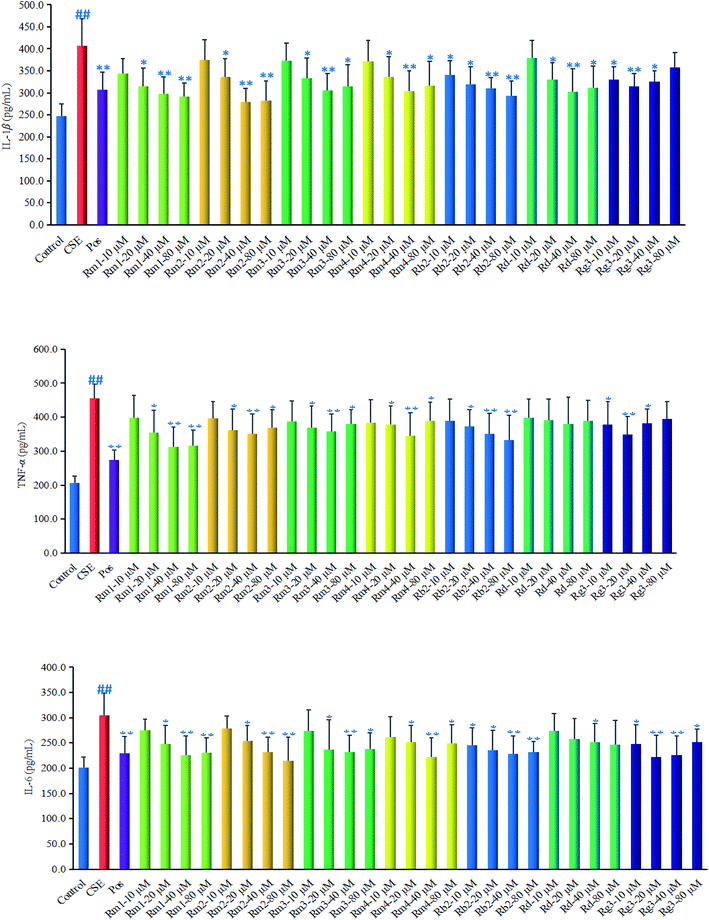
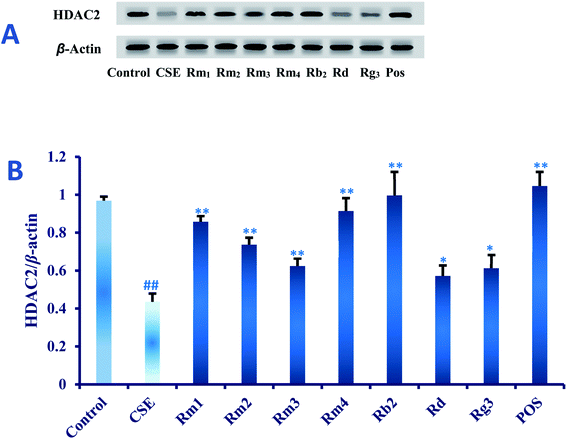
Ginsenoside Rg1 (ref. 11) and Rh2 (ref. 12) had been reported to significantly inhibit COPD-related inflammation. As part of an ongoing effort to search for anti-COPD agents from wild ginseng,23 compounds 1–7 were evaluated for their anti-inflammatory effects on CSE-induced A549 cells. Dexamethasone was selected as positive drug (Pos).32In order to explore the underlying mechanism of anti-inflammatory property of compounds 1–7, we investigated the effect of them on the protein expression of HDAC2 in CSE-exposed A549 cells. In the present study, after 18 hours of CSE exposure in A549 cells, the expression of HDAC2 protein in each group of 40 μM concentration was measured as shown in Fig. 5. The results showed that the level of HDAC2 protein in A549 cells decreased significantly (p < 0.01) after CSE 18 hours exposure. Interestingly, treatment with each ginsenoside could significantly activated the expression of HDAC2 (p < 0.01, p < 0.05) as compared to the CSE group (Fig. 5) in detail, which suggested that these compounds may up-regulated the expression of HDAC2 in CSE-exposed A549 cells.
4 Conclusions
In the present study, four minor previously undescribed ginsenosides, named as ginsenoside Rm1 (1), Rm2 (2), Rm3 (3) and Rm4 (4), along with five known ginsenosides, Rb2 (5), Rd (6), Rg3 (7), Rg1 (8) and Rh2 (9) were isolated from wild ginseng by a UPLC-QTOF-MS-guided fractionation procedure. Their structures were elucidated on the basis of spectroscopic and spectrometric data (1D and 2D NMR, IR, and HR-ESI-MS). The anti-COPD activities were investigated using CSE-stimulated A549 cells. Compounds 1–7 could ameliorate inflammatory reaction in vitro. Furthermore, the protective effect against CSE could be related to the HDAC2 pathway. The study provided some evidences to further elucidate the chemical composition of wild ginseng, and revealed that the compounds isolated from wild ginseng had the protective effect on the injury of cigarette smoke in vitro. The study provides a theoretical basis for the further utilization and development of the wild ginseng. It reminds us that it is meaningful to explore the protective effect of the wild ginseng against the COPD in vivo.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was financially supported by the Bethune Plan Research Project of Jilin University (Grant 2018B22) and the Graduate Innovation Fund of Jilin University (Grant 101832018C085).Notes and references
- D. Xiao, H. Yue and Y. Xiu, et al., Accumulation characteristics and correlation analysis of five ginsenosides with different cultivation ages from different regions, J. Ginseng Res., 2015, 39, 338 CrossRef CAS PubMed.
- M. Mizuno, J. Yamada and H. Terai, et al., Differences in immunomodulating effects between wild and cultured Panax ginseng, Biochem. Biophys. Res. Commun, 1994, 200, 1672 CrossRef CAS PubMed.
- H. L. Zhu, H. Q. Lin, J. Tan and C. Z. Wang, et al., UPLC-QTOF/MS-based nontargeted metabolomic analysis of mountain-and garden-cultivated Ginseng of different ages in Northeast China, Molecules, 2019, 24, 33 CrossRef PubMed.
- The Global Initiative for Chronic Obstructive Lung Disease (GOLD), Global Strategy for the Diagnosis Management and Prevention of COPD 2019, available from: http://goldcopd.org Search PubMed.
- P. R. Sandra, L. D. Puerto-Nevado and T. E. Raúl, et al., Role of Recently Migrated Monocytes in Cigarette Smoke-Induced Lung Inflammation in Different Strain of Mice, PLoS One, 2013, 8, e72975 CrossRef PubMed.
- J. L. Wright, M. Cosio and A. Churg, Animal Models of Chronic Obstructive Pulmonary Disease, Drug Res., 2011, 51, 1004 Search PubMed.
- N. Kim, K. Kim and B. Y. Choi, et al., Metabolomic Approach for Age Discrimination of Panax ginseng Using UPLC-Q-Tof MS, J. Agric. Food Chem., 2011, 59, 10435 CrossRef CAS PubMed.
- N. Kim, K. Kim and D. H. Lee, et al., Nontargeted metabolomics approach for age differentiation and structure interpretation of age-dependent key constituents in hairy roots of Panax ginseng, J. Nat. Prod., 2012, 75, 1777 CrossRef CAS PubMed.
- L. Wu, A. L. Zhang and Y. M. Di, et al., Panax ginseng therapy for chronic obstructive pulmonary disease: a clinical trial protocol and pilot study, Chin. Med., 2014, 9, 20 CrossRef PubMed.
- D. Gross, Z. Shenkman and B. Bleiberg, et al., Ginseng improves pulmonary functions and exercise capacity in patients with COPD, Monaldi Arch. Chest Dis., 2002, 57, 242 CAS.
- S. Guan, Q. Liu and F. Han, et al., Ginsenoside Rg1 ameliorates cigarette smoke-induced airway fibrosis by suppressing the TGF-β1/Smad pathway in vivo and in vitro, Biomed. Res., 2017, 2, 1 Search PubMed.
- W. Y. Bi, B. D. Fu and H. Q. Shen, et al., Sulfated Derivative of 20(S)-Ginsenoside Rh2 Inhibits Inflammatory Cytokines Through MAPKs and NF-kappa B Pathways in LPS-Induced RAW264.7, Macrophages, Inflammation, 2012, 35, 1659 CrossRef CAS PubMed.
- S. H. Jung, M. S. Woo and S. Y. Kim, et al., Ginseng saponin metabolite suppresses phorbol ester-induced matrix metalloproteinase-9 expression through inhibition of activator protein-1 and mitogen-activated protein kinase signaling pathways in human astroglioma cells, Int. J. Cancer, 2010, 118, 490 CrossRef PubMed.
- D. Malhotra, R. K. Thimmulappa and N. Mercado, et al., Denitrosylation of HDAC2 by targeting Nrf2 restores glucocorticosteroid sensitivity in macrophages from COPD patients, J. Clin. Invest., 2011, 121, 4289 CrossRef CAS PubMed.
- P. J. Barnes, Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease, J. Allergy Clin. Immunol., 2013, 131, 636 CrossRef CAS PubMed.
- A. Hakim, I. M. Adcock and O. S. Usmani, Corticosteroid resistance and novel anti-inflammatory therapies in chronic obstructive pulmonary disease, Drugs, 2012, 72, 1299 CrossRef CAS PubMed.
- J. P. Barnes, Development of new drugs for COPD, Curr. Med. Chem., 2013, 20, 1531 CrossRef PubMed.
- M. Yokode, T. Kita and H. Arai, et al., Cholesteryl ester accumulation in macrophages incubated with low density lipoprotein pretreated with cigarette smoke extract, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 2344 CrossRef CAS PubMed.
- Y. Yamaguchi, S. Kagota, J. Haginak and M. Kunitomo, et al., Participation of peroxynitrite in oxidative modification of LDL by aqueous extracts of cigarette smoke, FEBS Lett., 2002, 512, 218 CrossRef CAS PubMed.
- L. Chen, Q. Ge, G. T. Jin and H. Alkhouri, et al., Effects of cigarette smoke extract on human airway smooth muscle cells in COPD, Eur. Respir. J., 2014, 44, 634 CrossRef CAS PubMed.
- M. Bak, J. S. G. Hong and J. W. Lee, et al., Red ginseng marc oil inhibits iNOS and COX-2 via NFκB and p38 pathways in LPS-stimulated RAW 264.7 macrophages, Molecules, 2012, 17, 13769 CrossRef CAS PubMed.
- Z. Qi, W. Li and J. Tan, et al., Effect of ginsenoside Rh2 on renal apoptosis in cisplatin-induced nephrotoxicity in vivo, Phytomedicine, 2019, 61, 152862 CrossRef CAS PubMed.
- Z. Qi, Z. Li and X. W. Guan, et al., Four Novel Dammarane-Type Triterpenoids from Pearl Knots of Panax ginseng Meyer cv. Silvatica, Molecules, 2019, 24, 1159 CrossRef PubMed.
- J. G. Cho, M. K. Lee and J. W. Lee, et al., Physicochemical Characterization and NMR Assignments of Ginsenosides Rb1, Rb2, Rc, and Rd Isolated from Panax ginseng, J. Ginseng Res., 2010, 34, 113 CrossRef CAS.
- M. C. Yang, D. S. Seo and J. K. Hong, et al., Ginsenosides from the roots of Korean cultivated-wild ginseng, Nat. Prod. Sci., 2008, 14, 171 CAS.
- B. D. Fu, W. Y. Bi and C. L. He, et al., Sulfated derivatives of 20 (S)-ginsenoside Rh2 and their inhibitory effects on LPS-induced inflammatory cytokines and mediators, Fitoterapia, 2013, 84, 303 CrossRef CAS PubMed.
- H. X. Sun, Y. Chen and Y. Ye, Ginsenoside Re and Notoginsenoside R1: Immunologic Adjuvants with Low Haemolytic Effect, Chem. Biol., 2006, 3, 718 CAS.
- L. Tu, Y. Zhao and Z. Y. Yu, et al., Six New Dammarane Triterpenoids from Viburnum cylindricum, Helv. Chim. Acta, 2010, 91, 1578 CrossRef.
- C. Z. Gu, J. J. Lv and X. X. Zhang, et al., Minor dehydrogenated and cleavaged dammarane-type saponins from the steamed roots of Panax notoginseng, Fitoterapia, 2015, 103, 97 CrossRef CAS PubMed.
- M. Yoshikawa, T. Murakami and T. Ueno, et al., Bioactive saponins and glycosides. VIII. Notoginseng (1): new dammarane-type triterpene oligoglycosides, notoginsenosides-A, -B,-C, and-D, from the dried root of Panax notoginseng (Burk.) FH, Chen, Chem. Pharm. Bull, 1997, 45, 1039 CrossRef CAS PubMed.
- R. Teng, H. Li and J. Chen, et al., Complete assignment of 1H and 13C NMR data for nine protopanaxatriol glycosides, Magn. Reson. Chem., 2002, 40, 483 CrossRef CAS.
- C. H. Ma, L. He and Z. Wang, et al., Betulin inhibited cigarette smoke-induced COPD in mice, Biomed. Pharmacother., 2017, 85, 679 CrossRef CAS PubMed.
- P. J. Barnes and I. M. Adcock, Glucocorticoid resistance in inflammatory diseases, Lancet, 2009, 373, 1905 CrossRef CAS.
- P. J. Barnes, Role of HDAC2 in the Pathophysiology of COPD, Annu. Rev. Physiol., 2009, 71, 451 CrossRef CAS PubMed.
- J. A. Marwick, P. A. Kirkham and C. S. Stevenson, et al., Cigarette smoke alters chromatin remodeling and induces proinflammatory genes in rat lungs, Am. J. Respir. Cell Mol. Biol., 2004, 31, 633 CrossRef CAS PubMed.
- L. Zeng, J. C. Dong and W. Q. Yu, et al., Baicalin attenuates inflammation by inhibiting NF-κB activation in cigarette smoke induced inflammatory models, Pulm. Pharmacol. Ther., 2010, 23, 411 CrossRef CAS PubMed.
- F. M. Moodie, J. A. Marwick and C. S. Anderson, et al., Oxidative stress and cigarette smoke alter chromatin remodeling but differentially regulate NF-kB activation and proinflammatory cytokine release in alveolar epithelial cells, FASEB J., 2004, 18, 1897 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: 1H NMR, 13C NMR, HMQC, HMBC, HR-ESI-MS of compound 1–9. See DOI: 10.1039/c9ra06635g |
| This journal is © The Royal Society of Chemistry 2019 |