Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
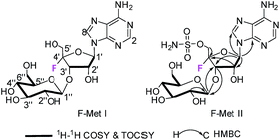
Two 3′-O-β-glucosylated nucleoside fluorometabolites related to nucleocidin in Streptomyces calvus†
Xuan
Feng
,
Davide
Bello
 ,
Phillip T.
Lowe
,
Phillip T.
Lowe
 ,
Joshua
Clark
and
David
O'Hagan
,
Joshua
Clark
and
David
O'Hagan
 *
*
School of Chemistry, University of St Andrews, North Haugh, St Andrews, Fife, KY16 9ST, UK. E-mail: do1@st-andrews.ac.uk
First published on 20th August 2019
Abstract
The antibiotic nucleocidin is a product of the soil bacterium Streptomyces calvus T-3018. It is among the very rare fluorine containing natural products but is distinct from the other fluorometabolites in that it is not biosynthesised from 5′-fluorodeoxyadenosine via the fluorinase. It seems to have a unique enzymatic fluorination process. We disclose here the structures of two 4′-fluoro-3′-O-β-glucosylated metabolites (F-Mets I and II) which appear and then disappear before nucleocidin production in batch cultures of S. calvus. Full genome sequencing of S. calvus T-3018 and an analysis of the putative biosynthetic gene cluster for nucleocidin identified UDP-glucose dependent glucosyl transferase (nucGT) and glucosidase (nucGS) genes within the cluster. We demonstrate that these genes express enzymes that have the capacity to attach and remove glucose from the 3′-O-position of adenosine analogues. In the case of F-Met II, deglucosylation with the NucGS glucosidase generates nucleocidin suggesting a role in its biosynthesis. Gene knockouts of nucGT abolished nucelocidin production.
Introduction
The antibiotic nucleocidin 1 is among the very rare fluorinated natural products.1 It is a 5′-O-sulfonyl antibiotic produced by the actinomycete bacterium Streptomyces calvus.2 Nucleocidin 1 is a member of a small class of such antibiotics which include the chlorine containing ascamycin 2 and dealanylascamycin 3, as shown in Fig. 1A.3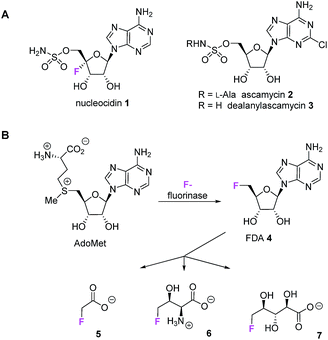 | ||
| Fig. 1 (A) Nucleoside antibiotics 1–3. (B) The fluorinated natural products 5–7 derive from the fluorinase via FDA 4, however this enzyme is not involved in nucleocidin 1 biosynthesis. | ||
The fluorine atom unique to nucleocidin 1 is located at C4′ of the ribose ring of the 5′-O-sulfamylated adenosine. The biosynthesis of nucleocidin is unknown. Only three other bacterial fluorometabolites have been identified: these are fluoroacetate 5,4,5 4-fluorothreonine 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4,5 and (2R,3S,4S)-trihydroxy-5-fluoropentanoic acid 7,6 however these metabolites are biosynthesised by pathways which are initiated by the C–F bond forming enzyme, adenosyl fluoride synthase (EC 2.5.1.63) also known as ‘fluorinase’.7 This enzyme combines fluoride ion and S-adenosyl-L-methionine to generate 5′-fluorodeoxy adenosine 4 (FDA) which is then biotransformed to the bacterial fluorometabolites 5–7 as summarised in Fig. 1B.1,8 However, full genome sequencing of the nucleocidin 1 producer S. calvus9 indicates that there is no related fluorinase gene present in S. calvus and the positioning of the fluorine at the C-4′ position of the ribose ring in nucleocidin 1 is inconsistent with an origin from FDA 4, suggesting a unique fluorination enzyme.
4,5 and (2R,3S,4S)-trihydroxy-5-fluoropentanoic acid 7,6 however these metabolites are biosynthesised by pathways which are initiated by the C–F bond forming enzyme, adenosyl fluoride synthase (EC 2.5.1.63) also known as ‘fluorinase’.7 This enzyme combines fluoride ion and S-adenosyl-L-methionine to generate 5′-fluorodeoxy adenosine 4 (FDA) which is then biotransformed to the bacterial fluorometabolites 5–7 as summarised in Fig. 1B.1,8 However, full genome sequencing of the nucleocidin 1 producer S. calvus9 indicates that there is no related fluorinase gene present in S. calvus and the positioning of the fluorine at the C-4′ position of the ribose ring in nucleocidin 1 is inconsistent with an origin from FDA 4, suggesting a unique fluorination enzyme.
For many years nucleocidin 1 production remained elusive because the publically available strains of S. calvus did not support its biosynthesis. Recently however, this was recognised to be the result of a bald gene (bld A) mutation which when reversed in S. calvus ATCC 13382, re-established nucleocidin production as well as an aerial mycelial phenotype.9,10 In this study we have used a producing strain of S. calvus T-3018 held by Pfizer, which does not have this mutation and is a competent nucleocidin 1 producer. In our efforts to explore the biosynthesis of nucleocidin 1 we recently reported the incorporation of isotopically labelled glycerols into 1 and demonstrated that both of the C5′-hydrogens of the ribose ring derive intact from the pro-R hydroxymethyl group of glycerol. This indicates that there is no oxidation at this carbon as glycerol is progressed along the pentose phosphate pathway and into the ribose moiety of nucleocidin.11 This places certain limitations on the biosynthesis of 1 by excluding late stage intermediates that might activate C4′ for fluorination, by oxidation at C5′.
When monitoring S. calvus T-3018 cultures by 19F{1H}-NMR to assess nucleocidin 1 production, periodically two unidentified fluorinated metabobiles (F-Mets I and II) appear.11 These compounds have also been observed in other studies on nucleocidin 1 production in S. calvus ATCC 13382![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9a and most recently during nucleocidin production in Streptomyces asteroporus DSM 41452.10
9a and most recently during nucleocidin production in Streptomyces asteroporus DSM 41452.10
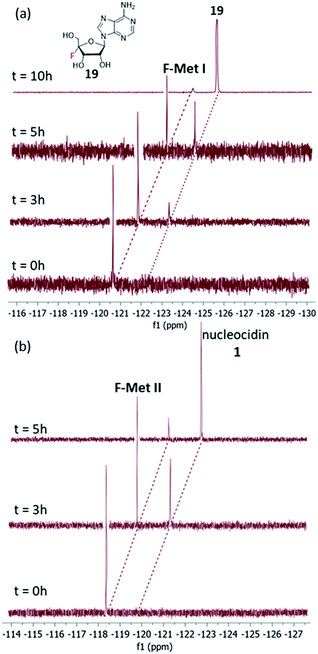
Our observations indicate that F-Mets I and II appear and disappear prior to the accumulation of nucleocidin 1 itself, suggesting an intimate association with antibiotic production. A typical time course profile from a S. calvus T-3018 fermentation is shown in Fig. 2 and it can be seen that F-Mets I and II accumulate between days 4 and 8 and then disappear between days 8–11 as nucleocidin 1 emerges. Clearly the structure of these compounds could offer an insight into the fluorometabolite pathway in S. calvus. We are now able to report their structures and have identified genes/enzymes which lead to their formation and in one case its conversion to nucleocidin 1.
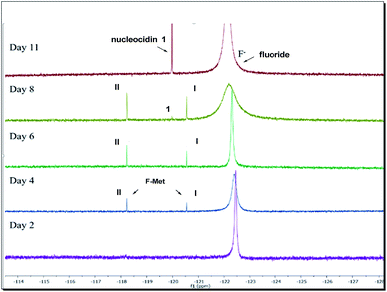 | ||
| Fig. 2 19F-NMR time course of nucleocidin 1 production in S. calvus showing the appearance and disappearance of F-Mets I and II over 11 days. | ||
Results and discussion
F-Mets I and II were isolated in low milligram amounts by preparative HPLC guided by MS–MS analysis after adsorption onto charcoal from 6–8 day cultures of S. calvus T-3018. High resolution mass spectrometry (HRMS) gave accurate masses and therefore elemental constitutions (F-Met I = 448.14673 amu; C16H23FN5O9. F-Met II = 527.11993 amu; C16H24FN6O11S). Although the titres were low, sufficient material was isolated to be able to record 1H- and 19F-NMR in each case and a 13C-NMR for F-Met II. Detailed 2D-NMR (COSY, TOCSY and HMBC) analyses indicated that the compounds were the 3′-O-β-glucosylated derivatives of 1 and that F-Met I is a 3′-O-β-glucosylated-4′-fluoroadenosine and F-Met II is a 3′-O-β-glucosylated nucleocidin. The NMR data are summarised in Table 1 alongside nucleocidin 1 for comparison. See ESI† for full details. The structure of F-Met II is illustrated in a book chapter from 1995, indicating a compound isolated from a Ciba-Geigy bacterium, Streptomyces strain R-156, however we cannot find any details of its isolation or characterisation beyond this anecdotal comment.12| Position | F-Met I (δF = −120.54 ppm) | F-Met II (δF = −118.23 ppm) | Nucleocidin 1 (ref. 9) (δF = −119.98 ppm) | ||
|---|---|---|---|---|---|
| δ H ppm | δ H ppm | δ C ppm | δ H ppm | δ C ppm | |
| 2 | 8.22 (s) | 8.36 (s) | — | 8.21(s) | 152.9 |
| 8 | 8.29 (s) | 8.38 (s) | 139.2 | 8.21(s) | 139.4 |
| 1′ | 6.37 (d, 2.0) | 6.45 (d, 0.9) | 90.9 | 6.39 (d, 1.9) | 91.2 |
| 2′ | 4.83 (dd, 5.6, 2.0) | 4.91 (dd, 5.9, 1.4) | 72.6 | 4.85 (dd, 6.5, 1.9) | 71.9 |
| 3′ | 5.16 (dd, 16.7, 5.9) | 5.35 (dd, 18.3, 6.0) | 76.8 | 5.10 (m) | 70.7 |
| 4′ | — | — | 114.7 | — | 115.9 |
| 5′ Ha | 3.76 (dd, 11.8, 4.8) | 4.42 (d, 9.1), | 66.5 | 4.35, 4.39 (m) | 66.9 |
| 5′ Hb | 3.85 (overlap with 6′′ Hb) | 4.46 (d, 8.3) | |||
| 1′′ | 4.70 (d, 7.7) | 4.76 (d, 7.7) | 102.9 | — | — |
| 2′′–5′′ | 3.39–3.47 (m) | 3.41–3.48 (m) | 69.9–77.0 | — | — |
| 6′′ Ha | 3.69 (dd, 11.8, 4.9) | 3.69 (m) | 61.3 | — | — |
| 6′′ Hb | 3.84 (overlap with 5′ Hb) | 3.86 (m) | |||
A putative gene cluster was previously identified by Zechel and Bechthold9 for nucleocidin 1 biosynthesis in S. calvus ATCC 13382 by homology to the gene cluster of the structurally related antibiotics ascamycins 2 and 3.13 A summary of the gene cluster is illustrated in Fig. 3 and more comprehensively in Fig. S36.†
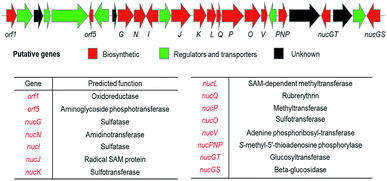 | ||
| Fig. 3 Organisation of the putative nucleocidin biosynthetic gene cluster in S. calvus. A fuller annotation and a comparison with the ascamycin 2 antibiotic gene cluster, can be found in the ESI Fig. S36.† | ||
The integrity of the gene cluster was established when correcting the mutation in a bld gene, which codes a Leu-tRNAUUA able to translate a rare TTA codon into leucine. Three genes in the nuc cluster (nucJ, orf4 and orf6) contain this rare TTA codon and when the mutation in the bldA gene was corrected to re-establish this Leu-tRNAUUA the production of nucleocidin was re-established. We have independently sequenced the S. calvus T-3018 genome (recently deposited in GenBank)14 and find that unlike S. calvus ATCC 13382, S. calvus T-3018 does not contain a bld gene mutation.
The genome of S. asterosporus DSM 41452 was recently sequenced10 and was shown to possess a similar cluster; when this organism was grown with fluoride it produced nucleocidin 1, adding further support that this biosynthetic gene cluster is responsible for nucleocidin biosynthesis.
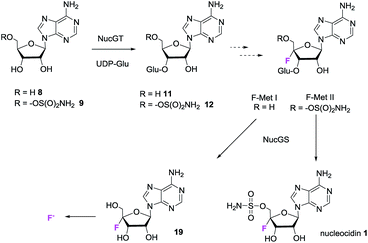
There is also a 3′-phosphoadenosine 5′-phosphosulfate synthases (PAPS) cassette within the genomes of these organisms, consistent with a requirement to assemble the sulfamoyl moiety of the antibiotic. From the S. calvus sequences, the putative nuc cluster predicts glycosyltransferase (nucGT) and β-glucosidase (nucGS) genes which sit at the end of the cluster and are located three genes apart (see Fig. S36† for details). In the light of the structures of F-Mets I and II, we were motivated to over-express these two candidate glucosylating genes to assay their products. This was achieved by PCR amplification from genomic DNA (S. calvus), insertion of the gene into a pEHISTEV plasmid and then over- expression in E. coli Rosetta cells.5,15 In each case soluble protein of the predicted molecular mass (35.5 kDa for NucGT and 53.6 kDa for NucGS) was obtained after purification on a nickel column (see ESI†). The proteins were unambiguously confirmed by MS–MS analyses. The over-expressed glycosyltransferase (NucGT), was explored as a catalyst for 3′-O-β-glucosylation of adenosine 8 and sulfamoyladenosine 916 as candidate substrates using the common glucose donor, UDP-glucose.17
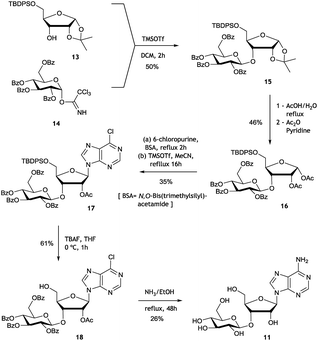
Both 8 and 9 were good substrates based on initial rates with a saturating concentration of UDP-glucose to generate 11 and 12 respectively (Fig. 4). Sulfamoyl adenosine 9 was the more efficient substrate. Adenosine-5′-sulfate 1018 was also examined as a substrate for NucGT however it was not processed by the enzyme, and presumably the formal charge of the sulfate is incompatible with enzyme binding. Additional support for the product structures was secured by an independent synthesis of 3′-O-β-gluco-adenosine 11 as illustrated in Fig. 5. The synthesis involved a β-glycoside specific coupling of protected ribose 1319 and activated glucose 1420 to generate 15. Introduction of the adenosine was then achieved via a 6-chloropurine strategy21 to generate 18, with global aminolysis completing the synthesis. Synthetic and enzymatically generated 11 were identical (see Fig. S34 and S35†). This synthesis also offers reinforcement of the structures of F-Mets I and II in terms of NMR signatures consistent with 3′-O-glucosylation and β-glycoside linkage.
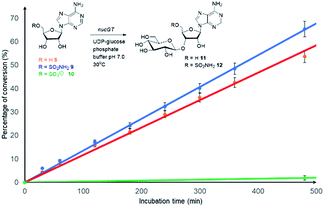 | ||
Fig. 4 Comparison of the initial rates (triplicates) of 8–10 with Nuc-GT. Rates 8 ( ), 9 ( ), 9 (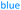 ) and 10 ( ) and 10 (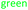 ). ). | ||
In order to assay the glucosidase (Nuc-GS) in the context of nucelocidin biosynthesis we required access to samples of F-Mets I and II. These are not readily synthesised, and thus analytical samples of each were secured by semi-preparative HPLC from S. calvus T-3018 fermentations arrested at day 8. Although only analytical amounts of material were isolated in each case, reactions could be conveniently monitored by 19F{1H}-NMR. Incubation of F-Met I with Nuc-GS resulted in its complete conversion to 4′-fluoroadenosine 19 as illustrated in Fig. 6a. This compound has previously been prepared and is reported to be stable in buffer for several days but it slowly hydrolyses to release fluoride.22
Incubation of F-Met I with NucGS gave a clear conversion to a new product with a signal at −122.5 ppm by 19F-NMR as illustrated in Fig. 6a, consistent with literature values for 19 and LC-MS data which also indicated the formation of 19. On the other hand, treatment of F-Met II with NucGS resulted in its conversion to nucleocidin 1, as illustrated in Fig. 6b. These in vitro biotransformations are consistent with the observed time course experiments illustrated in Fig. 2 and can be interpreted that F-Met I is deglucosylated to 19 and slowly decomposes to fluoride. If compound 19 survives the extraction after adsorption onto charcoal/Celite from the whole cells of S. calvus, then its presence would be disguised in the 19F NMR spectrum by the dominant fluoride ion peak at −122.5 ppm (see Fig. 2). For F-Met II, the action of NucGS generates nucleocidin 1 and it follows that NucGS and F-Met II could be involved in the final step in nucleocidin biosynthesis.
In order to further interrogate the roles of NucGS and NucGT in nucleocidin biosynthesis, knockout experiments were conducted for nucGT and nucGS (see Fig. S37 and S38†). In-frame deletion of nucGT completely abolished fluorometabolite production, consistent with a role in nucleocidin biosynthesis. A similar deletion of nucGS did not significantly affect the production of F-Mets I and II, consistent with their glucosylated status, however nucleocidin 1 production was not affected either. On further analysis of the S. calvus genome there are two additional β-glycosidase enzymes coded for elsewhere on the chromosome which have high (∼88%) amino acid sequence similarity to NucGS (see Fig. S39†) suggesting redundancy in terms of these enzymes with their capacity to deglucosylate F-Met II. We note that in the recently reported genome sequence of the nucleocidin producer, Streptomyces asteroporus DSM 41452![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 there are corresponding genes with a 100% amino acid similarity. Thus from the two organisms known to produce nucelocidin, both have these genes in their clusters, indicative of key roles in the biosynthesis.
10 there are corresponding genes with a 100% amino acid similarity. Thus from the two organisms known to produce nucelocidin, both have these genes in their clusters, indicative of key roles in the biosynthesis.
The previous isotopic labelling studies with glycerol11 indicate that C5′ of adenosine is not oxidised during the biosynthesis of 1, and this study suggests that the fluorination substrate is a 3′-O-β-glycosylated adenosine as summarised in Fig. 7.
It follows that an enzymatic fluorination mechanism involving oxidation of free alcohols at C3′ and/or C5′ seem unlikely, raising the prospect that fluorination occurs by direct C4′–H activation. One hypothesis would involve a C4′-centred oxocarbenium ion which is attacked by fluoride ion. Within the biosynthetic gene cluster are a number of genes encoding enzymes of unknown function and also a gene (nucJ) predicted to encode a radical SAM/iron–sulfur (Fe–S) cluster enzyme which may have the capacity to achieve such a two electron oxidation.
Conclusions
We have identified two novel fluorometabolites (F-Met I and F-Met II), which are 3′-O-glucosylated, 4′-fluoro-riboadenosines from S. calvus. The products of two genes (nucGS and nucGT) identified in the putative nucleocidin gene cluster were over-expressed and the corresponding proteins have the ability to glucosylate (NucGT) and deglucosylate (NucGS) these two fluorometabolites at the 3′-hydroxyl group of the ribose ring. In the case of F-Met II, deglucosyaltion generates nucleocidin 1. These observations support a role for F-Met I and F-Met II in the biosynthesis of nucleocidin. The identity of these metabolites is highly suggestive that one is a product of the unknown fluorination enzyme in S. calvus, and should provide a greater focus In efforts to identify this interesting enzyme.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
We thank the EPSRC Catalysis HUB (University of Manchester) for financial support. We also thank Roger Howard, Usa Reilly, Jeffery Janso and Alessandra Eustaquio at the Biocatalysis Technologies Laboratory, Pfizer, Groton, USA, for financial support, a producing strain of S. calvus, and supporting the research with genome sequencing.Notes and references
- D. O'Hagan and H. Deng, Chem. Rev., 2015, 115, 634–649 CrossRef PubMed.
- S. O. Thomas, V. L. Singleton, J. A. Lowery, R. W. Sharpe, L. M. Pruess, J. N. Porter, J. H. Mowat and N. Bohonos, Antibiot. Annu., 1956, 1956–1957 Search PubMed.
- (a) K. Isono, M. Uramoto, H. Kusakabe, N. Miyata and T. Koyama, J. Antibiot., 1984, 37, 670–672 CrossRef CAS PubMed; (b) E. Takahashi and T. Beppu, Biochem. J., 1982, 233, 459–463 Search PubMed.
- M. Sanada, T. Miyano, S. Iwadare, J. M. Williamson, B. H. Arison, J. L. Smith, A. W. Douglas, J. M. Leich and E. Inamine, J. Antibiot., 1986, 39, 259–265 CrossRef CAS PubMed.
- H. Deng, L. Ma, N. Bandaranayaka, Z. Qin, G. Mann, K. Kyeremeh, Y. Yu, T. Shephers, J. H. Naismith and D. O'Hagan, ChemBioChem, 2014, 15, 364–368 CrossRef CAS PubMed.
- L. Ma, A. Bartholomé, M. H. Tong, Z. Qin, Y. Yu, T. Shepherd, K. Kyeremeh, H. Deng and D. O'Hagan, Chem. Sci., 2015, 6, 1414–1419 RSC.
- (a) D. O Hagan, C. Schaffrath, S. L Cobb, J. T. G. Hamilton and C. D Murphy, Nature, 2002, 416, 279 CrossRef CAS PubMed; (b) C. Dong, F. L. Huang, H. Deng, C. Schaffrath, J. B. Spencer, D. O'Hagan and J. H. Naismith, Nature, 2004, 427, 561–565 CrossRef CAS.
- H. Deng, S. M. Cross, R. P. McGlinchey, J. T. G. Hamilton and D. O'Hagan, Chem. Biol., 2008, 15, 1268–1276 CrossRef CAS PubMed.
- (a) X. M. Zhu, S. Hackl, M. N. Thaker, L. Kalan, C. Weber, D. S. Urgast, E. M. Krupp, A. Brewer, S. Vanner, A. Szawiola, G. Yim, J. Feldmann, A. Bechthold, G. D. Wright and D. L. Zechel, ChemBioChem, 2015, 16, 2498–2506 CrossRef CAS PubMed; (b) L. Kalan, A. Gessner, M. N. Thaker, N. Waglechner, X.-M. Zhu, A. Szawiola, A. Bechthold, G. D. Wright and D. L. Zechel, Chem. Biol., 2013, 20, 1214–1224 CrossRef CAS PubMed.
- S. Zhang, D. Klementz, J. Zhu, R. Makirtynskyy, A. R. O. Pasternak, S. Günther, D. L. Zechel and A. Bechthold, J. Biotechnol., 2019, 292, 23–31 CrossRef CAS PubMed.
- (a) X. Feng, N. Al Maharik, A. Bartholomé, J. E. Janso, U. Reilly and D. O'Hagan, Org. Biomol. Chem., 2017, 15, 8006–8008 RSC; (b) A. Bartholomé, J. E. Janso, U. Reilly and D. O'Hagan, Org. Biomol. Chem., 2017, 15, 61–64 RSC.
- H. Kristinsson, K. Nebel, A. C. O'Sullivan, J. P. Pachlatko, and Y. Yamaguchi, ACS Symposium Series, Synthesis and Chemistry of Agrochemicals IV, 1995, pp. 206–219 Search PubMed.
- C. Zhao, J. Qi, W. Tao, L. He, W. Xu, J. Chen and Z. Deng, PLoS One, 2014, 9, e114722 CrossRef PubMed.
- Whole Genome Shotgun data for S. calvus T-3018 has been deposited at DDBJ/ENA/GenBank under the accession VCNP01000000.
- H. Liu and J. H. Naismith, Protein Expression Purif., 2009, 63, 102–111 CrossRef CAS PubMed.
- P. Mujumdar, S. Bua, C. T. Supuran, T. S. Peat and S.-A. Poulsen, Bioorg. Med. Chem. Lett., 2018, 28, 3009–3013 CrossRef CAS.
- L. L. Lairson, B. Henrissat, G. J. Davies and S. G. Withers, Annu. Rev. Biochem., 2008, 77, 521–555 CrossRef CAS PubMed.
- F. Egami and N. Takahashi, Bull. Chem. Soc. Jpn., 1955, 28, 666–668 CrossRef CAS.
- B. J. Beahm, K. W. Dehnert, N. L. Derr, J. Kuhn, J. K. Eberhart, D. Spillmann, S. L. Amacher and C. R. Bertozzi, Angew. Chem., Int. Ed., 2014, 53, 3347–3352 CrossRef CAS.
- W. Pilgrim and P. V. Murphy, J. Org. Chem., 2010, 75, 6747–6755 CrossRef CAS PubMed.
- H. Kristinsson, K. Nebel, A. C. O'Sullivan, F. Struber, T. Winkler and Y. Yamaguchi, Tetrahedron, 1994, 50, 6825–6838 CrossRef CAS.
- (a) S. Lee, C. Uttamapinant and G. L. Verdine, Org. Lett., 2007, 9, 5007–5009 CrossRef CAS PubMed; (b) D. Guillerm, M. Muzard, B. Allart and G. Guillerm, Bioorg. Med. Chem. Lett., 1995, 5, 1455 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc03374b |
| This journal is © The Royal Society of Chemistry 2019 |