Open Access Article
Open Access ArticleBiodegradable nanosyringes for intracellular amplification-based dual-diagnosis and gene therapy in single living cells†
Xiaotong
Shen
,
Ying
Zhang
,
Jianghui
Sun
,
Hua
Lu
,
Jin
Ouyang
 and
Na
Na
and
Na
Na
 *
*
Key Laboratory of Theoretical and Computational Photochemistry, College of Chemistry, Beijing Normal University, Beijing 100875, China. E-mail: nana@bnu.edu.cn
First published on 6th May 2019
Abstract
The efficient delivery of biomolecules into living cells as well as their easy biodegradation have been challenges for the application of intracellular amplification for sensitive multiple-diagnosis and gene therapy for cancer. Herein, new strategies of amplification-based dual-detection of cancer biomarkers (Let-7a miRNA and VEGF) and gene therapy for cancers are put forward. These are achieved through biodegradable nanosyringes (NSs), rigid and sharp in vitro but degradable in vivo, which are applied for the efficient loading, delivery and release of biomolecules (enzymes, nucleic acids, and even silencing RNA) into living cells. After penetrating cell membranes and escaping from endosomes through their rigid and sharp tips, NSs release biomolecules for fast and easy “one-step” rolling circle amplification (ring formation and amplification) in single living cells. Therefore, based on signals from two probes, FAM-Probe and Cy5-Probe, that selectively bind to amplification products, 100 aM of Let-7a and 100 fM of VEGF could be detected, which are much lower than reported values. Furthermore, siRNAs can also be delivered by NSs for gene therapy, and their therapeutic effect was evaluated by their in vivo antitumor efficacy in CCRF-CEM subcutaneous xenograft nude mice. Rigid in vitro and degradable in vivo, NSs show potential for achieving fast, sensitive and safe cancer diagnosis and efficient therapy.
Introduction
Fast and efficient cancer screening, diagnosis and treatment of cancer in the early stages,1 as well as cancer metastases and complications,2,3 make the highly sensitive detection of multiple cancer biomarkers and gene therapy in multiple types of cells more significant.4 Intracellular amplification (signals could be exponentially amplified) and gene therapy could be highly sensitive and effective methods for cancer diagnostics and therapeutics by detecting and treating RNA biomarkers in cells.5,6 However, because amplification and gene therapy require various molecules, such as ligase, polymerase, primers, and even silencing RNA for subsequent gene therapy, the efficient delivery of diagnostic and therapeutic biological molecules into living cells is gradually emerging as a fundamental challenge.7 For example, without tedious and complicated in vitro ring formation,8,9 modified rolling circle amplification (RCA) with intracellular ring formation by target RNA catalyzed by splintR ligase was reported for mRNA detection in fixed cells.10 However, through simple incubation, the efficiency of biomolecule delivery into living cells was not enough to support further amplification. Therefore, attaining efficient intracellular delivery of multiple biomolecules is key to realizing amplification-based, highly sensitive detection of mRNA biomarkers or modulation of gene expression for therapeutics.Additional advances in applying nanoparticles to cancer diagnostics or therapeutics have been reported;11,12 and the combination of therapy and imaging by nanoparticles has emerged as a new and efficient strategy to achieve precise treatment or personalized medicine.13,14 However, concerns about inefficient transfection, toxicity and degradation, limited site accessibility, and complicated modifications during the delivery process still limit further developments for their application in intracellular diagnostics and therapeutics.15–17 Recently, we designed unique silicon nanourchins for the delivery of biomolecules to achieve intracellular “one-step” reverse-transcription helicase-dependent isothermal amplification in living cells for mRNA detection.5 However, the spherical-like structure of these nanourchins could still be improved for better penetration of cell membranes. Nanoneedles, being rigid and durable with a conical body and sharp tips, could be ideal tools to penetrate cell membranes with minimal impact on their viability and metabolism for subsequent intracellular applications.18 In addition, combined with the favorable biocompatibility, low toxicity, and high loading capacity of silicon nanomaterials,19 silicon nanoneedles are developing into a very sophisticated and minimally invasive tool for in vivo delivery.7,20 In contrast, the reported intracellular delivery by silicon nanoneedles is mainly based on injection by vertical arrays21,22 or by relatively large-scale nanowires (being relatively long with less rigidity),23 which would result in low delivery efficiency and reactivity for effective intracellular detection. Therefore, the development of more sophisticated nanoneedles of an appropriate size for delivering biological molecules into cells would be significant for efficient diagnosis and therapeutic applications.
Here, new strategies of intracellular amplification-based dual-diagnosis and gene therapy are put forward, which are achieved through biodegradable nanosyringes (NSs). With their mesoporous structures and sharp tips, the NSs were applied for the efficient loading and delivery of biomolecules into cells for amplification-based dual-detection of cancer biomarkers, as well as gene therapy for cancer. With their high loading efficiency, efficient delivery, low toxicity and biodegradability, the NSs showed promise for application in highly sensitive dual-detection of cancer biomarkers and gene therapy, indicating significant potential for their use in cancer diagnosis and therapy.
Results and discussion
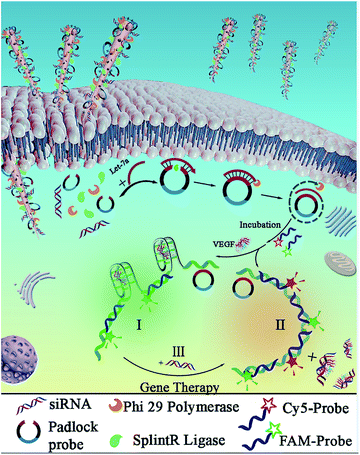
Design of nanosyringes for dual-diagnosis and gene therapy
In the experiment, biodegradable silicon nanosyringes (NSs) were prepared by both covalent binding and electrostatic self-assembly of biomolecules onto free-standing silicon nanoneedles. Upon recognition of the specific nucleic acid molecule, padlock probes, which consisted of two target-complementary segments, were designed. The ends of the probes were joined through the action of a ligase, creating circular DNA molecules catenated to the target sequence in order to provide highly specific detection with minimal background.24,25 Next, the delivery of the designed padlock probes, ligase and polymerase into living cells by NSs was achieved for improved RCA with intracellular ring formation. Subsequently, dual-detection of cancer biomarkers, as well as gene therapy by silencing RNA (small interfering RNA, siRNA), was carried out intracellularly. Based on the important role of highly sensitive miRNA detection in cells for the early diagnosis of cancers,26 Let-7a miRNA (a ubiquitous cancer biomarker) and vascular endothelial growth factor (VEGF) (involved in angiogenesis, inflammation, and wound healing, which is a special biomarker for human acute lymphoblastic leukemia),27 were selected as models for the fast diagnosis of cancer. Simultaneously, siRNA that blocks VEGF expression in cells was also transferred into living cells to inhibit tumor angiogenesis for gene therapy for acute lymphoblastic leukemia.As shown in Scheme 1, the NSs were prepared by the detachment and modification of short mesoporous silicon nanoneedles. The mesoporous silicon nanoneedles were synthesized through HF-assisted etching of silicon wafers by scratching the Si surface with Ag particles in an HF solution (Scheme 1A).28 Next, the free-standing silicon nanoneedles were scraped off after sharpening by KOH solution etching. Thereafter, the mesoporous silicon nanoneedles were functionalized with amino groups by (3-aminopropyl)triethoxysilane (APTES) both on the surface and inside the pores. Next, NSs were obtained by loading the enzymes (splintR ligase and phi29 DNA polymerase) with the assistance of 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC), as well as loading the designed padlock probes and siRNA by electrostatic self-assembly onto the nanoneedles (Scheme 1B). The sequences of the padlock probes and siRNA are listed in Table S1 (see ESI†).
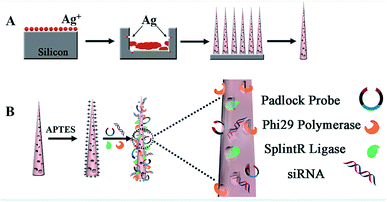 | ||
| Scheme 1 Illustration of the synthesis of NSs. (A) Synthesis of mesoporous silicon nanoneedles. (B) Loading of biomolecules onto the nanoneedles to obtain NSs/siRNA-loaded NSs. | ||
As shown in Scheme 2, after the delivery of biological molecules into the cells by NSs and the release of molecules from the endosomes, an improved “one-step” intracellular RCA was employed with Let-7a to promote ring formation catalyzed by splintR ligase, which was followed by the amplification catalyzed by phi29 polymerase. This intracellular ring formation and amplification process can be completed in “one-step” within ∼2 h, which is much faster than traditional RCA with an extra in vitro ligation, which takes ∼16 h. Here, the eleven 5′- and 3′-terminal bases of the padlock probe were designed to be complementary to half the sequence of a 22 nt miRNA target (Let-7a) for the subsequent RCA reactions, and an aptamer structure of VEGF was designed in the middle part of the padlock probe to form a G-quadruplex. Without using traditional nonspecific FL dyes, two selective probes, Cy5-Probe and FAM-Probe, were designed to improve sensitivity (the sequences are listed in Table S1†).
Therefore, for human acute lymphoblastic leukemia cells (CCRF-CEM) in the presence of both Let-7a and VEGF, the RCA products triggered by Let-7a would hybridize with the FAM-Probe combined with binding to VEGF, which would result in green emission for Let-7a detection (Scheme 2I). However, for nonlymphocytic leukemia cells without VEGF, the Cy5-Probe (with a sequence complementary to the VEGFaptamer) and the FAM-Probe would hybridize to the corresponding sites of the amplification products. This hybridization would result in increased red emission of the Cy5-Probe and finally result in orange emission combined with the green signals (Scheme 2II). Furthermore, the siRNA can also be transferred into cells to act as a reagent of gene therapy in order to block VEGF expression by hybridization with VEGF. Therefore, hybridization of the Cy5-Probe and FAM-Probe with RCA products would be initiated, resulting in the sensing of VEGF based on increased red emission (Scheme 2III).
Characterizations on NSs
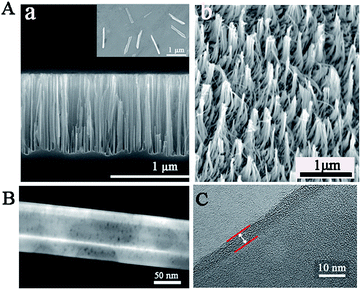
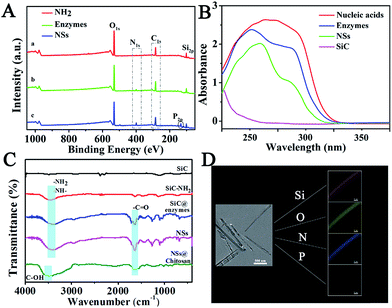
To confirm the successful synthesis of NSs, morphology characterization was employed by scanning electron microscope (SEM), transmission electron microscope (TEM), and high resolution transmission electron microscopy (HRTEM). As shown in Fig. 1, the NSs were approximately 1 μm in length (Fig. 1A) and 100 nm in diameter (Fig. 1B), with a modified layer approximately 5 nm thick on the surface of the NSs (Fig. 1C), which is in agreement with the results obtained from dynamic light scattering (DLS) measurements (Fig. S2†). In addition, the successful loading of enzymes and nucleic acids onto NSs was confirmed by X-ray photoelectron spectra (XPS), showing increased peaks at 284.5 eV (C1s) and 401.1 eV (N1s), as well as a new peak at 129.9 eV (P2p) in comparison to a nanoneedle without biomolecules loaded (Fig. 2A). The remarkable UV-vis absorption of the NSs at 260 nm and 320 nm further confirmed the loading of biomolecules onto the NSs, and these absorptions were attributed to the presence of nucleic acids and enzymes, respectively (Fig. 2B). The characteristic FT-IR peaks of biomolecules loaded onto the NSs were also recorded (Fig. 2C). Furthermore, as demonstrated by energy dispersive X-ray (EDX) analysis (Fig. 2D), the elements corresponding to enzymes and nucleic acids (O, N and P) were well distributed on the NSs. In addition, multipoint Brunauer–Emmett–Teller (BET) analysis was employed to obtain typical adsorption/desorption isotherms for mesoporous nanoneedle powders (Fig. S1A, see ESI†), showing a mean surface area of 182 m2 g−1 and a pore size of 3.2 nm. In addition, the mass loss tested by thermal gravity analysis (TGA) also demonstrated the successful formation of biomolecule-loaded NSs (Fig. S1B†). Therefore, the biomolecule-loaded NSs were successfully synthesized and available for subsequent applications.In vitro analysis performance by NSs
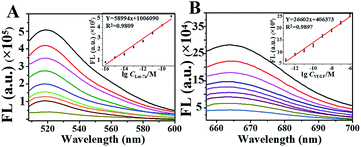
The feasibility of using NSs for amplification to detect both Let-7a and VEGF was examined by in vitro testing in an enzyme reaction buffer (splintR ligase reaction buffer + phi29 DNA polymerase reaction buffer, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). As demonstrated, we obtained significant green emission at 520 nm (Fig. 3A) for the detection of Let-7a and red emission at 665 nm for VEGF sensing (Fig. 3B), which increased as the concentration of the target increased. The emissions showed good linearities for detecting VEGF (Y = 58994x + 1
1). As demonstrated, we obtained significant green emission at 520 nm (Fig. 3A) for the detection of Let-7a and red emission at 665 nm for VEGF sensing (Fig. 3B), which increased as the concentration of the target increased. The emissions showed good linearities for detecting VEGF (Y = 58994x + 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 006
006![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 090, R2 = 0.9809) and Let-7a (Y = 26602x + 406
090, R2 = 0.9809) and Let-7a (Y = 26602x + 406![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 373, R2 = 0.9897), with limits of detection (LOD) of 100 fmol L−1 and 100 amol L−1, respectively which were much lower than reported values.29 In addition, the RCA products were subjected to agarose gel electrophoresis, yielding the same products as traditional RCA (Fig. S3†). Therefore, the feasibility of the developed NSs-based RCA has been confirmed, and this method can potentially be used for highly sensitive miRNA detection in a single cell.
373, R2 = 0.9897), with limits of detection (LOD) of 100 fmol L−1 and 100 amol L−1, respectively which were much lower than reported values.29 In addition, the RCA products were subjected to agarose gel electrophoresis, yielding the same products as traditional RCA (Fig. S3†). Therefore, the feasibility of the developed NSs-based RCA has been confirmed, and this method can potentially be used for highly sensitive miRNA detection in a single cell.
Cell toxicity and biodegradability of NSs
Toxicity and the biodegradability were examined before biological applications. The standard CCK-8 assay (Fig. S4†) demonstrated more than 82.7% cell viability for NSs at concentrations ranging from 0.5 to 50 μg mL−1 after incubation for 24 h, and 90% cell viability was demonstrated for 30 μg mL−1 NSs, which was applied for the subsequent cell imaging. To test their biodegradability, the NSs were incubated in cell-culture medium at 37 °C for different times. As demonstrated, NSs progressively dissolved over time, and their structures began to collapse after 1 h, ultimately losing their shape after 4 h, and only their solid stump remained after 16 h (Fig. S5A†). In addition, the gradually fading colors of the NSs solutions with time (Fig. S5B†) also confirmed their dissolution and biodegradation. Significantly, this unique degradation behavior enabled the release of loaded biomolecules, which matched with the subsequent intracellular amplifications for cell imaging. Therefore, these NSs would be temporarily present in cells and completely dissolved after completing their missions.Cell uptake experiments
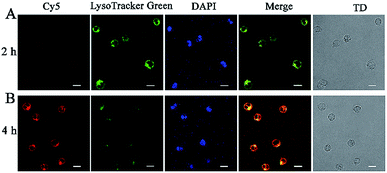
To confirm the release of NSs from endosomes for subsequent RCA reactions, Cy5-siRNA labelled silicon nanoneedles were prepared for incubation with CCRF-CEM (human acute lymphoblastic leukemia) cells for 2 h. A co-staining experiment was further performed by staining lysosomes with LysoTracker Green and the cell nucleus with 4′,6-diamidino-2-phenylindole (DAPI). Demonstrated by the overlap of red (Cy5-siRNA) and green (Lysosome) FL signals (Fig. 4A), Cy5-siRNA were localized to lysosomes after incubation for 2 h. Although the red emission was not significant, this finding may still indicate the successful cellular internalization of NSs through endocytosis.30 However, after 4 h of incubation, weaker FL signals from LysoTracker Green were observed in combination with significantly increased red emission from the increased amount of Cy5-siRNA in the cells (Fig. 4B). This finding suggested the spread of LysoTracker Green from acidic lysosomes to the neutral cytoplasm, because lysosome staining was employed in acidic conditions.9 Therefore, the efficient translocation or release of NSs from lysosomes was achieved.31 In addition, the colocalization coefficients (Pearson's correlation coefficient) were 0.96 for early endosomes and lysosomes. This result indicated an endocytic pathway for cellular uptake of NSs with efficient escape of the delivered proteins from endo-lysosomes into the cytoplasm (Fig. S6†).Feasibility and reliability of NS-based RCA
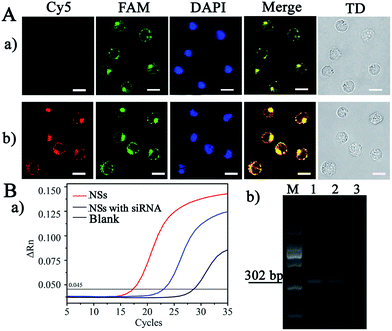
Subsequently, the feasibility of applying the NS-based RCA to cell imaging was evaluated by the detection of endogenous VEGF, whose concentration was controlled by introducing siRNA to block VEGF expression for gene therapy, as an example. The hematopoietic cells of human acute lymphoblastic leukemia cells (CCRF-CEM), with high expression of VEGF during the hematopoietic process, were selected for the imaging. As demonstrated in Fig. 5A-a, significant green FL emission was observed at cytosolic locations in the cells, and this signal was generated from the hybridization of FAM-Probe and the RCA products triggered by Let-7a (the process is shown in Scheme 2I). However, with the addition of siRNA for VEGF interference (Fig. 5A-b), a remarkable increase in red emission from the cytosolic locations was observed, due to the competitive hybridization of the Cy5-Probe rather than VEGF (the process is shown in Scheme 2III). Therefore, the present NSs-based RCA can be used for intracellular imaging for miRNA and VEGF sensing.Furthermore, traditional RT-PCR was employed to demonstrate the reliability of the present amplification method. As shown in Fig. 5B-a, a lag in time corresponding to the maximum slope in the FL curve was demonstrated by the point of inflection (blue line, Fig. 5B-a) compared with that of CCRF-CEM cells without siRNA interference (red line, Fig. 5B-a). Therefore, we obtained decreased VEGF-mRNA concentrations after gene interference by siRNA, as demonstrated by the dynamic correlation between FL intensity and the relative VEGF-mRNA concentration in the cells. This finding is also in accordance with the imaging performed after NS-based intracellular RCA (Fig. 5A). In addition, the corresponding RT-PCR products were subjected to agarose gel electrophoresis, which resulted in the same products at 302 bp. Therefore, the present NS-based RCA method is reliable and feasible for intracellular imaging of VEGF.
Real cell imaging and distribution of NSs in organs
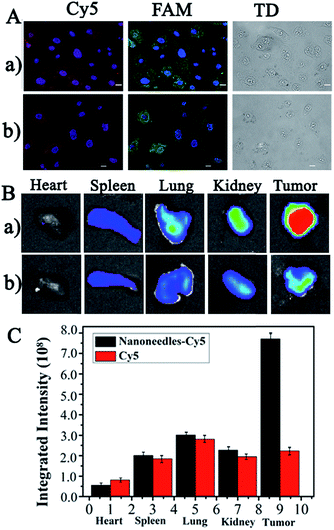
The real cell imaging of common HepG2 cells, with high Let-7a expression but low VEGF expression,32,33 was employed to further verify the reliability of the present NS-based method. As shown in Fig. 6A, significant red and green emission were observed in HepG2 cells without (Fig. 6A-a) and with siRNA treatment (Fig. 6A-b); these emissions were generated from the hybridization of the Cy5-Probe and the FAM-Probe at the corresponding sites of amplification products by Let-7a (Scheme 2II). The similar FL images with and without siRNA further confirmed the low expression of VEGF in HepG2 cells. Therefore, based on the VEGF information, this NS-based RCA method can be used for the discrimination of CCRF-CEM cells from HepG2 cells and can also be used for the evaluation of gene therapy by siRNA interference.For in vivo detection, the distribution of NSs present in organs was employed to verify whether the NSs can be successfully accumulated in the tumor site after blood circulation for efficient cancer diagnostic and therapy. In the experiment, Cy5-DNA-labelled NSs and dissociative Cy5-DNA were prepared for tail vein injection into HepG2 subcutaneous xenograft nude mice. The mice were sacrificed after 5 h of administration, and the main organs and tumors were excised for ex vivo imaging and semi-quantification. As shown in Fig. 6B and C, much greater FL signals at the tumor area on the NSs treated mice were shown via the enhanced permeability and retention effect (EPR effect), which was due to the leaky vasculature in the solid tumor tissues compared with healthy vessels in normal organs. Therefore, the present NSs would contribute greatly to higher therapeutic efficiency and reduced side effects.
Evaluation of gene therapy of NSs by in vivo imaging
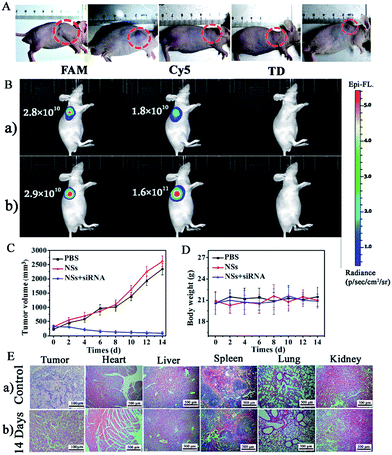
Finally, an in vivo gene therapy evaluation of siRNA in CCRF-CEM subcutaneous xenograft nude mice was employed with the injection of siRNA-loaded NSs to interfere with the expression of VEGF in the mice tumors. As shown in Fig. 7A, by the injection of siRNA-loaded NSs (1.0 mg mL−1, dispersed in PBS buffer) directly into the tumor sites of mice, we observed obvious suppression of the tumor in two days, and the size of the tumor decreased gradually, as shown by the smaller tumor size of approximately 91 mm3 on the 14th day. In contrast, without siRNA, the size of the tumor increased with time, showing sizes of 2350 mm3 and 2621 mm3 on the 14th day for the PBS-injection group and the NSs-injection group, respectively (Fig. 7C). A comparison of tumor size for the different groups is shown in Fig. 7C.In addition, in vivo optical imaging of mice without and with the injection of siRNA was carried out. As shown in Fig. 7B-a, without siRNA-loaded NS injection, we recorded significant emission from the FAM-Probe and a quite low signal from the Cy5-Probe caused by the presence of both Let-7a and VEGF at the tumor site. However, 14 days after the injection of siRNA to interfere with VEGF expression in mice with tumors, we observed a comparable FAM-Probe signal but a dramatically increased Cy5-Probe signal. This difference was due to the suppression of VEGF expression (Scheme 2III), which confirmed the therapeutic effect of the siRNA (Fig. 7B-b).
During the siRNA gene therapy, we did not observe obvious changes in the body weights of the mice, indicating that the effects on the mice were lower (Fig. 7D). This finding was further confirmed by the H&E staining of tissue sections from the untreated and siRNA-treated groups. As demonstrated in Fig. 7E, significant tumor cell apoptosis could be observed in tumor sections after injection with siRNA-loaded NSs. However, no obvious difference between the two groups was observed in other tissues, such as heart, liver, spleen, lung, or kidney tissues. Therefore, gene therapy by siRNA-loaded NSs was verified and was determined to be safe with few side-effects on other tissues.
Conclusions
In conclusion, biodegradable NSs were synthesized for efficient intracellular delivery of enzymes, nucleic acids, and even gene silencing regents for both dual-diagnosis and gene therapy for cancer. Being rigid and sharp in vitro as well as degradable in vivo, the NSs can support the efficient loading of biomolecules, penetrate cell membranes and be released from endosomes for a fast and simple “one-step” intracellular RCA (including both ring formation and amplification) in living cells. With the present NSs, highly sensitive dual-detection of cancer biomarkers and gene therapy were successfully achieved, in single living cells and mice with tumors, showing significant potential in cancer diagnosis and therapy.Ethical statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Beijing Normal University and Experiments were approved by the Animal Ethics Committee of Beijing Normal University.Conflicts of interest
There are no conflicts to declare.Acknowledgements
N. N., X. S., Y. Z., J. S., H. L. gratefully acknowledge financial support provided by the National Natural Science Foundation of China (Grant No. 21675015 and 21874012). J. O. is thankful for financial support provided by the National Natural Science Foundation of China (Grant No. 21675014). All the authors are thankful for financial support provided by the Fundamental Research Funds for the Central Universities.References
- J. Condeelis and J. E. Segall, Nat. Rev. Cancer, 2003, 3, 921–930 CrossRef CAS PubMed.
- R. A. Patchell, Cancer Treat. Rev., 2003, 29, 533–540 CrossRef PubMed.
- S. Valastyan and R. A. Weinberg, Cell, 2011, 147, 275–292 CrossRef CAS PubMed.
- A. K. Shalek, J. T. Robinson, E. S. Karp, J. S. Lee, D.-R. Ahn, M.-H. Yoon, A. Sutton, M. Jorgolli, R. S. Gertner, T. S. Gujral, G. MacBeath, E. G. Yang and H. Parka, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1870–1875 CrossRef CAS PubMed.
- X. T. Shen, Y. Wang, Y. Zhang, J. Ouyang and N. Na, Adv. Funct. Mater., 2018, 28, 1803286 CrossRef.
- Z. Wu, G. Q. Liu, X. L. Yang and J. H. Jiang, J. Am. Chem. Soc., 2015, 137, 6829–6836 CrossRef CAS PubMed.
- C. Chiappini, E. D. Rosa, J. O. Martinez, X. Liu, J. Steele, M. M. Stevens and E. Tasciotti, Nat. Mater., 2015, 14, 532–539 CrossRef CAS PubMed.
- R. Deng, L. Tang, Q. Tian, Y. Wang, L. Lin and J. Li, Angew. Chem., 2014, 126, 2421–2425 CrossRef.
- K. Ren, Y. Liu, J. Wu, Y. Zhang, J. Zhu, M. Yang and H. Ju, Nat. Commun., 2016, 7, 13580 CrossRef CAS PubMed.
- R. J. Deng, K. X. Zhang, Y. P. Sun, X. J. Ren and J. H. Li, Chem. Sci., 2017, 8, 3668–3675 RSC.
- X. D. Meng, W. H. Dai, K. Zhang, H. F. Dong and X. J. Zhang, Chem. Sci., 2018, 9, 1184–1190 RSC.
- Z. J. Gu, L. Yan, G. Tian, S. J. Li, Z. F. Chai and Y. L. Zhao, Adv. Mater., 2013, 25, 3758–3779 CrossRef CAS PubMed.
- Y. F. Ma, J. Huang, S. J. Song, H. B. Chen and Z. J. Zhang, Small, 2016, 12, 4936–4954 CrossRef CAS PubMed.
- Z. M. Ying, Y. Y. Yuan, B. Tu, L. J. Tang, R. Q. Yu and J. H. Jiang, Chem. Sci., 2019, 10, 4828–4833 RSC.
- Y. L. Zhong, X. T. Sun, S. Y. Wang, F. Peng, F. Bao, Y. Y. Su, Y. Y. Li, S.-T. Lee and Y. He, ACS Nano, 2015, 9, 5958–5967 CrossRef CAS PubMed.
- F. Y. Wang, Q. D. Yang, G. Xu, N.-Y. Lei, Y. K. Tsang, N. B. Wong and J. C. Ho, Nanoscale, 2011, 3, 3269–3276 RSC.
- J. Wen, K. Yang, F. Y. Liu, H. J. Li, Y. Q. Xu and S. G. Sun, Chem. Soc. Rev., 2017, 46, 6024–6045 RSC.
- A. K. Shalek, J. T. Robinson, E. S. Karp, J. S. Lee, D.-R. Ahn, M.-H. Yoon, A. Sutton, M. Jorgolli, R. S. Gertner, T. S. Gujral, G. MacBeath, E. G. Yang and H. Park, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1870–1875 CrossRef CAS PubMed.
- J. H. Park, L. Gu, G. von Maltzahn, E. Ruoslahti, N. B. Sangeeta and M. J. Sailor, Nat. Mater., 2009, 8, 331–336 CrossRef CAS PubMed.
- R. Elnathan, B. Delalat, D. Brodoceanu, H. Alhmoud, F. J. Harding, K. Buehler, A. Nelson, L. Isa, T. Kraus and N. H. Voelcker, Adv. Funct. Mater., 2015, 25, 7215–7225 CrossRef CAS.
- R. Kawamura, M. Miyazaki, K. Shimizu, Y. Matsumoto, Y. R. Silberberg, R. R. Sathuluri, M. Iijima, S. Kuroda, F. Iwata, T. Kobayashi and C. Nakamura, Nano Lett., 2017, 17, 7117–7124 CrossRef CAS PubMed.
- Z. Huang, H. Fang and J. Zhu, Adv. Mater., 2007, 19, 744–748 CrossRef CAS.
- F. Peng, Y. Y. Su, X. P. Wei, Y. M. Lu, Y. F. Zhou, Y. L. Zhong, S.-T. Lee and Y. He, Angew. Chem., Int. Ed., 2013, 52, 1457–1461 CrossRef CAS PubMed.
- J. Banér, M. Nilsson, M. Mendel-Hartvig and U. Landegren, Nucleic Acids Res., 1998, 26, 5073–5078 CrossRef PubMed.
- M. Nilsson, H. Malmgren, M. Samiotaki, M. Kwiatkowski, B. P. Chowdhary and U. Landegren, Science, 1994, 265, 2085–2088 CrossRef CAS PubMed.
- K. B. Reddy, Cancer Cell Int., 2015, 15, 38 CrossRef PubMed.
- C. Giampietro, G. Deflorian, S. Gallo, A. D. Matteo, D. Pradella, S. Bonomi, E. Belloni, D. Nyqvist, V. Quaranta, S. Confalonieri, G. Bertalot, F. Orsenigo, F. Pisati, E. Ferrero, G. Biamonti, E. Fredrickx, C. Taveggia, C. D. R. Wyatt, M. Irimia, P. P. D. Fiore, B. J. Blencowe, E. Dejana and C. Ghigna, Nat. Commun., 2015, 6, 8479 CrossRef CAS PubMed.
- Y. He, Y. L. Zhong, F. Peng, X. P. Wei, Y. Y. Su, S. Su, W. Gu, L. S. Liao and S.-T. Lee, Angew. Chem., Int. Ed., 2011, 50, 3080–3083 CrossRef CAS PubMed.
- R. Freeman, J. Girsh, A. F. Jou, J. A. A. Ho, T. Hug, J. Dernedde and I. Willner, Anal. Chem., 2012, 84, 6192–6198 CrossRef CAS PubMed.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Chem. Soc. Rev., 2017, 46, 4218–4244 RSC.
- J. Mosquera, I. García and L. M. Liz-Marzan, Acc. Chem. Res., 2018, 51, 2305–2313 CrossRef CAS PubMed.
- W. Fiedler, U. Graeven, S. Ergun, S. Verago, N. Kilic, M. Stockschlader and D. Hossfeld, Blood, 1997, 89, 1870–1875 CAS.
- X. Y. Wen, S. Y. Wu, Z. Q. Li, Z. Q. Liu, J. J. Zhang, G. F. Wang, Z. H. Jiang and S. G. Wu, Phytother. Res., 2009, 23, 778–784 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, additional data and graphs. See DOI: 10.1039/c9sc01894h |
| This journal is © The Royal Society of Chemistry 2019 |